Abstract
The focal adhesion protein vinculin (1066 residues) can be separated into a 95-kDa head and a 30-kDa tail domain. Vinculin's lipid binding sites localized on the tail, helix 3 (residues 944–978) and the unstructured C-terminal arm (residues 1052–1066, the so-called lipid anchor), influence focal adhesion turnover and are important for cell migration and adhesion. Using magnetic tweezers, we characterized the cell mechanical behavior in mouse embryonic fibroblast (MEF)-vin(−/−) cells transfected with EGFP-linked-vinculin deficient of the lipid anchor (vinΔC, residues 1–1051). MEF-vinΔC cells incubated with fibronectin-coated paramagnetic beads were less stiff, and more beads detached during these experiments compared to MEF-rescue cells. Cells expressing vinΔC formed fewer focal contacts as determined by confocal microscopy. Two-dimensional traction measurements showed that MEF-vinΔC cells generate less force compared to rescue cells. Attenuated traction forces were also found in cells that expressed vinculin with point mutations (R1060 and K1061 to Q) of the lipid anchor that impaired lipid binding. However, traction generation was not diminished in cells that expressed vinculin with impaired lipid binding caused by point mutations on helix 3. Mutating the src-phosphorylation site (Y1065 to F) resulted in reduced traction generation. These observations show that both the lipid binding and the src-phosphorylation of vinculin's C-terminus are important for cell mechanical behavior.
Introduction
Adhesion of cells to extracellular matrix (ECM) proteins or to neighboring cells is a prerequisite for their survival and proliferation. The contact to ECM proteins triggers biochemical signals inside the cell and initiates protein synthesis and reorganization of the cytoskeleton (1). An important group of adhesive receptors are the integrins, comprising part of the series of focal adhesions (FAs) that link the extracellular matrix to the actin cytoskeleton of the cell. FAs themselves consist of several proteins—including talin, zyxin, paxillin, FA-kinase, and vinculin—that enable the cell to generate mechanical forces for adhesion and migration (1). The maturation of FAs is believed to be force-dependent; however, the mechanism is still not clear (2,3).
The 116-kDa vinculin with its 95-kDa head and 30-kDa tail domain is one of the first proteins involved in FA formation (4). It exists in two states—the open and closed conformations (5). In the closed or autoinhibited state, the vinculin head masks the binding sites for paxillin, actin, and phospholipids on the vinculin tail. Activating vinculin frees the binding sites on the tail (5,6). In the open or activated state, vinculin localizes to FAs and connects the actin-cytoskeleton via talin with the integrin receptor (5,6). Förster resonance energy transfer experiments with a vinculin construct carrying a yellow fluorescent protein in the polylinker region and a cyan fluorescent protein at the C-terminus of vinculin-tail revealed that concurrent binding of talin and actin enables the displacement of the head from the tail and the activation of vinculin (7,8). Talin interacts with the vinculin-head (residues 1−258), which lowers the affinity to its tail domain (9–11). Phospholipids such as phosphatidylinositol 4–5 bisphosphate have also been reported to induce a conformational shift in vinculin and to activate it (9–11).
Cells deficient of vinculin are still able to form focal contacts but spread poorly on ECM- coated surfaces (12–16). They are more motile and less adhesive than wild-type cells (12). Stable retransformation of the vinculin-tail in F9 mouse carcinoma vinculin (−/−) cells improved adhesion and decreased cell motility (17). Reintroduction of ∼8% intracellular vinculin is sufficient to recover the wild-type phenotype (17). In magnetic twisting, magnetic tweezer, and atomic-force-microscopic measurements, F9 vinculin (−/−) cells showed a lower shear modulus compared to F9 wild-type cells suggesting that vinculin operates as an intracellular mechano-coupler (16,18–20). It is also believed that phosphorylation of vinculin is important for the mechano-coupling function of vinculin (21). Removing the phosphorylation site at position Y822 caused an upregulation of p-ERK, which resulted in the reduction of cell migration (21). Furthermore, c-src-dependent vinculin phosphorylation at position Y100 and Y1065 affects cell spreading and migration, indicating that the phosphorylation of vinculin stabilizes the active or open conformation (22). In vitro experiments showed that the presence of acidic lipid vesicles elevates the src-dependent phosphorylation of vinculin (23,24). We then hypothesize that there is a direct link between lipid binding and mechanical function such as cell migration, and that this link is affected by src-dependent phosphorylation.
Three regions on the 30-kDa tail domain have been identified as candidates for lipid-binding: 1), helix 3 (residues 935–978); 2), helix 5 (residues 1020–1040); and 3), the lipid anchor (residues 1052–1066) (25,26). Pull-down assays with artificial lipid membranes showed that, in contrast to full-length vinculin-tail (Vtail), a vinculin-tail variant with six mutated surface-exposed basic residues in helix 3 (i.e., K952, K956, R963, and R966 to Q) and the C-terminal lipid anchor (i.e., R1060 and K1061 to Q) as well as a variant lacking the last 15 amino acids (VtailΔC) were both unable to interact with acidic phosphatidylserine or phosphatidylinositol vesicles under physiological conditions (25–27). The actin binding of these vinculin mutants, however, was not affected (25–27). Detailed studies showed lipid insertion behavior of helix 3 and the unstructured C-terminus of Vtail (28,29). Furthermore, it was reported that cells transfected with a vinculin variant including these six point mutations (vinculin-LD; K925, K956, R963, R966, R1060, and K1061 to Q) show a reduced focal adhesion turnover rate, impaired cell adhesion on different extracellular substrates, and decreased cell motility (25). Cells expressing vinculin without the lipid anchor (vinculinΔC) had the same reduced FA adhesion turnover rate and decreased cell motility (26). These results imply that the membrane interaction of vinculin's lipid anchor influences cell mechanical behavior.
In this study, we determined the effect of the lipid anchor (residues 1052–1066, vinculinΔC) on cell stiffness, binding strength to ECM-coated surfaces, and traction generation. Magnetic tweezer experiments showed that mouse embryonic fibroblasts (MEF)-vin(−/−) cells transiently transfected with enhanced green fluorescent protein (EGFP)-labeled vinculinΔC are less stiff than MEF-wild-type and MEF-vin(−/−) cells retransfected with full-length (EGFP)-vinculin (= MEF-resc). Measuring the binding strength of fibronectin(FN)-coated beads attached to integrin receptors, we found that more beads detached during force application from MEF-vin(−/−) cells retransfected with vinculinΔC compared to the MEF-resc cells. Actomyosin-driven contractile forces of different cells were determined using two-dimensional-traction microscopy. MEF-vinculinΔC cells developed less force and formed fewer FAs than MEF-resc cells. Two-dimensional-traction measurements using various vinculin mutants deficient in lipid binding revealed that not only the lipid anchor but also the src-phosphorylation at Y1065 affects cellular force generation.
Materials and Methods
Cell lines
Mouse embryonic fibroblast (MEF) vinculin (−/−) and wild-type were a kind gift of Dr. E.D. Adamson (Burnham Institute, La Jolla, CA). These cell lines were generated and characterized by Xu et al. (15).
Cell culture
MEF cell lines were maintained in low-glucose (1 g/L) Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (low endotoxin), 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin (Dulbecco's modified Eagle's medium, complete medium; Biochrom, Berlin, Germany) and kept at 37°C with 5% CO2.
Cloning and expression of vinculin
Mouse vinculin-encoding cDNA was kindly provided by Dr. E.D. Adamson. To distinguish transfected from untransfected cells, an EGFP cassette was cloned into pcDNA3.1 eukaryotic expression vector (Invitrogen, Karlsruhe, Germany), using NheI and XhoI restriction sites. The polymerase-chain-reaction-amplified vinculin wild-type (1–1066) and VinΔC (1–1051) constructs were C-terminally fused to the EGFP cassette using AflII and XbaI restriction sites. The fused EGFP constructs are driven by a CMV promoter (Clontech, Mountain View, CA). EGFP-linked vinculin-LD, vinculin-helix 3 (H3), and vinculin-C-terminal (CT) constructs were kindly provided by Dr. Wolfgang H. Ziegler (IZKF; University of Leipzig, Leipzig, Germany), which were previously generated and described by Chandrasekar et al. (25). In brief, the following lipid binding variants of vinculin were used: vinculin-H3 with point mutations on K925, K956, R963, and R966 to Q; vinculin-CT (C-terminal arm) with point mutations on R1060 and K1061 transferred to Q; and vinculin-LD with point mutations on K925, K956, R963, R966, R1060, and K1061 to Q. The exchange of tyrosine (Y) on position 1065 to phenylalanine (F) was done by site-directed mutagenesis. To transfect cells, 2 × 105 cells were seeded in a 30-mm ∅ tissue culture dish for 24 h. The cells were then transfected with 4-μg DNA using Lipofectamine (Invitrogen). Before fluorescence microscopy and tweezer measurements, 1 × 105 cells of the transfected cells were reseeded in a 30-mm-diameter dish or on FN-coated coverslips, respectively.
Magnetic tweezers
The principle of the magnetic tweezer device used is described in Kollmannsberger and Fabry (30). In brief, superparamagnetic 4.5-μm epoxylated beads (Invitrogen) were coated with fibronectin (100 μg/mL; Roche Diagnostics, Mannheim, Germany) in phosphate-buffered saline (PBS) at 4°C for 24 h. Beads were washed in PBS and stored at 4°C. Before measurements, fibronectin-coated beads were sonicated, added to cells (1 × 105 beads/dish), and incubated for 30 min at 5% CO2 and 37°C. A magnetic field with a high-field gradient was generated using a solenoid (250 turns of ∅ 0.5-mm copper wire, solenoid length = 2 cm, mean solenoid diameter 1 cm) with a needle-shaped core (HyMu80 alloy; Carpenter, Reading, PA). The needle tip was placed at a distance of 20−30 μm from a bead bound to a cell, using a motorized micromanipulator (Injectman NI-2; Eppendorf, Hamburg, Germany). Bright-field images of the cell, bead, and needle tip were taken by a charge-coupled device camera (ORCA ER; Hamamatsu, Hamamatsu City, Japan) at a rate of 40 frames/s. The bead position was tracked on-line using an intensity-weighted center-of-mass algorithm. A preset force was maintained by continuously updating the solenoid current or by moving the solenoid such that the needle-tip to bead distance was kept constant. Measurements on multiple beads per well were performed at 37°C for 1 h, using a heated microscope stage on an inverted microscope at 40× magnification (NA 0.6) under bright-field illumination. Transfected MEF-vin(−/−) cells were identified in fluorescence mode using an EGFP-filter.
To ensure that cells had not experienced any significant forces resulting from previous measurements, the needle was moved by at least 0.5 mm between two measurements. The bead as well as the needle tip position and the solenoid current were continuously recorded at a rate of 40 s−1. Image acquisition was triggered and synchronized with the solenoid current generator, using a custom-made C2+ program run on a PC equipped with an AD-DA board (NI-6052E; National Instruments, Austin, TX).
Force protocol and data analysis
When a force step with an amplitude, ΔF, was applied to a bead, it moved with a displacement, d(t), toward the needle tip. The ratio, d(t)/ΔF, defines the so-called creep-response, J(t) (31). The creep-response of cells followed a power law,
| (1) |
where t0 is a reference time that was arbitrarily set to 1 s (32–34). The parameter J0 (units of μm/nN) characterizes the elastic cell properties and corresponds to a compliance (i.e., inverse of stiffness). The exponent β is a measure for the viscoelasticity and varies between β = 0 (purely elastic solid) and β = 1 (purely viscous liquid) (32). Typical values for β in our measurements were between 0.1 and 0.3.
Bead detachment
To determine the bead-binding strength to the cell, a staircaselike force protocol was applied during 10 s. With every increase of force, some beads were not able to sustain the higher force and detached from the cell. The fraction of beads that detached during the measurements was then used to quantify the binding strength to the cell. Since beads were coated with an extracellular matrix protein (fibronectin), the bead-binding strength is a measure for the adhesion strength of cells to extracellular-matrix coated surfaces.
Traction microscopy
Gels for traction experiments were cast on rectangular 75 × 25 mm nonelectrostatic silane-coated glass slides (Menzel, Braunschweig, Germany) according to the procedure described by Pelham and Wang (35). Gels with 6.1% acrylamide and 0.24% bis-acrylamide were used. The Young's modulus of the gels was measured with a magnetically driven plate rheometer and found to be 13 kPa. Red fluorescent 1-μm carboxylated beads (Molecular Probes, Eugene, OR) were suspended in the gels and centrifuged at 300g toward the gel surface during polymerization at 4°C. The beads served as markers for gel deformation. The surface of the gel was activated with sulfo-SANPAH (Pierce Biotechnology, Rockford, IL) and coated with bovine collagen G (Biochrom) at 50 μg/mL. The cell suspension added to the gel was contained in a silicone ring (Flexiperm; In Vitro, Göttingen, Germany) attached to the glass slide.
Cell tractions were computed from the displacement field of the gel surface that occurred when the cells were detached from the substrate adding trypsin (36). Local gel deformations were computed by comparing an image pair taken before and after trypsin treatment using a difference-with-interpolation method (37). To compute the tractions, the unconstrained Fourier transform approach of Butler et al. was followed (36). To characterize the contractile forces of each cell by a single number, the elastic strain energy stored in the polyacrylamide gel due to the cell tractions was calculated as the product of local tractions and gel deformations, integrated over the spreading area of the cell (36).
Cell fixation and fluorescence microscopy
The transfected cells were trypsinized 24 h after transfection, and 1 × 105 cells were reseeded on fibronectin-coated coverslips (FN, 25 μg/mL; Roche Diagnostics). After 24 h of incubation, the cells were washed with PBS and fixed in 3% paraformaldehyde (Sigma-Aldrich, Munich, Germany) for 20 min at room temperature. For staining the actin cytoskeleton, cells were incubated for 15 min with Alexa-phalloidin (Sigma-Aldrich). Note, that Alexa phalloidin was dissolved in lyso-phosphocholine for better diffusion into the cell. This is a softer approach of cell permeabilization than Triton X-100. To stain for focal adhesions, MEF-wild-type and MEFvin(−/−) cells were incubated with monoclonal antibodies against vinculin (Sigma, Deisenhofen, Germany) and paxillin (BD Bioscience, Heidelberg, Germany), respectively. After washing with PBS, the cover slides were mounted with MOWIOL (Kuraray, Frankfurt, Germany).
To visualize the linkage between FAs and the actin cytoskeleton, a fluorescence microscope (Leica DM16000B; Leica, Solms, Germany) was used. Images were taken using a 40 × objective. The contacts between FN-coated beads and focal adhesions inside the cells were visualized by confocal microscopy (LSM 510, 25 × oil immersion, NA = 0.8; Zeiss, Jena, Germany). The calculation of the spreading area ensured that only cells of similar size were used for FA determination. The LSM-Image Browser software (Zeiss) was used to calculate the spreading area of the cells.
Results
Localization of EGFP-vinculin constructs in MEF cells
The interaction between the integrin-linked focal adhesions and the ECM is necessary for cell survival and is mainly driven by transmembrane receptors, like integrins (38). Here, we examined the localization of various vinculin constructs linked to EGFP expressed in MEF-vin(−/−) cells. Fig. 1 shows images of MEF-wt (wild-type), MEF-vin(−/−) (vinculin knock-out), MEF-resc (rescue), and MEF-vinΔC (transfected with vinculin lacking the lipid anchor; residues 1–1051) cells. The focal adhesions (green) in MEF-wt and MEF-vin(−/−) cells were stained with antibodies against paxillin, and focal adhesions in MEF-resc and MEF-vinΔC cells were visualized using the EGFP molecule attached to the different vinculin peptides. The position of the 4.5-μm FN-coated beads (red, arrow) was determined by confocal microscopy, which were in close proximity of the FA components such as paxillin or vinculin.
Figure 1.
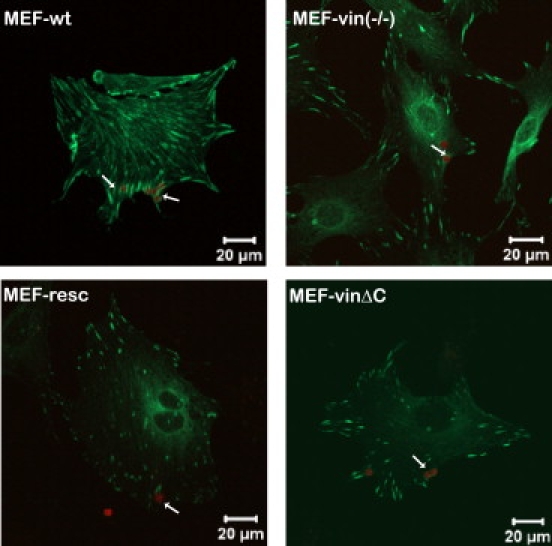
Confocal microscopic images show colocalization of the autofluorescent beads (red) with focal adhesions (green). Transfected and nontransfected MEF-vin(−/−) cells were seeded on fibronectin (FN)-coated glass slides and allowed to adhere overnight. Before fixation, the cells were incubated with FN-coated beads for 1 h. The FAs in MEF-wt and MEF-vin(−/−) cells were stained with antibodies against paxillin; in MEF-vin(−/−) cells transfected with vinculin (MEF-resc) and vinculinΔC (MEF-vinΔC), focal adhesion were determined using the N-terminal EGFP label. The arrows mark the FN-coated beads, which are in close proximity to the focal adhesion proteins, vinculin and paxillin. Note that the focal plane of the confocal micrograph was adjusted to the beads and not to the basal cell surface.
Cell stiffness and binding strength
The expression of vinculinΔC has been reported to result in reduced FA turnover and cell migration as well as altered mechanical properties (26). The effect of the lipid anchor on cell mechanics was measured using the magnetic tweezer device. For stiffness measurements, we applied forces of 1 nN to FN-coated beads. We used MEF-wt and MEF-vin(−/−) cells, both of which are stable cell lines, and full-length vinculin (MEF-resc) or vinculinΔC (MEF-vinΔC) were then transiently transfected in MEF-vin(−/−) cells. MEF-wt and MEF-resc cells showed similar stiffness, whereas MEF-vin(−/−) and MEF-vinΔC cells revealed significantly reduced stiffness values of ∼48% and ∼26%, respectively, compared to MEF-wt cells (100%) (Fig. 2).
Figure 2.
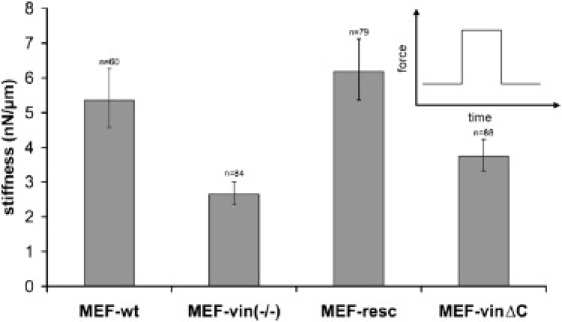
Stiffness of the transfected and nontransfected MEF cells were determined at 1 nN force. The n indicates the number of cells (equals the number of beads) measured in these experiments. Error bars denote the mean ± SE. (Inset) Force-versus-time protocol of these measurements.
To test whether the binding strength to fibronectin-coated beads differed between vinculin mutant cell lines, we increased the externally applied force over 10 s in a staircaselike manner from 0.5 to 10 nN (Fig. 3 A). With increasing force, the bead detachment increased. At 10 nN force, ∼20% and ∼14% of the beads detached from MEF-wt and MEF-resc, respectively, while ∼37% of the beads detached during force application from MEF-vin(−/−) cells. MEF-vinΔC showed similarly high levels of detached beads at 10 nN (Fig. 3 B). The fraction of beads that detached at a given force level can be regarded as a direct measure of the adhesion strength (yielding force) of the force-transmitting elements and bonds among the FN-coated beads, integrins, focal adhesion proteins, and the cytoskeleton. These results imply that the lipid anchor of vinculin may influence the adhesion strength between the cytoskeleton and ECM-coated surfaces.
Figure 3.
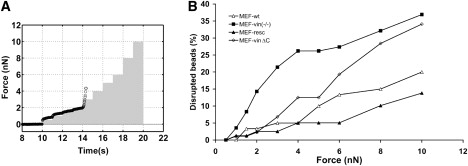
(A) A staircaselike force protocol was applied during 10 s (solid bars). With every increase of force, some beads detached from the cell; the displacement of a bead that detaches at a force of 3 nN is shown as an example (solid circle). (B) The percentage of detached beads versus pulling force in MEF wild-type and vinculin mutant cells is used to quantify the binding strength to the cell.
FAs per cell
It has been reported that focal adhesion maturation depends on the internal tension and the adhesion strength of the cell to the extracellular matrix (2,3,39). We then determined the number of focal adhesions in MEF cell lines of similar spreading area (Fig. 4). MEF-wt and MEF-resc cells showed similar numbers of focal adhesions, whereas MEF-vin(−/−) cells transfected with vinculinΔC showed ∼23% and MEF-vin(−/−) cells ∼40% fewer focal adhesions compared to MEF wild-type and rescue cells (Fig. 4). The size of the FAs in these different cell lines was similar.
Figure 4.
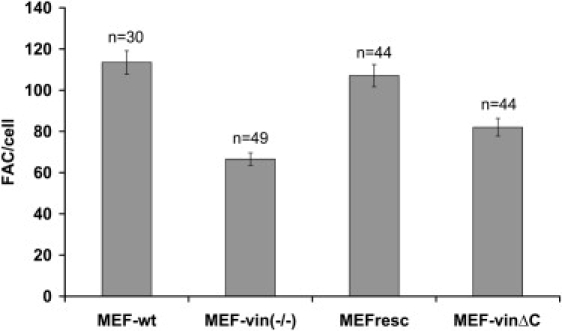
Numbers of focal adhesions (FAs) per cell determined from confocal microscopic images. Only cells of similar spreading area (∼4000 μm2) were analyzed, showing ∼115 FAs in MEF-wt, 65 FAs in MEF-vin(−/−), 110 FAs in MEF-resc, and 82 FAs in MEF-vinΔC cells. Note that n is the number of cells measured, and that the error bars denote the mean ± SE.
Two-dimensional-traction microscopic measurements
To confirm the relationship between focal adhesion formation and cellular contractile force generated by actomyosin motor activity that is transmitted via the focal adhesions, we used traction microscopy on ECM-coated PAA gels. No difference in the distribution of the actin cytoskeleton between the different cell lines was found (Fig. 5 A, right images). MEF-vin(−/−) cells generated ∼80% and MEF-vinΔC mutants generated ∼41% less tractions compared to the MEF-wild-type and rescue cells (100%) (Fig. 5 B). These results suggest that vinculin's lipid anchor is important for force generation during cell adhesion.
Figure 5.
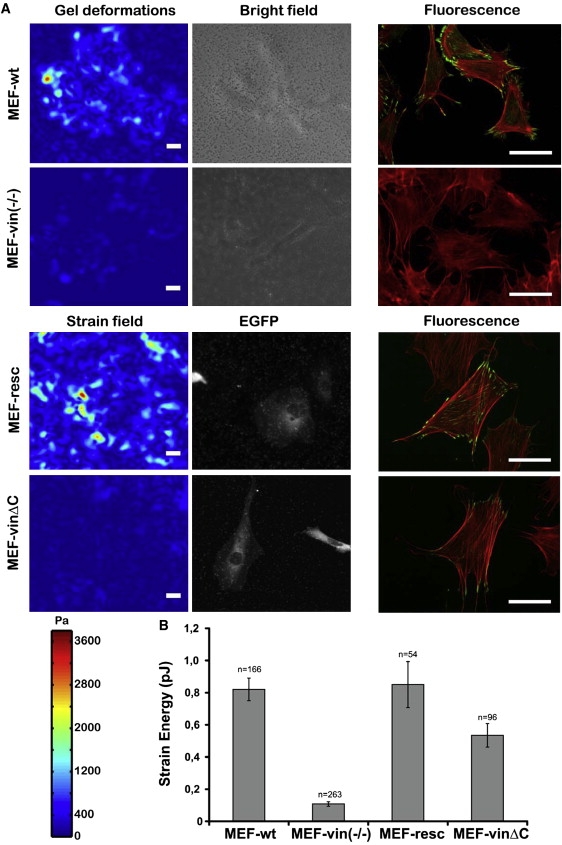
Two-dimensional-traction microscopy measurements for MEF-wt, MEF-vin(−/−), MEF-resc, and MEF-vinΔC cells. (A) Gel deformation and bright-field and fluorescent images of various MEF cells. The gel deformation is color-coded. Vinculin was stained with a monoclonal antibody in MEF-wt and MEF-vin(−/−). For visualization of the vinculin constructs in MEF-resc and MEF-vinΔC, the EGFP label was used. The actin cytoskeleton was stained with TRITC-Phalloidin. The scale bar is 50 μm. (B) Plot of the average elastic strain energy stored in the extracellular matrix due to cellular tractions. Note that n indicates the number of cells measured, and that the error bars denote the mean ± SE.
Previously, it has been shown that the mutation of basic residues on the surface of the vinculin-tail in helix 3 (H3; K952, K956, R963, and R966 to Q) and in the C-terminal lipid anchor (CT; R1060 and K1061 to Q) result in impaired lipid membrane interaction (25). To test the influence of the lipid-binding sites on cell mechanical behavior, we performed additional two-dimensional-traction microscopy measurements with these vinculin mutants. MEF-vin(−/−) cells were transfected with EGFP-linked vinculin-H3, vinculin-CT, and vinculin-LD constructs. All constructs localized in focal adhesions (Fig. 6 A). There was no detectable difference in FA size and distribution. The two-dimensional-traction measurements expressed as relative strain energy compared to MEF-resc cells are shown in Fig. 6 B. MEF-vin(−/−) cells transfected with a lipid-binding deficient variant of vinculin (MEF-LD) or with a vinculin variant with mutations in the lipid anchor region (MEF-CT) both showed a reduction in strain energy of ∼50% compared to MEF-resc cells. However, MEF-vin(−/−) cells carrying vinculin with the mutated H3 domain, which also influences the lipid binding according to Chandrasekar et al. (25), showed similar strain energy levels as MEF-resc cells, suggesting that only the change of vinculin's lipid anchor localized at the C-terminus affects force generation in MEF cells.
Figure 6.
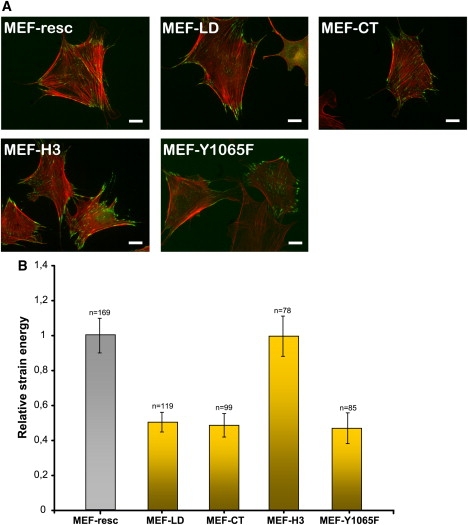
(A) Images of MEF-vin(−/−) cells transfected with EGFP-labeled vinculin-LD (MEF-LD), vinculin-CT (MEF-CT), and vinculin-H3 (MEF-H3). The actin cytoskeleton was stained with TRITC-Phalloidin. The scale bar is 20 μm. (B) Strain energy of MEF-LD, MEF-CT, MEF-H3, and MEF-Y1065F cells was normalized to MEF-resc (100%). Note that n indicates the number of cells measured, and that the error bars denote the mean ± SE.
Within the lipid anchor there is also a src-dependent phosphorylation position at residue 1065 (22,40). The mutation of this tyrosine phosphorylation site results in impaired cell spreading and migration, which suggests that the src-phosphorylation may affect cell mechanical behavior. The vinculinY1065F mutant showed a decreased force generation in comparison to rescue cells (Fig. 6 B). Note that differential scanning calorimetric experiments using lipid vesicles consisting of 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dimyristoyl-sn-glycero-3-phosphorylserine in the presence of vinculin's last 21 amino acids indicate that the Y1065 to F mutant has similar lipid binding compared to Y1065 wild-type (Volker Wirth, personal communication, 2009).
Discussion
Mechanical tension between the interconnected assemblies of the extracellular matrix and the cytoskeleton plays a critical role in determining cell behavior and structure. The forces generated by and in a cell have been shown to regulate many biological functions (41). Here, we examined the involvement of vinculin interaction with the lipid membrane in MEF cells and the effect of this interaction on cell stiffness, binding strength, and stress-stiffening.
The MEF-wt phenotype was restored when MEF-vin(−/−) cells were transiently transfected with EGFP-labeled vinculin, indicating that the N-terminal EGFP label has no obvious effect on the localization or regulation of the vinculin molecule in MEF cells. These so-called MEF-resc cells also showed almost identical mechanical properties to their wild-type counterparts. Therefore, the MEF-vin(−/−) cells are suitable, and are a useful model system for characterizing different vinculin mutants. Magnetic tweezer measurements showed that the cell stiffness of MEF-vin(−/−) cells decreased significantly and returned to wild-type level in MEF-resc cells (42). However, MEF-vin(−/−) cells, when transiently transfected with EGFP-vinculinΔC, were less stiff than MEF-resc cells. Previously, it was shown that the vinculin-tail lacking the lipid anchor exhibited impaired interactions with acidic phospholipid vesicles, while keeping the secondary structure and the actin-binding properties intact (26,27). A direct involvement of the lipid anchor in membrane binding was shown by differential scanning calorimetry (29). These findings, together with the diminished stiffness of MEF-vinΔC cells, indicate that the linkage of vinculin's lipid anchor with the cell membrane is important for cell mechanical function. Support for this assumption also comes from our bead-binding strength measurements. With increased pulling force, more beads detached from MEF-vinΔC cells than from MEF-wt or rescue cells. At high forces, MEF-vinΔC cells displayed almost the same level of detached beads as vinculin knockout cells, suggesting that the lipid anchor is necessary for effective adhesion to ECM-coated surfaces.
The reduced binding strength of MEF-vinΔC cells may also influence the tractions generated by these cells. The strain energy generated by MEF-vin(−/−) cells was approximately eightfold lower, and the strain energy generated by MEF-vinΔC cells was approximately twofold lower compared to MEF-wt or MEF-resc cells. This observation indicates that the lipid anchor plays an important role for internal force generation and effective cell adhesion to ECM-coated surfaces. Cell adhesion forces are believed to be due to the actin-ECM connection via the focal adhesion complex and the myosin II-driven force development (2,43). Blocking the contractility with substances such as ML-7, BDM, or KT5926, or by overexpression of peptides like caldesmon (which inhibits actin-dependent myosin II ATPase activity) leads to the dissolution of focal contacts (39,44). The reduced numbers of focal adhesions in MEF-vinΔC compared to MEF-wt and rescue cells might be a secondary effect due to the reduced force generation. Therefore, we tested the hypothesis that the lipid anchor is necessary for force generation and the maintenance of cellular prestress, which prevents the dissolution of the focal adhesions in cells. Further experiments with other vinculin variants deficient in membrane binding revealed that only the lipid anchor, and not helix 3 (H3), affects cellular force generation. The relative strain energy of 1), MEF-vinΔC; 2), MEFvin-LD; or 3), MEFvin-CT variants were all decreased to similarly low levels, whereas the MEF-H3 cells demonstrate strain energies comparable to MEF-resc cells. This suggests that only the membrane association of the lipid anchor (residue 1052−1066) is important for force generation, whereas the lipid binding of MEF-H3 has no influence on cell mechanical behavior. Actin binding to vinculin tail is not impaired in MEF-vinΔC cells (25,26), but so far, to our knowledge, this has not been unequivocally tested in MEFvin-LD or MEF-CT cells; therefore, a possibility remains that their lower tractions are the result of impaired actin-vinculin binding.
It was reported that the tyrosine phosphorylation sites (residues Y100 and Y1065) on vinculin are targeted by c-src kinase (22,23). When the phosphorylation is prevented, the interaction of vinculin with the Arp2/3 subunit p34Arc is inhibited, which results in impaired cell spreading and migration (40). Further, it was shown that phospholipids stimulate the phosphorylation of vinculin by src kinase (23,24). In this study, we have attempted to describe the mechanical influence of phosphorylation at position Y1065. Using two-dimensional-traction microscopy, we found that MEF-Y1065F cells generated ∼50% less strain energy compared to MEF-resc cells, which is similar to MEF-vinΔC, MEF-LD, and MEF-CT cells (Fig. 6, A and B). These first results are intriguing, but further studies that are more detailed are needed to characterize the molecular mechanism of vinculin's phosphorylation at position 1065 and 100 in mechanical terms.
In summary, it is conceivable that the anchorage of vinculin to lipid membranes by the C-terminus alters its conformation, which could enhance its activation and FA recruitment (9,10,24). Crystal structure data of vinculin show that the lipid anchor region is not buried inside the vinculin molecule when it is in a closed conformation (45,46). Data from our laboratory confirm an interaction of the hydrophobic part of lipid membranes with vinculin's C-terminal peptide (29). This conformational switch may be important for the src-dependent phosphorylation. Phosphorylation of vinculin's Y1065 is essential for the binding of the p34 subunit of Arp2/3 to vinculin and consequently for the reinforcement of the focal adhesion complex as well as the mechanical linkage to the actin cytoskeleton. Only vinculin carrying the full lipid anchor with an intact src-phosphorylation site may enable the cell to generate sufficient prestress for adequate adhesion.
Acknowledgments
We thank Dr. E. D. Adamson for providing the MEF cells, Dr. W. H. Ziegler for supplying the EGPF-Vinculin-LD, -H3, and -CT constructs, and D. Paranhos-Zitterbart for help with traction microscopic measurements. We also thank U. Scholz for administrative support, B. Reischl, C. Albert, and N. Bonakdar for technical support, and Dr. B. Hoffmann for stimulating discussions.
This work was supported by grants from Bayerisch-Französisches Hochschulzentrum, Deutscher Akademischer Austausch Dienst, Bavaria California Technology Center, Deutsche Forschungsgemeinschaft, and Deutsche Krebshilfe.
References
- 1.Bershadsky A.D., Ballestrem C., Carramusa L., Zilberman Y., Gilquin B. Assembly and mechanosensory function of focal adhesions: experiments and models. Eur. J. Cell Biol. 2006;85:165–173. doi: 10.1016/j.ejcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Bershadsky A.D., Balaban N.Q., Geiger B. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003;19:677–695. doi: 10.1146/annurev.cellbio.19.111301.153011. [DOI] [PubMed] [Google Scholar]
- 3.Möhl C., Kirchgessner N., Schäfer C., Küpper K., Born S. Becoming stable and strong: the interplay between vinculin exchange dynamics and adhesion strength during adhesion site maturation. Cell Motil. Cytoskeleton. 2009;66:350–364. doi: 10.1002/cm.20375. [DOI] [PubMed] [Google Scholar]
- 4.Goldmann W.H. Kinetic determination of focal adhesion protein formation. Biochem. Biophys. Res. Commun. 2000;271:553–557. doi: 10.1006/bbrc.2000.2653. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler W.H., Liddington R.C., Critchley D.R. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Critchley D.R. Focal adhesions—the cytoskeletal connection. Curr. Opin. Cell Biol. 2000;12:133–139. doi: 10.1016/s0955-0674(99)00067-8. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Cohen D.M., Choudhury D.M., Kioka N., Craig S.W. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 2005;169:459–470. doi: 10.1083/jcb.200410100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen D.M., Kutscher B., Chen H., Murphy D.B., Craig S.W. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 2006;281:16006–16015. doi: 10.1074/jbc.M600738200. [DOI] [PubMed] [Google Scholar]
- 9.Gilmore A.P., Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R.P., Craig S.W. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem. Biophys. Res. Commun. 1995;210:159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- 11.Steimle P.A., Hoffert J.D., Adey N.B., Craig S.W. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J. Biol. Chem. 1999;274:18414–18420. doi: 10.1074/jbc.274.26.18414. [DOI] [PubMed] [Google Scholar]
- 12.Coll J.L., Ben-Ze'ev A., Ezzell R.M., Rodriguez Fernandez J.L., Baribault H. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc. Natl. Acad. Sci. USA. 1995;92:9161–9165. doi: 10.1073/pnas.92.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldmann W.H., Schindl M., Cardozo T.J., Ezzell R.M. Motility of vinculin-deficient F9 embryonic carcinoma cells analyzed by video, laser confocal, and reflection interference contrast microscopy. Exp. Cell Res. 1995;221:311–319. doi: 10.1006/excr.1995.1380. [DOI] [PubMed] [Google Scholar]
- 14.Ezzell R.M., Goldmann W.H., Wang N., Parasharama N., Ingber D.E. Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell Res. 1997;231:14–26. doi: 10.1006/excr.1996.3451. [DOI] [PubMed] [Google Scholar]
- 15.Xu W., Baribault H., Adamson E.D. Vinculin knockout results in heart and brain defects during embryonic development. Development. 1998;125:327–337. doi: 10.1242/dev.125.2.327. [DOI] [PubMed] [Google Scholar]
- 16.Mierke C.T., Kollmannsberger P., Paranhos-Zitterbart D., Smith J., Fabry B. Mechano-coupling and regulation of contractility by the vinculin tail domain. Biophys. J. 2008;94:661–670. doi: 10.1529/biophysj.107.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W., Coll J.L., Adamson E.D. Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells; effects on cell locomotion by domain deleted vinculin. J. Cell Sci. 1998;111:1535–1544. doi: 10.1242/jcs.111.11.1535. [DOI] [PubMed] [Google Scholar]
- 18.Goldmann W.H., Galneder R., Ludwig M., Xu W., Adamson E.D. Differences in elasticity of vinculin-deficient F9 cells measured by magnetometry and atomic force microscopy. Exp. Cell Res. 1998;239:235–242. doi: 10.1006/excr.1997.3915. [DOI] [PubMed] [Google Scholar]
- 19.Goldmann W.H., Galneder R., Ludwig M., Kromm A., Ezzell R.M. Differences in F9 and 5.51 cell elasticity determined by cell poking and atomic force microscopy. FEBS Lett. 1998;424:139–142. doi: 10.1016/s0014-5793(98)00155-0. [DOI] [PubMed] [Google Scholar]
- 20.Alenghat F.J., Fabry B., Tsai K.Y., Goldmann W.H., Ingber D.E. Analysis of cell mechanics in single vinculin-deficient cells using a magnetic tweezer. Biochem. Biophys. Res. Commun. 2000;277:93–99. doi: 10.1006/bbrc.2000.3636. [DOI] [PubMed] [Google Scholar]
- 21.Subauste M.C., Pertz O., Adamson E.D., Turner C.E., Junger S. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J. Cell Biol. 2004;165:371–381. doi: 10.1083/jcb.200308011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z., Izaguirre G., Lin S.Y., Lee H.Y., Schaefer E. The phosphorylation of vinculin on tyrosine residues 100 and 1065, mediated by SRC kinases, affects cell spreading. Mol. Biol. Cell. 2004;15:4234–4247. doi: 10.1091/mbc.E04-03-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S., Richert N., Pastan I. Phospholipids stimulate phosphorylation of vinculin by the tyrosine-specific protein kinase of Rous sarcoma virus. Proc. Natl. Acad. Sci. USA. 1982;79:4628–4631. doi: 10.1073/pnas.79.15.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito S., Werth D.K., Richert N.D., Pastan I. Vinculin phosphorylation by the SRC kinase. Interaction of vinculin with phospholipid vesicles. J. Biol. Chem. 1983;258:14626–14631. [PubMed] [Google Scholar]
- 25.Chandrasekar I., Stradal T.E., Holt M.R., Entschladen F., Jockusch B.M. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005;118:1461–1472. doi: 10.1242/jcs.01734. [DOI] [PubMed] [Google Scholar]
- 26.Saunders R.M., Holt M.R., Jennings L., Sutton D.H., Barsukov I.L. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 2006;85:487–500. doi: 10.1016/j.ejcb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Bakolitsa C., de Pereda J.M., Bagshaw C.R., Critchley D.R., Liddington R.C. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 1999;99:603–613. doi: 10.1016/s0092-8674(00)81549-4. [DOI] [PubMed] [Google Scholar]
- 28.Johnson R.P., Niggli V., Durrer P., Craig S.W. A conserved motif in the tail domain of vinculin mediates association with and insertion into acidic phospholipid bilayers. Biochemistry. 1998;37:10211–10222. doi: 10.1021/bi9727242. [DOI] [PubMed] [Google Scholar]
- 29.Diez G., List F., Smith J., Ziegler W.H., Goldmann W.H. Direct evidence of vinculin tail-lipid membrane interaction in beta-sheet conformation. Biochem. Biophys. Res. Commun. 2008;373:69–73. doi: 10.1016/j.bbrc.2008.05.182. [DOI] [PubMed] [Google Scholar]
- 30.Kollmannsberger P., Fabry B. High-force magnetic tweezers with force feedback for biological applications. Rev. Sci. Instrum. 2007;78:114301–114306. doi: 10.1063/1.2804771. [DOI] [PubMed] [Google Scholar]
- 31.Fung Y.C. Elasticity of soft tissues in simple elongation. Am. J. Physiol. 1967;213:1532–1544. doi: 10.1152/ajplegacy.1967.213.6.1532. [DOI] [PubMed] [Google Scholar]
- 32.Fabry B., Maksym G.N., Shore S.A., Moore P.E., Panettieri R.A. Time course and heterogeneity of contractile responses in cultured human airway smooth muscle cells. J. Appl. Physiol. 2001;91:986–994. doi: 10.1152/jappl.2001.91.2.986. [DOI] [PubMed] [Google Scholar]
- 33.Lenormand G., Millet E., Fabry B., Butler J.P., Fredberg J.J. Linearity and time-scale invariance of the creep function in living cells. J. R. Soc. Interface. 2004;1:91–97. doi: 10.1098/rsif.2004.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desprat N., Richert A., Simeon J., Asnacios A. Creep function of a single living cell. Biophys. J. 2005;88:2224–2233. doi: 10.1529/biophysj.104.050278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pelham R.J., Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler J.P., Tolic-Norrelykke I.M., Fabry B., Fredberg J.J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 2002;282:C595–C605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 37.Raupach C., Zitterbart D.P., Mierke C.T., Metzner C., Muller F.A. Stress fluctuations and motion of cytoskeletal-bound markers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:011918. doi: 10.1103/PhysRevE.76.011918. [DOI] [PubMed] [Google Scholar]
- 38.Geiger B., Bershadsky A., Pankov R., Yamada K.M. Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 39.Balaban N.Q., Schwarz U.S., Riveline D., Goichberg P., Tzur G. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 40.Moese S., Selbach M., Brinkmann V., Karlas A., Haimovich B. The Helicobacter pylori CagA protein disrupts matrix adhesion of gastric epithelial cells by dephosphorylation of vinculin. Cell. Microbiol. 2007;9:1148–1161. doi: 10.1111/j.1462-5822.2006.00856.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang N., Butler J.P., Ingber D.E. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 42.Klemm A.H., Diez G., Alonso J.L., Goldmann W.H. Comparing the mechanical influence of vinculin, focal adhesion kinase and p53 in mouse embryonic fibroblasts. Biochem. Biophys. Res. Commun. 2009;379:799–801. doi: 10.1016/j.bbrc.2008.12.124. [DOI] [PubMed] [Google Scholar]
- 43.Geiger B., Bershadsky A. Exploring the neighborhood: adhesion-coupled cell mechanosensors. Cell. 2002;110:139–142. doi: 10.1016/s0092-8674(02)00831-0. [DOI] [PubMed] [Google Scholar]
- 44.Helfman D.M., Levy E.T., Berthier C., Shtutman M., Riveline D. Caldesmon inhibits nonmuscle cell contractility and interferes with the formation of focal adhesions. Mol. Biol. Cell. 1999;10:3097–3112. doi: 10.1091/mbc.10.10.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borgon R.A., Vonrhein C., Bricogne G., Bois P.R., Izard T. Crystal structure of human vinculin. Structure (Camb) 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Bakolitsa C., Cohen D.M., Bankston L.A., Bobkov A.A., Cadwell G.W. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]


