Abstract
The association of CD4, a glycoprotein involved in T cell development and antigen recognition, and CCR5, a chemotactic G protein-coupled receptor, which regulates trafficking and effector functions of immune cells, forms the main receptor for the human immunodeficiency virus HIV. We observed that the vast majority of CCR5 is maintained within the intracellular compartments of primary T lymphocytes and in a monocytic cell line, contrasting with its relative low density at the cell surface. The CCR5-CD4 association, which occurs in the endoplasmic reticulum, enhanced CCR5 export to the plasma membrane in a concentration–dependent manner, whereas inhibition of endogenous CD4 with small interfering RNAs decreased cell surface expression of endogenous CCR5. This effect was specific for CCR5, as CD4 did not affect cell distribution of CXCR4, the other HIV co-receptor. These results reveal a previously unappreciated role of CD4, which contributes to regulate CCR5 export to the plasma membrane.
Keywords: Animals; Antigens, CD4; metabolism; Antigens, Surface; metabolism; CHO Cells; Cell Compartmentation; physiology; Cells, Cultured; Cricetinae; Cricetulus; Endoplasmic Reticulum; metabolism; Hela Cells; Humans; Intracellular Space; metabolism; Protein Binding; Protein Transport; Receptors, CCR5; metabolism
Introduction
The CC chemokine receptor 5 (CCR5) is a seven membrane-spanning domain, G protein-coupled receptor (GPCR), which regulates chemotaxis and effector functions of T-lymphocytes, macrophages, and dendritic cells (reviewed in 1). Upon activation by its cognate chemokines, CCL5 (former Regulated upon Activation, Normal T cell Expressed and Secreted, RANTES), CCL3 and CCL4 (former macrophage inflammatory proteins 1α and 1β) 2, CCR5 leads to cellular signaling through G proteins 3 as well as G protein-independent pathways 4. In addition to its physiological functions, CCR5 is also the main coreceptor of the human immunodeficiency virus (HIV), in association with CD4 5, a cell-surface glycoprotein that participates in molecular complexes involved in both T cell development and antigen recognition by T cells. CD4 is a single-membrane spanning domain receptor interacting with class II major histocompatibility complex molecules 6 and with its cognate ligand IL-16. So far, no specific physiological function has been attributed to the CCR5-CD4 complex.
In the context of immune cell chemotaxis in vivo, recent studies have shown that the sustained (hours) lymphocyte motility within lymph nodes, is driven by the lymphoid chemokine-rich environment and mediated by the activation of chemokine receptors 7. Paradoxically, studies in reconstituted models have shown that stimulated chemokine receptors, including CCR5, are rapidly desensitized (seconds to minutes) via phosphorylation and β-arrestin translocation before being internalized and eventually degraded 1,8–10. In fact, an adaptive mechanism to escape inactivation and maintain prolonged reactivity in vivo upon sustained stimulation has been reported for other GPGRs, based on the retention of important stocks of functional receptors within intracellular compartments (reviewed in 11). Initially described for protease-activated receptors (such as the thrombin receptor 12), this phenomenon was subsequently reported for dopamine D1 receptors in tubular renal cells 13, and for δ-opioid receptors in neurons 14. The list of GPCRs displaying a predominant intracellular localization is rapidly growing 11. A common feature for most of these prevalently intracellular GPCRs is their capacity of interacting with a vast array of membrane-associated or cytoplasmic proteins that facilitate their translocation to the cell surface 15. Often, these proteins possess their own functions, in addition to aiding GPCR expression at the cell surface 11. Among them, type-I single-transmembrane domain proteins with a large N-terminal extracellular domain and a short C-terminus are particularly frequent. This group includes Receptor-activity-modifying-proteins (RAMPs), which facilitate cell surface expression of a Class-B GPCRs 16; the Receptor Transporting Proteins (RTPs) and REEP1 (Receptor Expression Enhancing Protein 1), which permit functional cell surface targeting of odorant 17 and taste receptors 18; the M10 MHC class-I molecules which are necessary for the correct trafficking of pheromone receptors to the plasma membrane 19; the MC2R accessory protein (MRAP), which interacts with the melanocortin 2 receptor, regulating its trafficking from the endoplasmic reticulum (ER) to the cell surface 20.
In the current study we investigated the subcellular distribution of endogenous CCR5 receptor in human T lymphocytes and in a monocytic cell line and found that this receptor is predominantly intracellular. We also report that CCR5 and CD4 can associate within intracellular compartments and that this association enhances the export of CCR5 to the cell surface.
Material and Methods
Cell lines and transfections
THP-1 cells were a kind gift from Philip Strange lab (Reading, UK). Cell culture media were from GIBCO and chemicals from SIGMA unless otherwise specified. CHO-K1 cells were cultured in F12 medium, HeLa P4R5 in Dulbecco modified minimal essential medium (DMEM) and THP-1 cells in RPMI 1640, in an atmosphere of 5% CO2. Media were supplemented with 10% fetal bovine serum (FBS), 100 U/ml Penicillin and 100 μg/ml Streptomycin. Transfections of CHO-K1 and HeLa P4R5 cells were performed using GeneJuice-Transfection Reagent (Novagen).
Plasmid expression constructs
Constructs for BRET experiments were described previously 21. Briefly, CCR5 and CXCR4 coding regions were amplified from their respective cDNAs using appropriate sense and antisense primers, containing unique Hind III or Bam H1 sites, respectively. The fragments were then subcloned in frame between the Hind III and Bam H1 sites of a plasmid for YFP (Clontech) or for a humanized form of Renilla Luciferase (Biosignal; PerkinElmer Life Sciences). The plasmid for CCR5ΔCter-YFP was obtained by the amplification of a shorter fragment (corresponding to met1-gly301 CCR5) using a more proximal antisense primer. For CD4-Luc and CD4-YFP constructs, the stop codon of the CD4 sequence was suppressed and replaced by a restriction site using site directed mutagenesis (QuickChange site-directed mutagenesis kit, Stratagene). The coding regions of all constructs were entirely sequenced. The GABABR1-YFP and OBR-YFP constructs were kind gifts from Michel Bouvier (Montreal, Canada) and Ralf Jockers (Paris, France), respectively. The constructs for GFP-tagged wild-type VPU and phosphorylation mutant VPU2/6 22 were kind gifts from Florence Margottin (Paris, France). The plasmid for ER-DsRed was purchased from Clontech BD Biosciences.
Antibodies
Mouse monoclonal antibodies were as follows: 2D7 anti-human CCR5 (555991, BD Pharmingen) directed against the second extracellular loop of CCR5, OKT4 anti-human CD4 (for FACS and immunofluorescence, 140048, eBioscience), 12G5 anti-CXCR4 (ab 21555, Abcam), 1F6 anti-human CD4 (for immunoblot experiments, NCL-L-CD4-1F6, Novocastra), anti-GFP (11-814-460-001, Roche). Rat monoclonal antibodies were as follows: 1/85a Alexa-FluorR 647-conjugated anti-human CCR5 (313712, Biolegend), which recognizes the N-terminal extremity of CCR5 23, Alexa-FluorR 647-conjugated rat IgG2a, k isotype control (400526, Biolegend), 3F10 anti-hemagglutinin (anti-HA) (1-867-423, Roche). Rabbit polyclonal antibodies were as follows: anti-giantin (ab24586, Abcam) anti-TGN46 (ab16052, Abcam), anti-GPR78/BiP (ab21685, Abcam), anti-Phospho-p44/42 (Thr202/Tyr204) MAP Kinase (9101, Cell Signalling), anti ERK 1-2 (06-182, Cell Signalling), Cy3-conjugated anti-goat immunoglobulin (305-167-003, Jackson Immunoresearch), peroxidase-conjugated anti-mouse IgG (315-035-003, Jackson Immunoresearch), peroxidase-conjugated anti-goat IgG (28169, Anaspec). Goat polyclonal antibodies were as follows: anti-calnexin (C-20) (sc-6465, Santa Cruz Biotechnology), anti-actin (I-19) (sc-1616, Santa Cruz Biotechnology), peroxidase-conjugated anti-rabbit IgG (111-035-003 Jackson Immunoresearch), Cy5-conjugated anti-mouse IgG (610-110-121, Rockland). Donkey polyclonal antibodies were as follow: Cy3-conjugated anti-rabbit IgG (711-167-003 Jackson Immunoresearch), Cy3-conjugated anti-rat IgG (712-166-153 Jackson Immunoresearch), Alexa-FluorR 488-conjugated anti-mouse IgG (H+L) (A-21202, Molecular Probes, Invitrogen).
Human T lymphocyte preparation and activation
Human T-cells were isolated from the peripheral blood of healthy volunteers by Ficoll density gradient centrifugation, followed by negative depletion on magnetic beads (Human T lymphocyte Enrichment set-DM, Becton Dickinson). T-cells were activated by incubation for 48 hours with anti-CD3 and anti-CD28 antibody-coated beads (Dynabeads Human CD3/CD28 T Cell Expander Kit, Invitrogen), at a bead-to-cell ratio of 1:3, in RPMI 1640 medium supplemented with pooled human AB serum and antibiotics.
FACS analysis
The proportion of surface CCR5, and CD4 on resting or activated T lymphocytes, THP-1 cells and CHO-K1 cells expressing exogenous receptors was determined by analyzing in parallel, intact and permeabilized cells. For total staining, 1×106 cells were fixed in 1% paraformaldehyde for 20 min, washed with PBS and permeabilized for 1h at 4°C in PBS, containing 2% FBS and 0,2% Triton-X-100, as described 24. Cells were then incubated on ice for 1h in 100μl of PBS, 2% FBS with the 1/85a Alexa Fluor 647-conjugated anti-human CCR5 antibody (1:50) or the OKT4 anti-human CD4 (1:100) and control isotypes at the same dilutions. For CD4, this step was followed by 1h incubation with Cy5-conjugated anti-mouse IgG (1:100). For extracellular staining, cells were incubated on ice for 1h with PBS, 2% FBS without Triton-X-100 and subsequently incubated with the appropriate antibodies, as above. After washing with PBS, cells were fixed in 2% paraformaldheyde and processed on a Cytomycs FC500 FACS analyzer (Beckman Coulter).
To study the impact of CD4 co-expression on CCR5 targeting at the cell surface, CHO-K1 cells were co-transfected with plasmids coding for CCR5-Luc (at fixed concentration) and CD4-YFP (at increasing concentrations). 36h after transfection, cells were incubated on ice for 1h with 0.5μg/ml of the 2D7 anti-human CCR5 antibody, followed by incubation with Cy5-conjugated anti-mouse IgG (1:100).
To determine the effect of CD4 co-expression on CXCR4 targeting at the cell surface, CHO-K1 were co-transfected with plasmids coding for CXCR4-Luc (at fixed concentration) and CD4-YFP (at increasing concentrations). FACS experiments were conducted, as described above for CCR5, by incubating cells for 1h with 0.5μg/ml 12G5 anti-human CXCR4 antibody, followed by incubation with Cy5-conjugated anti-mouse IgG (1:100).
The level of endogenous CCR5 at the cell surface of THP-1 cells after CD4 silencing was measured with the 1/85a Alexa Fluor 647-conjugated anti-CCR5 antibody as described above. The proportion of surface CCR5 was evaluated by analyzing both intact and permeabilized cells.
For each analysis, 10000 cells were sorted and the data analyzed using the Cytomics RXP software.
Immunofluorescence
CHO-K1 cells were seeded on glass cover slips in six-well plates (2×105 per well), transfected with the indicated plasmids, and processed for immunofluorescence, 48h after transfection. Subsequently cells were fixed in 4% paraformaldehyde for 20 min at 4°C or in methanol for 5 min at −20°C, according to the antibody used. After quenching with 50mM NH4Cl in PBS for 15 min, cells were washed with PBS and then incubated with PBS-bovine serum albumin (BSA) 1% containing or not 0,1% saponin and labeled with the indicated antibodies.
The subcellular localization of endogenous CCR5 and CD4 in T lymphocytes and THP-1 cells was analyzed by immunofluorescence in cells maintained in suspension. After fixation in 4% paraformaldehyde for 20 min at room temperature and quenching (as described above) cells were incubated with PBS-BSA1% or PBS-BSA1% containing 0.05% of Tween 20, for permeabilization 25. Cells were then incubated for 1h with the 2D7 anti-human CCR5 or the OKT4 anti-human CD4 antibodies. Following incubation with appropriate secondary antibodies, cells were washed with PBS and mounted on slides.
All images were obtained using a confocal Leica DMIRE2 microsccope and analyzed using the Metamorph 7 software.
Immunoprecipitation and immunoblotting
Cells growing in 100mm dishes were lysed in 50mM HEPES, pH 7.4, 250mM NaCl, 2mM EDTA, 0.5% NP40, 10% glycerol, 0.5% deoxycholic acid (lysis buffer), containing Complete™ protease inhibitors (Roche), on ice for 1h. After centrifugation at 13000×g for 20min, the supernatant was collected and protein concentration determined (BCA protein assay, Pierce). Supernatants were incubated with anti-GFP antibody (2μg) for 3h at 4°C and then incubated with protein G-Sepharose beads for 1h at 4°C. After centrifugation, the immuno-bead-bound material was washed three times with lysis buffer. The immunoprecipitates were resolved in 10% SDS-PAGE gels. After transfer on nitrocellulose, blots were probed with the appropriate antibodies, all used at 1:1000 dilution. Immunoblots were revealed by luminescence (ECL, Amersham Bioscience).
ERK activation studies
CHO-K1 grown in 6-well plates were transfected with the plasmid encoding CCR5-Luc with or without the plasmid for CD4-YFP. After overnight starvation, cells were detached with EDTA 10mM in PBS, centrifuged (1400 ×g for 5 min) and resuspended in Hank’s-balanced salt solution. Aliquots were subjected to luciferase activity measurements to determine average CCR5-Luc concentration. After addition of 5μM coelenterazine h (Interchim), emitted light was measured in a Mithras LB940 reader (Berthold Instruments). Other aliquots of cells were stimulated with MIP-1β (Preprotech) for various periods of time, chilled on ice, centrifuged (8000 ×g for 1min at 4°C) and analyzed for ERK activation as described 26. Briefly, after resuspension in ice-cold 50 mM Tris-HCl pH 6.8, containing 2.5 mM orthovanadate, 2.5 mM EDTA and a cocktail of protease inhibitors, cells were lysed in 2X Laemmli sample buffer. Proteins were denatured (5min at 95°C) separated by 10% SDS-PAGE and transferred on nitrocellulose membranes. Membranes were probed with Phospho-p44/42 MAP kinase or MAP kinase 1-2 antibodies and autoradiograms quantified using the Image J 1.38X software.
BRET assays
CHO-K1 cells (5×105 per well of a 6-well plates) were transfected with 30ng of plasmid DNA coding for the BRET donor (CD4-Luc) and increasing amounts of BRET acceptor plasmids (CCR5-YFP, CCR5ΔCter-YFP, GABABR1-YFP; 10–1000ng per well) or YFP (0.5–100ng per well). 24h after transfection, cells were washed in PBS, detached using PBS-EDTA 10mM, centrifuged (1400 ×g for 5min), resuspended in Hank’s-balanced salt solution and distributed in 96-well plates (Perkin-Elmer plates; 1×105 cells per well). After addition of the luciferase substrate, coelenterazine h (5μM final concentration), luminescence and fluorescence were measured simultaneously (at 485 and 530nm, respectively) in a Mithras LB940 plate reader. The BRET ratio was calculated as: [(emission at 530nm/emission at 485 nm) − (background at 530nm/background at 485nm)], where background corresponds to signals in cells expressing the Rluc fusion protein alone under the same experimental conditions. For better readability, results were expressed in milli-BRET units (mBRET), 1 mBRET corresponding to the BRET ratio multiplied by 1000. BRET ratios were plotted as a function of [(YFP-YFP0)/YFP0]/[Rluc/Rluc0], where YFP is the fluorescence signal at 530nm after excitation at 485 nm, and Rluc the signal at 485 nm after addition of coelenterazine h. YFP0 and Rluc0 correspond to the same values in cells expressing the Rluc fusion protein alone. In some experiments, cells were incubated 14h before the BRET assay with Brefeldin A at 2μg/ml.
RNA interference
THP-1 cells were transfected using the AMAXA™ nucleofector system, in Amaxa solution V (program U010), with 0.5μM of siRNA oligonucleotides (S1: 5′-GAACUGACCUGUACAGCUUUU-3′; S2: 5′-GAGCGGAUGUCUCAGAUCAUU-3′; S3: 5′-UAACUAAAGGUCCAAUCCAAUU-3′) targeting human CD4 or an unrelated oligonucleotide (5′-UAGCGACUAAACACAUCAA-3′, all from Dharmacon). For each experiment 2 rounds of transfection were performed: 2×106 cells were transfected a first time, cultured for 72h, resuspended and transfected again using the same protocol. Assays were conducted 7 days after the first transfection. For RNA interference experiments on human T cells, 4×106 freshly purified T lymphocytes were transfected using the AMAXA™ nucleofector system, in Amaxa solution T (program U14) with 1μM of S1 and S3 oligonucleotides, as indicated above. After nucleofection, cells were cultured in RPMI medium supplemented with 10% FBS, antibiotics and Interleukin-7 (5ng/ml) for 72h and then activated for 48h with anti-CD3 and anti-CD28 antibody-coated beads as described above.
Statistical Analysis
Data are expressed as a mean value ± SEM. Data were analyzed by using Student’s t test and p < 0.05 was considered statistically significant.
Results
CCR5 is predominantly retained within intracellular compartments
To determine the cellular distribution of CCR5, both purified T lymphocytes from healthy donors and cells of the human THP-1 monocytic cell line, expressing endogenous CCR5 were analysed by FACS (Figure 1A, and Sup. Table 1). In freshly prepared unstimulated T lymphocytes, the proportion of surface CCR5 was very low, ≈95% of total receptors being intracellular, as measured by fluorescence analysis of intact or permeabilized cells with a CCR5-specific antibody. As shown in previous studies 27, surface CCR5 increased after stimulation with anti-CD3 and anti-CD28 antibodies (from 5% to ≈11%). In THP-1 cells, CCR5 was also principally present within intracellular compartments, with about 90% of the receptor being found within internal compartments. These observations were obtained with 2 different antibodies recognizing distinct epitopes (Sup. Table 1). The same procedure was used to measure in parallel the distribution of the CD4 molecule. As expected, in both T lymphocytes and monocytes, CD4 was predominantly expressed at the cell surface (up to 92% in activated T lymphocytes). To confirm these findings, immunofluorescence confocal microscopy studies were conducted on the same cells subjected (P) or not (NP) to membrane permeabilization (Figure 1B). Whereas the CD4 staining was comparable in permeabilized and non-permeabilized cells, the major proportion of CCR5 appeared to be intracellular in both T lymphocytes and THP-1 cells. The Jurkat T cell line, which does not express CCR5, was used as negative control for staining. Colocalization studies were also conducted and demonstrated that endogenous CCR5 colocalized with both Golgi (Giantin) end ER (Bip) markers in THP-1 cells, although additional subcellular localizations cannot be excluded. CCR5 distribution was finally analyzed in transiently transfected CHO cells, which will be used in this study as a reconstitution model. As found for the endogenous receptor in T lymphocytes and THP-1 cells, in CHO cells exogenous CCR5 was predominantly intracellular under our experimental conditions, whether wild-type receptor, or a form tagged at the C-terminus with a yellow variant of the green fluorescent protein (YFP), were expressed (Sup. Figure 1 and Sup. Table 1). Confocal microscopy analyses of transfected CHO cells confirmed that both wild-type CCR5 and CCR5-YFP colocalized with a panel of both Golgi and ER markers (Sup Figures 2). From the data above it appears that both endogenous and exogenous CCR5 are prone to intracellular retention.
Figure 1. Subcellular distribution of the human CCR5 receptor.
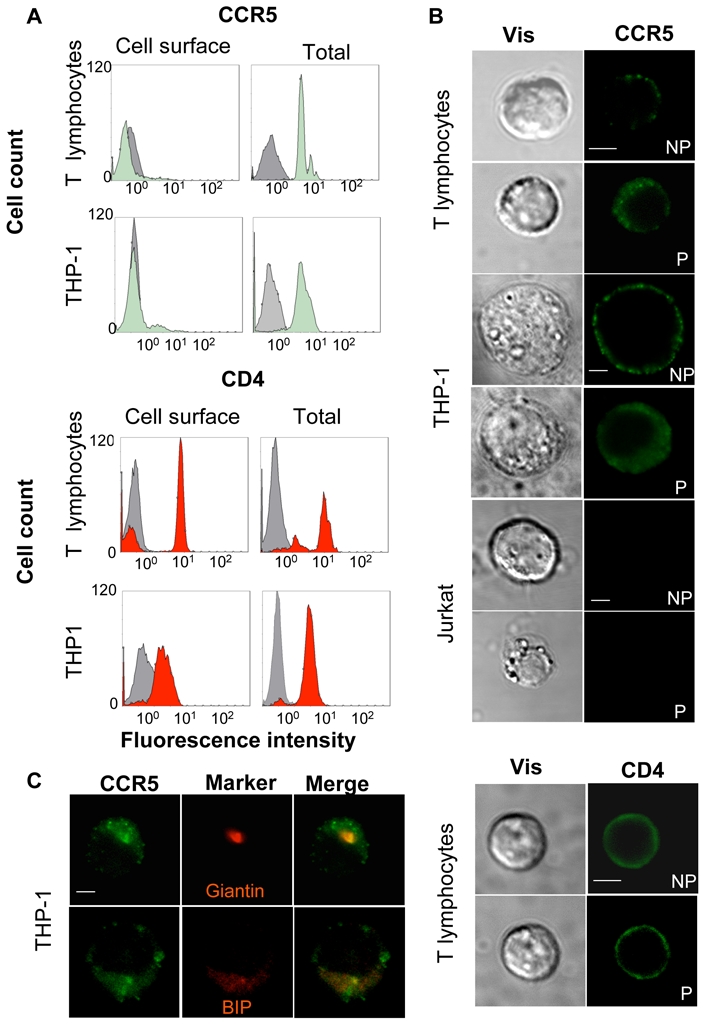
(A) FACS analyses showing human resting T lymphocytes from healthy donor or THP-1 monocytes were stained for surface (left panels) and total (after cell permeabilization, right panels) CCR5 (upper panels) or CD4 (lower panels), using 1/85a anti-human CCR5 antibody or OKT4 anti-human CD4, respectively, and analysed by flow cytometry. Green and red histograms correspond to specific labelling for CCR5 and CD4, respectively. Gray histograms are the isotypic control for each antibody. (B) Human resting T lymphocytes, THP-1 and Jurkat cells (negative control) were stained for surface (non-permeabilized, NP) and total (permeabilized P) CCR5 or CD4, using 2D7 anti-human CCR5 or OKT4 anti-human CD4 antibodies. Images were obtained by confocal microscopy. Scale bar: 10μM. (C) THP-1 cells, expressing endogenous receptor, were stained with the 2D7 anti-human CCR5 antibody and antibodies directed against Bip, an ER marker, or Giantin, a Golgi marker (see methods), and analysed by confocal microscopy. Colocalization of CCR5 (green) with Bip or Giantin (both in red) is shown in orange (Merge). Scale bar: 10μM.
CCR5 and CD4 associate in the ER
As indicated in the introduction, the cell surface expression of many predominantly intracellular GPCRs can be markedly enhanced by the association with other proteins, which interact with the receptors in the ER and/or the Golgi apparatus, and remain associated with them, once the receptors have reached the cell surface 11,15. We thus investigated whether CD4, in addition to its proper immunological functions, might also interact with CCR5 in the ER and facilitate its export to the cell surface. For this purpose, we made use of a natural mutant of CCR5, lacking its C-terminal intracellular tail 28, that is completely retained in the ER 29, as supported by the complete absence of cell-surface detection by antibodies in immunofluorescence and FACS experiments (Figure 2A,B). CHO cells were transiently transfected with a construct encoding CD4, in addition to plasmids for either wild type or truncated CCR5, harbouring a N-terminal HA epitope and a C-terminal YFP (HA-CCR5-YFP and HA-CCR5ΔCter-YFP constructs, respectively). In these cells, similar amounts of CD4 were co-immunoprecipitated with both the full length and truncated forms of CCR5 (Figure 2D) and both HA-CCR5ΔCter-YFP and HA-CCR5-YFP colocalized with CD4 within intracellular compartments (Figure 2C). We confirmed that the CD4-CCR5 association in CHO cells, evidenced by co-IP studies, was not due to non-specific hydrophobic interaction, by showing that mixing cell-lysates expressing either molecule did not allow co-immunoprecipitation (Supp. Figure 3). CD4–CCR5 interaction was further examined in live CHO cells using bioluminescence resonance energy transfer (BRET). CD4 fused to Renilla luciferase (CD4-Luc) was expressed in the presence of increasing concentrations of BRET acceptors, consisting of either HA-CCR5-YFP or HA-CCR5ΔCter-YFP. In both cases, hyperbolic saturation curves were obtained, indicating specific interactions (Figure 2E). BRET signals markedly depend on the relative distance of donor and acceptor 30, explaining why maximal BRET values were different in wild-type CCR5 and the truncated mutant. In contrast, the value of the YFP:Luc ratio, for which half-maximal BRET was obtained, was comparable, indicating that these two forms of CCR5 display the same propensity to associate with CD4. Free YFP and the GABABR1 receptor fused to YFP (GABABR1-YFP) were used as negative controls. Although GABABR1 is constitutively retained in the ER 31 like HA-CCR5ΔCter-YFP, only a weak non-saturable interaction with CD4-Luc could be measured. Finally, in the presence of free YFP, only a linear non-specific BRET signal was obtained. Bystander BRET signals may occur upon random collisions in cells expressing non-physiological high levels of BRET pairs 30. To rule out this possibility, we verified that the levels of BRET donor and acceptor proteins at BRET saturating concentration were comparable or below those of endogenous CD4 and CCR5 in human monocytic THP-1 cells (Sup. Figure 4). To confirm that the measured BRET signals are a result of interactions occurring in the ER, cells were pre-treated with brefeldin A (BFA), a drug, which induces Golgi apparatus disassembly and redistribution of Golgi-derived vesicles to the ER. The Golgi apparatus was indeed dismantled after BFA treatment (Figure 3A), and FACS analysis could not detect almost any surface receptor in cells expressing HA-CCR5-YFP (Figure 3B). Despite these effects, BRET values remained comparable whether or not cells were treated with BFA (Figure 3C). Together, these data are consistent with the hypothesis that CCR5 and CD4 associate within intracellular compartments, likely in the ER, before they are exported to the cell surface.
Figure 2. CCR5-CD4 association in the ER.
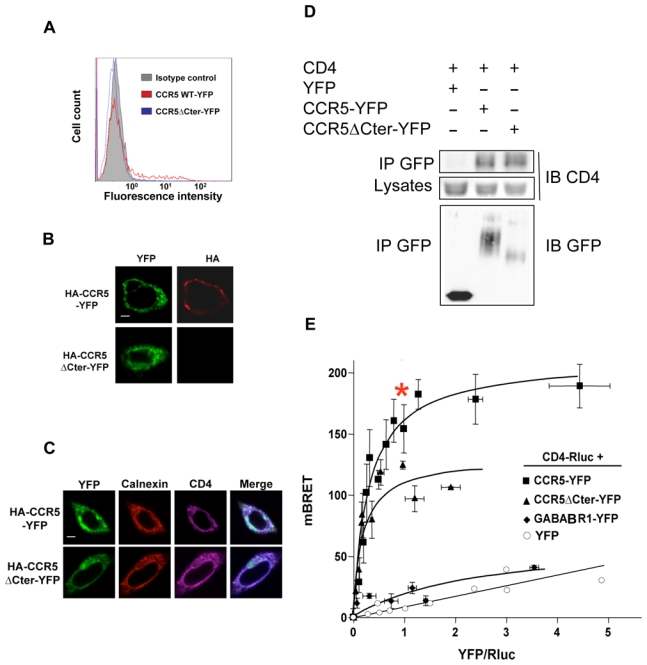
(A) CHO-K1 cells transfected with HA-tagged CCR5-YFP or CCR5ΔCter-YFP were analyzed for surface expression by FACS using the 1/85a Alexa-fluorR-647-conjugated anti-human CCR5, or (B) processed for immunofluorescence using the 3F10 anti-HA antibody. (C) Subcellular colocalization of CD4 with wild-type HA-CCR5-YFP and HA-CCR5ΔCter-YFP. CHO-K1 cells were co-transfected with CD4 and HA-CCR5-YFP or HA-CCR5ΔCter-YFP, permeabilized and stained for CD4 and calnexin using a monoclonal mouse anti-CD4 (OKT4) and goat anti-calnexin polyclonal antibody, respectively. After incubation with the appropriate Cy5 or Cy3-labeled secondary antibody, cells were examined with a confocal microscope. Note the substantial amount of intracellular CD4, likely due to the absence in CHO-K1 cells of the tyrosine kinase lck, which is known to stabilize CD4 at the cell surface of leukocytes. Scale bar: 10μM. (D) Co-immunoprecipitation experiments of CD4 with both wild-type and mutant CCR5ΔCter. CHO-K1 cells were co-transfected with plasmids coding for CD4 and HA-CCR5-YFP or HA-CCR5ΔCter or free YFP. After receptor precipitation with a monoclonal anti-GFP antibody, the presence of co-precipitated CD4 was revealed by immunoblot. (E) BRET analysis of CD4 interaction with wild-type and mutant CCR5. CHO-K1 cells were co-transfected with plasmids coding for CD4-Rluc (the BRET donor) and increasing concentrations of CCR5-YFP, CCR5ΔCter-YFP (the BRET acceptors) or GABABR1-YFP or free YFP (negative controls). Energy transfer was measured after addition of the membrane permeable luciferase substrate coelenterazine h. The BRET signal was determined by calculating the ratio of light emitted at 530nm over the light emitted at 485 nm as described in Material and Methods. BRET specificity was further controlled by showing that the BRET signal remained stable upon increasing the amounts of BRET pairs at a fixed ratio of donor: acceptor (data not shown). Maximal BRET signals were significantly different in cells expressing HA-CCR5ΔCter-YFP and HA-CCR5-YFP, likely because the variable length of the two C-terminal tails, to which the BRET acceptor was fused, affected the energy transfer. In contrast, the value of the YFP: Luc ratio, for which half-maximal BRET obtained, was comparable, indicating that these two forms of CCR5 display the same propensity to associate with CD4. The asterisk indicates the experimental conditions used for quantification of CD4 and CCR5 (see Sup. Figure 3A).
Figure 3. CCR5-CD4 association is not affected by Brefeldin A treatment.
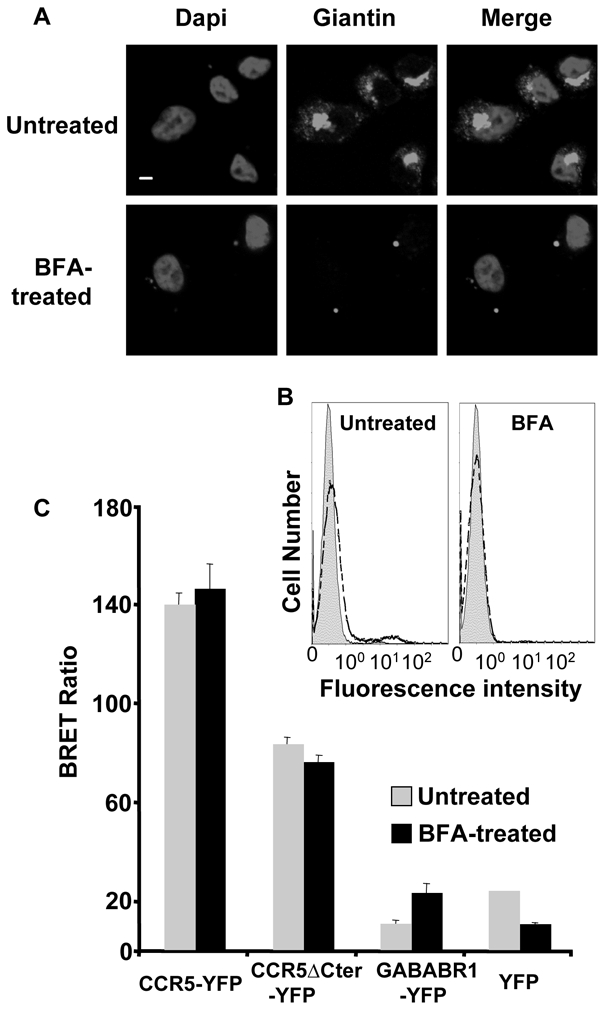
CHO-K1 cells were transfected with the plasmids coding for CD4-Luc and HA-CCR5-YFP or HA-CCR5ΔCter-YFP at concentrations yielding sub-maximal BRET (indicated by an asterisk in figure 2E). Fourteen hours before BRET analysis cells were subjected or not to a Brefeldin A (BFA) treatment (see methods). (A) BFA-treated and untreated cells were stained for the Golgi marker Giantin; DAPI was added to label nuclei. Scale bar: 10μM. (B) FACS analysis of CCR5 cell surface expression in cells treated or not with BFA. The grey filled histogram is the isotypic control, the dotted open histogram corresponding to the specific signal (C) BRET analysis of CD4-CCR5 (wild-type and truncated) interaction in CHO-K1 cells after BFA treatment. Procedure and BRET controls were as in Figure 2E.
Association with CD4 in the ER enhances CCR5 export to the cell surface
We next examined whether CD4 might enhance the proportion of surface CCR5 in a reconstituted CHO-K1 cell model. A fixed concentration of a plasmid coding for luciferase-tagged CCR5 (CCR5-Luc) was co-transfected with increasing amounts of a CD4-YFP construct. Surface CCR5 values, measured by FACS with an anti-CCR5 antibody in cells co-expressing CD4-YFP, were plotted as a function of the CD4-YFP-associated fluorescence (Figure 4). In co-transfected cells, surface CCR5 progressively increased with increasing levels of CD4-YFP to reach a maximum, corresponding to an augmentation of more than 100%, and then returned gradually to basal values for higher levels of CD4-YFP (Figure 4B). The descending portion of the curve was not due to a decreased number of CD4 molecules that could reach the cell surface, since in the same cells, surface CD4 increased linearly with increasing amounts of total CD4-YFP (Sup. Figure 4B). Throughout the experiments we verified that the amount of CCR5-Luc remained constant by measuring luciferase-associated activity. In control experiments, with either free YFP or with the single transmembrane domain leptin receptor fused to YFP (OBR-YFP), no enhancement of surface CCR5 was observed (Figure 4A and B). As for BRET experiments, the effect of CD4 on surface CCR5 expression was achieved at physiological concentrations of the two proteins (Sup. Figure 4A). HIV can use a second chemokine co-receptor, in conjunction with CD4, to infect target cells 32. This receptor, CXCR4, also displays a predominant intracellular distribution in human T lymphocytes and THP-1 monocytes (reference 33 and data not shown) and can form stable complexes with CD4, which are detectable by co-immunoprecipitation experiments (Figure 4C and 34). However, contrasting with what was observed for CCR5, its surface expression was not positively modulated by CD4 in transfected cells (Figure 4D).
Figure 4. Specific modulation of CCR5 cell surface expression by CD4.
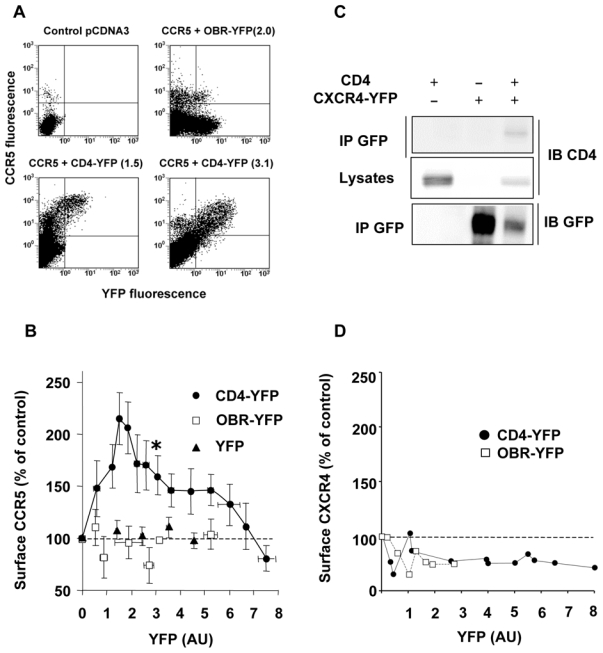
(A) CHO-K1 cells were co-transfected with fixed concentrations of a plasmid encoding CCR5-Luc and increasing concentrations of the CD4-YFP plasmid or controls. CCR5 surface expression was determined by FACS analysis using a mouse anti-human CCR5 (2D7) as primary antibody followed by a Cy5-conjugated anti-mouse antibody. Representative experiments are shown. Above each panel, the YFP-associated signal (between parenthesis) of the indicated YFP-fused protein is shown, using the same units as in panel B. (B) Results of 14 independent experiments, each representing 8–10 individual transfections. The luciferase signal was used to monitor the total amount of CCR5. Experiments in which the luciferase signal was different from the average by more than 10% were discarded. Surface CCR5 values, represented as the percentage (± SEM of 4–6 grouped values) of control cells expressing CCR5 alone, were plotted as a function of the YFP signal, reflecting the concentration of CD4-YFP, of control leptin receptor fused to YFP (OBR-YFP) or free YFP. The asterisk indicates the experimental conditions used for quantification of CD4 and CCR5 (see Sup Figure 3A). (C) Co-immunoprecipitation analysis of CD4 - CXCR4 interaction. CHO-K1 cells were co-transfected with plasmids coding for CD4 and CXCR4-YFP and processed for immunoprecipitation as described in Figure 2D. (D) Absence of CXCR4 cells surface modulation by increasing concentrations of CD4. The experiment was conducted as described in panel A, using a mouse anti-CXCR4 antibody (12G5) followed by a Cy5-conjugated anti-mouse antibody. Each point corresponds to an individual transfection.
The functional significance of the enhancement of surface CCR5, due to the presence of CD4, was estimated by measuring ERK1-2 activation. CHO cells expressing identical amounts of CCR5-Luc, determined by comparing luciferase activity, and a concentration of CD4-YFP corresponding to its maximal effect on CCR5 export, were stimulated or not with non-saturating concentrations of MIP-1β for increasing periods of time up to 10 min (Figure 5). The maximal activation of the MAP kinase doubled in cells co-expressing CD4 and, in addition, it was also reached much faster (compare lanes 3 (time=1min) in Figure 5A and corresponding histograms in Figure 5B).
Figure 5. CD4 expression enhances CCR5-dependent ERK activation.
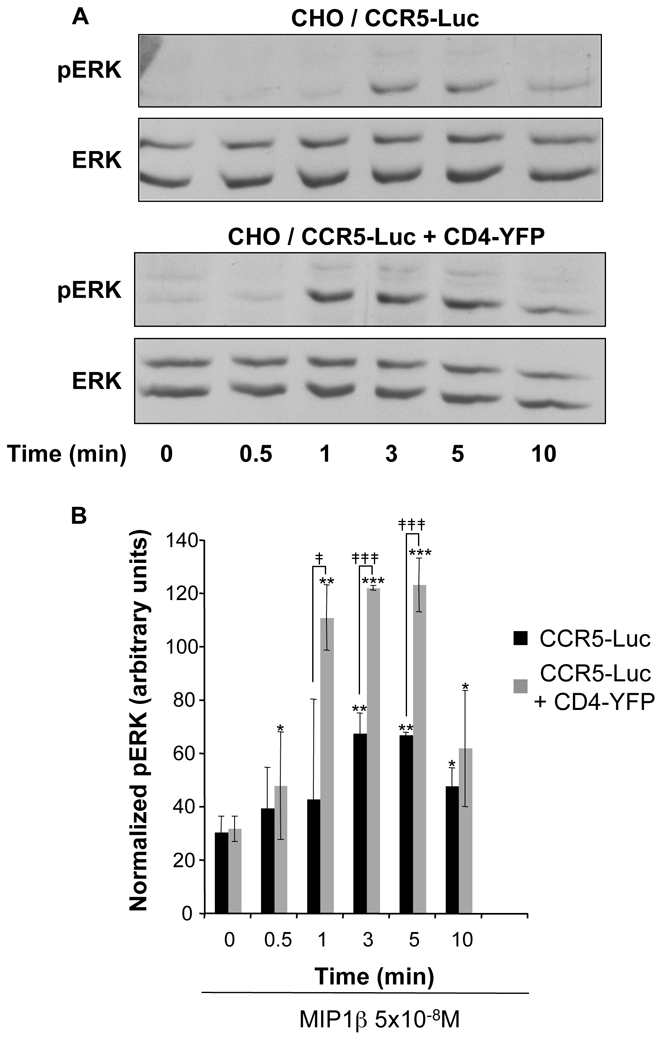
(A) CHO-K1 cells were transfected with the plasmid for CCR5-Luc with or without the plasmid for CD4-YFP at concentrations, which correspond to the asterisk in Figure 3B. Cells were serum-starved overnight and then stimulated with MIP1β (5×10−8M) for the indicated durations of time. The luciferase signal was measured to monitor the total amount of CCR5. Immunoblotting was performed with the indicated antibodies. Equal amounts of lysates were loaded in each lane and a representative immunoblot of 3 independent experiments is shown. Total ERK represents the loading control. (B) Quantification of phosphorylated ERK 1-2 (normalized with total ERK) from 3 independents experiments. * indicate significant difference from untreated cells; ‡ indicate significant difference between cells expressing or not CD4; * and ‡: p<0.05, **: p<0.01, *** and ‡‡‡: p<0.001.
We next examined whether the effect of CD4 on CCR5 surface targeting also occurs in cells expressing endogenous receptors. We reasoned that a substantial decrease of CD4 molecules in these cells might affect the density of CCR5 at the cell surface. A panel of 3 interfering RNAs (siRNAs) was used, which targeted the CD4 sequence. THP-1 cells were submitted to 2 consecutive nucleofections and tested after a week to achieve sufficient inhibition of CD4 synthesis and decay of CCR5 molecules already present at the cell surface before the effect of siRNAs. A significant reduction of 70–80% of total CD4 was obtained with 2 specific siRNAs, compared to the control (Figure 6A,B). In the cells showing reduced CD4, surface CCR5 was significantly decreased by 25–50%, compared to the control and to cells expressing the ineffective siRNA (Figure 6C,D). Unexpectedly, cells treated with the 2 siRNAs active on CD4 expression showed a significant enhancement of 20–60% of total CCR5 (Sup Figure 5) suggesting a possible feedback regulation. These results support the hypothesis that CD4 molecules can modulate the proportion of surface CCR5 in cells expressing both molecules. In a physiopathological context, the HIV-1-encoded Vpu protein was reported to induce rapid degradation of CD4 shortly after its synthesis 35 via the ER-associated protein degradation pathway 36. In line with our findings above, the specific targeting of CD4 molecules to the proteasome by overexpressed Vpu should affect the surface expression of CCR5. A stable Hela cell line, expressing both CCR5 and CD4, was transiently transfected with either wild type GFP-tagged Vpu or a GFP-fused mutant Vpu that has lost its capacity of down-regulating CD4 36 or free GFP (Sup Figure 6). After transfection, surface expression of CCR5 was quantified by FACS in cells expressing comparable levels of green fluorescence. Confirming the hypothesis, expression of Vpu-GFP induced comparable and significant reduction of both total CD4 and surface CCR5, whereas the control mutant Vpu was ineffective.
Figure 6. CD4 silencing negatively modulates CCR5 surface expression in THP-1 and primary T cells.
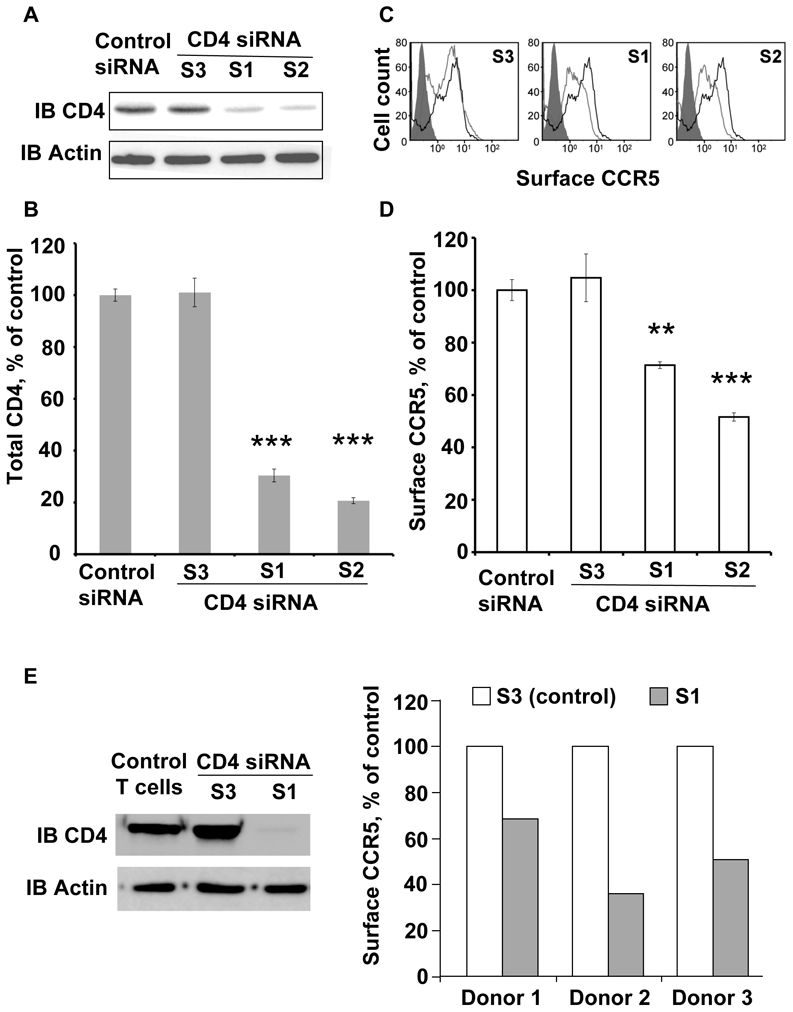
(A) Western blot analysis of CD4 from THP-1 cells nucleofected with siRNAs directed against CD4 (S1, S2 and S3) or scrambled control. CD4 was detected using the 1F6 anti-human CD4 antibody. (B) Quantification of CD4 after treatment with indicated siRNAs (n=3, in triplicates). Asterisks indicate statistical significance (p<0.001) with cells treated with control siRNA. (C) FACS analysis of cell surface CCR5 using the 1/85a Alexa-fluor 647-conjugated anti-human CCR5. Grey open histograms of S1, S2 and S3-treated cells are compared to the control (scramble siRNA) histogram in black. Grey filled histogram corresponds to isotypic control. (D) The effect of CD4 inhibition on surface CCR5 was measured as follows: the proportion of surface CCR5 was calculated by FACS (by comparing the signal in unpermeabilzed and permeabilized cells) in 3 independent experiments, in triplicates (**p<0.01; ***p<0.001); the values were compared to the proportion of surface CCR5 in THP-1 nucleofected with control scramble siRNA. (E) Left panel: Representative western blot analysis of CD4 from donor-purified, activated (see methods) primary T cells, nucleofected with 2 siRNAs targeting CD4 sequences (S1 and S3); note that, as for THP-1 cells, the S3 siRNA did not reduce CD4 concentration in T cells. Right panel: effect of CD4 inhibition on surface CCR5 in T cells from 3 donors. The proportion of surface CCR5 was calculated by FACS, by comparing the signal in unpermeabilzed and permeabilized cells, and values obtained in cells nuleofected with the S1 siRNA were compared to the proportion of surface CCR5 in cells nucleofected with the ineffective S3 siRNA.
Finally, the contribution of CD4 to the cell surface expression of CCR5 was explored in primary human cells. T cells were purified from PBMCs, nucleofected with appropriate siRNAs, activated to enhance the cell surface expression of CCR5 and analyzed for both total CD4 content and proportion of cell surface CCR5 (Figure 6E). In line with the results obtained with THP-1 cells, for all 3 tested donors, the siRNA-mediated reduction of CD4 was associated with a decreased proportion of CCR5 at the cell surface compared to the control, ranging from 30 to 65%. Primary monocytes purified from one donor were also examined with the same assay and displayed a 75% reduction in the proportion of surface CCR5 (not shown).
Discussion
Although previously unreported, the predominant intracellular localization observed here for CCR5 was described for other GPCRs and might constitute a more general feature than previously appreciated. The physiological significance of this phenomenon may vary as a function of the receptor and the tissue where it occurs. For example, thrombin receptors are irreversibly activated by cleavage, internalized and degraded in lysosomes, whereas intracellular receptors are protected from activation by the protease. Replenishment of plasma membrane thrombin receptors is thus correlated with recovery of thrombin responsiveness 12. In neurons, only a small fraction of δ-opioid receptors is localized at the neuronal plasma membrane 37, consistent with their low physiological involvement in acute pain response 38. Cell surface translocation of δ-opioid receptors from intracellular compartments might account for the enhanced effect of drugs, specifically targeting this receptor, during chronic pain 37. Similarly, what we describe here for CCR5 may represent a mechanism for maintaining a sustained sensitivity of leukocytes to chemokine guidance within tissues.
From a mechanistic point of view, two important issues must be addressed to fully understand this phenomenon, namely, what are the molecular tethers, which retain receptors within intracellular compartments, and what are the intracellular events that allow their release. So far, only two proteins directly and specifically involved in the control of GPCR cell surface export via intracellular retention have been identified. The second extracellular loop of the protease-activated PAR2 receptor was shown to interact with the N-terminal domain of the Golgi-resident type I transmembrane protein p24A, and to be retained in the Golgi because of this interaction. Upon activation of cell surface PAR2, the small G protein ARF1 is recruited in its GDP-bound form, to Golgi membranes, where a specific exchange factor activates ARF1. This process results in the dissociation of PAR2 from p24A and receptor sorting to the plasma membrane 39. In a different context, during development, a GPCR-retaining protein was reported to control the surface receptor availability of Frizzled, a GPCR, which promotes caudalizing signals. This ER-resident protein, Shisa, is specifically expressed in head ectoderm, where it binds to and inhibits cell surface trafficking of Frizzled. Shisa-mediated receptor retention thus constitutes a mechanism to control head-tail polarity 40. Other recent examples of regulatory retaining proteins have been reported for growth-factor receptors 41, the gamma secretase complex 42 or the glutamate transporter 43, suggesting that this field may rapidly evolve.
A clear picture of the cellular mechanisms, which allow the release of intracellular GPCRs and their cell surface translocation is still missing. Various cellular signals were reported to elicit “acute” effects, such as the rise in intracellular calcium by either release from intracellular stores or direct opening of ion channels, in the case of δ-opioid receptors 44. Similarly, in tubular renal cells, dopamine D1 receptors can be recruited from cytosolic stores to the plasma membrane upon agonist activation of cell surface receptors 13 or via atrial natriuretic peptide-dependent heterologous activation 45. Interestingly, the cell surface targeting of dopamine D1 receptors is also regulated via the interaction with DRIP78, a putative two-transmembrane domain protein, resident of the ER, which binds to a FXXXFXXXF motif found in the C-terminus of the receptor 46. The potential link between signal-mediated export of dopamine D1 receptors and interaction with DRIP78 has not been established yet. Nevertheless, DRIP78 is an example of an emerging paradigm in the field, namely that GPCR translocation to the plasma membrane from intracellular compartments often requires their interaction with non-conventional chaperones and escorts proteins. In addition to classical chaperones, such as calnexin, calreticulin and Bip, which regulate folding and export of proteins engaged in the secretory pathway 47, a vast array of membrane-associated or cytoplasmic proteins has been identified, which interact with GPCRs within intracellular compartments and facilitate their cell surface expression. The list of these proteins, which also include RAMPs, RTPs, REEP, MRAP, and M10 MHC class-I molecules, already mentioned in the introduction, is rapidly growing 11. Based on the data reported in the present study the CD4 molecule may be a new member of this list.
CCR5-CD4 association has been known for a long time, as these molecules combine to form a receptor for HIV at the cell surface of mononuclear blood cells 5. Although still a matter of debate, studies based on different experimental approaches have suggested the existence of a physical, constitutive, virus-independent, CCR5-CD4 association at the cell surface of both transfected 34,48 and primary cells 49,50. The association of HIV co-receptors is likely mediated by interactions involving the extracellular globular domain of CD4 and the extracellular regions of CCR5 34,48,51, but it has not yet been determined whether these interactions occur once the HIV co-receptors have reached the plasma membrane or if they are established within intracellular compartments before their export to the cell surface. Our data strongly support the hypothesis that CCR5-CD4 interaction occurs within intracellular compartments, likely in the ER.
Facilitating the export of CCR5 to the cell surface is clearly not the principal biological function of CD4 and it is possible that the surface expression of CCR5 is enhanced by the interaction with other proteins, which eventually interact with CCR5 along the biosynthetic pathway. In addition, it remains to be established whether CCR5 might also be rapidly mobilized by cell signalling events, as for the dopaminergic and opioid receptors mentioned above. From a mechanistic point of view, CD4-CCR5 association in the ER might facilitate further interactions with components of the biosynthetic pathway, likely via the carboxyterminal tail of CCR5. Indeed, although not involved in the interaction with CD4, the CCR5 tail was reported to contain a bipartite motif critical for CCR5 cell surface expression 52, and we confirmed that the CCR5ΔCter mutant was completely retained within intracellular compartments, even in the presence of excess CD4 (not shown). Interestingly, the effect of CD4 on surface CCR5 was concentration-dependent, displaying a bell-shaped pattern comparable to that observed for calnexin and the ER-membrane-associated protein, DRIP78 on the maturation and trafficking of dopaminergic receptors 46,53. The existence of an optimal concentration of chaperones or escort proteins likely reflects the requirement of a specific stoichiometry between these proteins, the receptors, and other components of the biosynthetic machinery. In the case of CD4, an alternative possible explanation may be that, at high concentrations, CD4 itself can saturate the export machinery.
For many years, no functional explanation was proposed of how HIV may have selected a composite receptor to infect human immune cells. Recently, a plausible explanation for the formation of the second major HIV receptor, constituted by CD4 and CXCR4 on T lymphocytes, came from the observation that CXCR4 associates with the T cell receptor to signal in T cells 54. Thus, the stable proximity of CXCR4 and CD4 at the cell surface of lymphocytes likely arises from the bridging of these proteins via a common interactor, the T cell receptor. Interestingly, based on our findings, a different explanation emerges for CCR5: the physiological constitutive association of CCR5 and CD4 in the ER of blood mononuclear cells, and the subsequent translocation of the resulting complexes at the plasma membrane might be promoted by the escort properties of CD4 for CCR5.
Supplementary Material
Acknowledgments
The authors thank Jean Liotard for his skilled technical assistance and Pierre Bourdoncle for his valuable assistance in cell-imaging experiments. This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale (INSERM), the Centre National de la Recherche Scientifique (CNRS), the Université Paris Descartes, Sidaction and the Fondation de France.
Footnotes
Authors’ Contributions
LA performed research, analyzed data and wrote the article; MGHS analyzed data and wrote the article; HS and AT performed research; GB contributed new reagents and analyzed data; CLJ designed the research, performed research and analyzed data; SM designed the research, analyzed data wrote the article.
References
- 1.Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal. 2004;16:1201–1210. doi: 10.1016/j.cellsig.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Raport CJ, Gosling J, Schweickart VL, Gray PW, Charo IF. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta, and MIP-1alpha. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 3.Aramori I, Ferguson SS, Bieniasz PD, Zhang J, Cullen B, Cullen MG. Molecular mechanism of desensitization of the chemokine receptor CCR-5: receptor signaling and internalization are dissociable from its role as an HIV-1 co-receptor. EMBO J. 1997;16:4606–4616. doi: 10.1093/emboj/16.15.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Corno M, Liu QH, Schols D, et al. HIV-1 gp120 and chemokine activation of Pyk2 and mitogen-activated protein kinases in primary macrophages mediated by calcium-dependent, pertussis toxin-insensitive chemokine receptor signaling. Blood. 2001;98:2909–2916. doi: 10.1182/blood.v98.10.2909. [DOI] [PubMed] [Google Scholar]
- 5.Alkhatib G, Combadiere C, Broder CC, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 6.Doyle C, Strominger JL. Interaction between CD4 and class II MHC molecules mediates cell adhesion. Nature. 1987;330:256–259. doi: 10.1038/330256a0. [DOI] [PubMed] [Google Scholar]
- 7.Asperti-Boursin F, Real E, Bismuth G, Trautmann A, Donnadieu E. CCR7 ligands control basal T cell motility within lymph node slices in a phosphoinositide 3-kinase-independent manner. J Exp Med. 2007;204:1167–1179. doi: 10.1084/jem.20062079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Signoret N, Oldridge J, Pelchen-Matthews A, et al. Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol. 1997;139:651–664. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohout TA, Nicholas SL, Perry SJ, Reinhart G, Junger S, Struthers RS. Differential desensitization, receptor phosphorylation, beta-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J Biol Chem. 2004;279:23214–23222. doi: 10.1074/jbc.M402125200. [DOI] [PubMed] [Google Scholar]
- 10.Vroon A, Heijnen CJ, Kavelaars A. GRKs and arrestins: regulators of migration and inflammation. J Leukoc Biol. 2006;80:1214–1221. doi: 10.1189/jlb.0606373. [DOI] [PubMed] [Google Scholar]
- 11.Achour L, Labbe-Juillie C, Scott MGH, Mariullo S. An escort for G Protein Coupled Receptors to find their path: implication for regulation of receptor density at the cell surface. Trends Pharmacol Sci. 2008;29:528–535. doi: 10.1016/j.tips.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 12.Hein L, Ishii K, Coughlin SR, Kobilka BK. Intracellular targeting and trafficking of thrombin receptors. A novel mechanism for resensitization of a G protein-coupled receptor. J Biol Chem. 1994;269:27719–27726. [PubMed] [Google Scholar]
- 13.Brismar H, Asghar M, Carey RM, Greengard P, Aperia A. Dopamine-induced recruitment of dopamine D1 receptors to the plasma membrane. Proc Natl Acad Sci U S A. 1998;95:5573–5578. doi: 10.1073/pnas.95.10.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cahill CM, McClellan KA, Morinville A, et al. Immunohistochemical distribution of delta opioid receptors in the rat central nervous system: evidence for somatodendritic labeling and antigen-specific cellular compartmentalization. J Comp Neurol. 2001;440:65–84. doi: 10.1002/cne.1370. [DOI] [PubMed] [Google Scholar]
- 15.Dong C, Filipeanu CM, Duvernay MT, Wu G. Regulation of G protein-coupled receptor export trafficking. Biochim Biophys Acta. 2007;1768:853–870. doi: 10.1016/j.bbamem.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLatchie LM, Fraser NJ, Main MJ, et al. RAMPs regulate the transport and ligand specificity of the calcitonin- receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 18.Behrens M, Bartelt J, Reichling C, Winnig M, Kuhn C, Meyerhof W. Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem. 2006;281:20650–20659. doi: 10.1074/jbc.M513637200. [DOI] [PubMed] [Google Scholar]
- 19.Loconto J, Papes F, Chang E, et al. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- 20.Metherell LA, Chapple JP, Cooray S, et al. Mutations in MRAP, encoding a new interacting partner of the ACTH receptor, cause familial glucocorticoid deficiency type 2. Nat Genet. 2005;37:166–170. doi: 10.1038/ng1501. [DOI] [PubMed] [Google Scholar]
- 21.Issafras H, Angers S, Bulenger S, et al. Constitutive agonist-independent CCR5 oligomerization and antibody-mediated clustering occurring at physiological levels of receptors. J Biol Chem. 2002;277:34666–34673. doi: 10.1074/jbc.M202386200. [DOI] [PubMed] [Google Scholar]
- 22.Estrabaud E, Le Rouzic E, Lopez-Verges S, et al. Regulated degradation of the HIV-1 Vpu protein through a betaTrCP-independent pathway limits the release of viral particles. PLoS Pathog. 2007;3:e104. doi: 10.1371/journal.ppat.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller A, Kelly E, Strange PG. Pathways for internalization and recycling of the chemokine receptor CCR5. Blood. 2002;99:785–791. doi: 10.1182/blood.v99.3.785. [DOI] [PubMed] [Google Scholar]
- 24.Fukuda S, Bian H, King AG, Pelus LM. The chemokine GRObeta mobilizes early hematopoietic stem cells characterized by enhanced homing and engraftment. Blood. 2007;110:860–869. doi: 10.1182/blood-2006-06-031401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schnoor M, Cullen P, Lorkowski J, et al. Production of type VI collagen by human macrophages: a new dimension in macrophage functional heterogeneity. J Immunol. 2008;180:5707–5719. doi: 10.4049/jimmunol.180.8.5707. [DOI] [PubMed] [Google Scholar]
- 26.Paruch S, El-Benna J, Djerdjouri B, Marullo S, Perianin A. A role of p44/42 mitogen-activated protein kinases in formyl-peptide receptor-mediated phospholipase D activity and oxidant production. Faseb J. 2006;20:142–144. doi: 10.1096/fj.05-3881fje. [DOI] [PubMed] [Google Scholar]
- 27.Moriuchi M, Moriuchi H. Octamer transcription factors up-regulate the expression of CCR5, a coreceptor for HIV-1 entry. J Biol Chem. 2001;276:8639–8642. doi: 10.1074/jbc.M008391200. [DOI] [PubMed] [Google Scholar]
- 28.Ansari-Lari MA, Liu XM, Metzker ML, Rut AR, Gibbs RA. The extent of genetic variation in the CCR5 gene. Nat Genet. 1997;16:221–222. doi: 10.1038/ng0797-221. [DOI] [PubMed] [Google Scholar]
- 29.Shioda T, Nakayama EE, Tanaka Y, et al. Naturally occurring deletional mutation in the C-terminal cytoplasmic tail of CCR5 affects surface trafficking of CCR5. J Virol. 2001;75:3462–3468. doi: 10.1128/JVI.75.7.3462-3468.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marullo S, Bouvier M. Resonance Energy Transfer approaches in Molecular Pharmacology and beyond. Trends Pharmacol Sci. 2007;28:362–365. doi: 10.1016/j.tips.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Marshall FH, Jones KA, Kaupmann K, Bettler B. GABAB receptors - the first 7TM heterodimers. Trends Pharmacol Sci. 1999;20:396–399. doi: 10.1016/s0165-6147(99)01383-8. [DOI] [PubMed] [Google Scholar]
- 32.Bleul CC, Farzan M, Choe H, et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 33.Ding Z, Issekutz TB, Downey GP, Waddell TK. L-selectin stimulation enhances functional expression of surface CXCR4 in lymphocytes: implications for cellular activation during adhesion and migration. Blood. 2003;101:4245–4252. doi: 10.1182/blood-2002-06-1782. [DOI] [PubMed] [Google Scholar]
- 34.Xiao X, Wu L, Stantchev TS, et al. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci U S A. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willey RL, Maldarelli F, Martin MA, Strebel K. Human Immunodeficiency Virus type 1 Vpu protein induces rapid degradation of CD4. J Virol. 1992;66:7193–7200. doi: 10.1128/jvi.66.12.7193-7200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Margottin F, Bour SP, Durand H, et al. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- 37.Cahill CM, Holdridge SV, Morinville A. Trafficking of delta-opioid receptors and other G-protein-coupled receptors: implications for pain and analgesia. Trends Pharmacol Sci. 2007;28:23–31. doi: 10.1016/j.tips.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Kieffer BL. Opioids: first lessons from knockout mice. Trends Pharmacol Sci. 1999;20:19–26. doi: 10.1016/s0165-6147(98)01279-6. [DOI] [PubMed] [Google Scholar]
- 39.Luo W, Wang Y, Reiser G. p24A, a type I transmembrane protein, controls ARF1-dependent resensitization of protease-activated receptor-2 by influence on receptor trafficking. J Biol Chem. 2007;282:30246–30255. doi: 10.1074/jbc.M703205200. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell. 2005;120:223–235. doi: 10.1016/j.cell.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 41.Couturier C, Sarkis C, Seron K, et al. Silencing of OB-RGRP in mouse hypothalamic arcuate nucleus increases leptin receptor signaling and prevents diet-induced obesity. Proc Natl Acad Sci U S A. 2007;104:19476–19481. doi: 10.1073/pnas.0706671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spasic D, Raemaekers T, Dillen K, et al. Rer1p competes with APH-1 for binding to nicastrin and regulates gamma-secretase complex assembly in the early secretory pathway. J Cell Biol. 2007;176:629–640. doi: 10.1083/jcb.200609180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggiero AM, Liu Y, Vidensky S, et al. The endoplasmic reticulum exit of glutamate transporter is regulated by the inducible mammalian Yip6b/GTRAP3-18 protein. J Biol Chem. 2008;283:6175–6183. doi: 10.1074/jbc.M701008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scott L, Kruse MS, Forssberg H, Brismar H, Greengard P, Aperia A. Selective up-regulation of dopamine D1 receptors in dendritic spines by NMDA receptor activation. Proc Natl Acad Sci U S A. 2002;99:1661–1664. doi: 10.1073/pnas.032654599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holtback U, Brismar H, DiBona GF, Fu M, Greengard P, Aperia A. Receptor recruitment: a mechanism for interactions between G protein-coupled receptors. Proc Natl Acad Sci U S A. 1999;96:7271–7275. doi: 10.1073/pnas.96.13.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bermak JC, Li M, Bullock C, Zhou QY. Regulation of transport of the dopamine D1 receptor by a new membrane- associated ER protein. Nat Cell Biol. 2001;3:492–498. doi: 10.1038/35074561. [DOI] [PubMed] [Google Scholar]
- 47.Ellgaard L, Helenius A. ER quality control: towards an understanding at the molecular level. Curr Opin Cell Biol. 2001;13:431–437. doi: 10.1016/s0955-0674(00)00233-7. [DOI] [PubMed] [Google Scholar]
- 48.Gaibelet G, Planchenault T, Mazeres S, et al. CD4 and CCR5 constitutively interact at the plasma membrane of living cells: A confocal fret-based approach. J Biol Chem. 2006;281:37921–37929. doi: 10.1074/jbc.M607103200. [DOI] [PubMed] [Google Scholar]
- 49.Lapham CK, Zaitseva MB, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 50.Singer S, Scott S, Kawka DW, et al. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J Virol. 2001;75:3779–3790. doi: 10.1128/JVI.75.8.3779-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Staudinger R. Interaction of soluble CD4 with the chemokine receptor CCR5. Biochem Biophys Res Commun. 2003;307:1066–1069. doi: 10.1016/s0006-291x(03)01315-9. [DOI] [PubMed] [Google Scholar]
- 52.Venkatesan S, Petrovic A, Locati M, Kim YO, Weissman D, Murphy PM. A membrane proximal basic domain and cysteine cluster in the C-terminal tail of CCR5 constitute a bipartite motif critical for cell surface expression. J Biol Chem. 2001;276:40133–40145. doi: 10.1074/jbc.M105722200. [DOI] [PubMed] [Google Scholar]
- 53.Free RB, Hazelwood LA, Cabrera DM, et al. D1 and D2 dopamine receptor expression is regulated by direct interaction with the chaperone protein calnexin. J Biol Chem. 2007;282:21285–21300. doi: 10.1074/jbc.M701555200. [DOI] [PubMed] [Google Scholar]
- 54.Kumar A, Humphreys TD, Kremer KN, et al. CXCR4 physically associates with the T cell receptor to signal in T cells. Immunity. 2006;25:213–224. doi: 10.1016/j.immuni.2006.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


