Abstract
We determined the 1.6-Å resolution crystal structure of a conserved hypothetical 29.9-kDa protein from the SIGY–CYDD intergenic region encoded by a Bacillus subtilis open reading frame in the YXKO locus. YXKO homologues are broadly distributed and are by and large described as proteins with unknown function. The YXKO protein has an α/β fold and shows high structural homology to the members of a ribokinase-like superfamily. However, YXKO is the only member of this superfamily known to form tetramers. Putative binding sites for adenosine triphosphate (ATP), a substrate, and Mg2+-binding sites were revealed in the structure of the protein, based on high structural similarity to ATP-dependent members of the superfamily. Two adjacent monomers contribute residues to the active site. The crystal structure provides valuable information about the YXKO protein's tertiary and quaternary structure, the biochemical function of YXKO and its homologues, and the evolution of its ribokinase-like superfamily.
Keywords: Bacillus subtilis YXKO, UPF0031 family, Kinase, Structural genomics
1. Introduction
The far-reaching goal of structural genomics programs is to map the entire protein folding space. These pilot projects exploit the available genome sequence information to select targets whose structures can be determined. Among genes coding for proteins that show no significant sequence similarity to proteins with known structure, there is a large group of conserved “hypothetical” open reading frames (ORFs)2 (Vitkup et al., 2001). Many of these ORFs include members from diverse organisms and can be grouped into large families. Their incidence in many branches of life suggests that they are important; however, their functions cannot be revealed through sequence analysis. Selection of such targets for structural genomics studies is important because the 3D structure of these proteins may offer insight not only into their folds but also into their biochemical or biophysical functions (Zarembinski et al., 1998)—provided that a structural homologue is already known (Martinez-Cruz et al., 2002)—and allow this knowledge to be extended to their sequence homologues. It is anticipated that as the number of nonredundant protein structures grows in the Protein Data Bank (PDB), this approach will be more successful in defining the functions of “hypothetical” proteins.
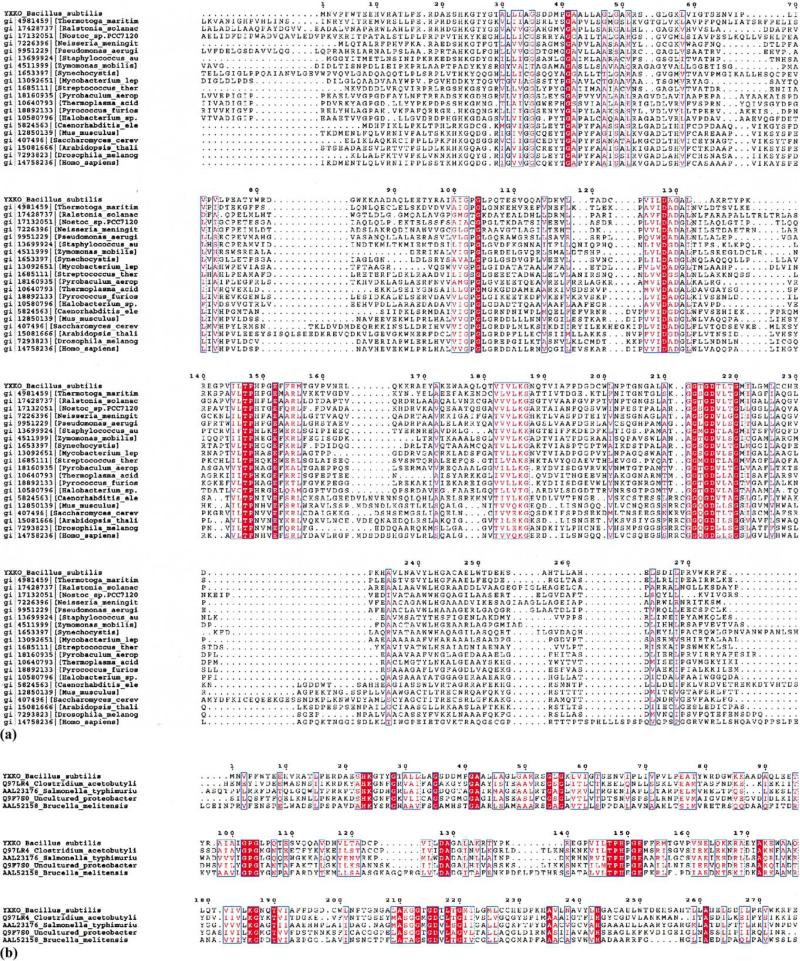
For this work we have selected, from a large set of structural genomics targets of the gram-positive bacterium Bacillus subtilis (http://www.mcsg.anl.gov), a conserved hypothetical 29.9-kDa protein from the SIGY–CYDD intergenic region encoded by the B. subtilis ORF in the YXKO locus (DNA bases 50795–51625) (Yoshida et al., 1996). [The code “YXKO” will be used for this protein (aka the Midwest Center for Structural Genomics APC234) throughout this text.] Sequence similarity searches of the National Center for Biotechnology Information nonredundant database, using BLAST (Altschul et al., 1997) and FASTA (Pearson, 2000), and the SWISS-PROT (Bairoch and Apweiler, 2000) and Protein Information Resource (Wu et al., 2002) databases resulted in 69 protein homologues of YXKO with statistically significant E values (E < 10–5). These proteins are broadly distributed and are found in 42 species of bacteria, 14 species of archaea, and 13 eukaryotes, including human, and are described as proteins with unknown function. A few are classified as probable sugar kinases (Fig. 1).
Fig. 1.
Multiple sequence alignment of YXKO using ClustalW. (a) Proteins from the ribokinase-like family, (b) probable bacterial kinases (YXKO subfamily). The residues strictly conserved are colored white and placed in red boxes; the residues highly conserved and with similar properties are in red.
These proteins belong to the Pfam (Bateman et al., 1999) UPF0031 family, which includes 62 uncharacterized bacterial proteins and contains two Prosite sequence patterns [UPF0031_1, (SAV)-(IVW)-(LVA)-(LIV)-G-(PNS)-G-L-(GP)-x-(DENQT), and UPF0031_2,(GA)-G-x-G-D-(TV)-(LT)-(STA)-G-x-(LIVM)]. The structures of all these proteins are unknown. YXKO has distant sequence homology with the ThiK protein (PDB entry 1EKK) deposited in the PDB (discussed later in this paper).
We have determined the 1.6-Å resolution crystal structure of YXKO from B. subtilis. The protein shows structural homology to members of the ribokinase-like carbohydrate kinase superfamily, as defined by SCOP (Murzin et al., 1995) and CATH (Orengo et al., 1997). Putative ATP- and substrate-binding sites in the YXKO structure were assigned and they reveal high structural similarity within ATP-dependent ribokinases (RKs). Further elucidation of YXKO's detailed biochemical function will require a combination of enzymatic, mutational, and structural studies; however, the crystal structure for YXKO provides important insight into the function of this protein and others in the UPF0031 family.
2. Materials and methods
2.1. Cloning, expression, and protein purification
The ORF of a B. subtilis YXKO protein (29.9 kDa) in the SIGY–CYDD intergenic region was amplified from genomic DNA with Pfx DNA polymerase using conditions and reagents provided by the vendor (Invitrogen). The gene was cloned into a pMCSG7 vector (Stols et al., 2002) using a modified ligation-independent cloning protocol (Dieckman et al., 2002). This process generated an expression clone producing a fusion protein with an N-terminal His6 tag and a TEV protease recognition site (ENLYFQ ↓ S). The fusion protein was overproduced in an Escherichia coli BL21 derivative harboring a plasmid encoding three rare E. coli tRNAs [Arg (AGG/AGA) and Ile (ATA)].
A selenomethionine (Se-Met) derivative of the expressed protein was prepared as described by Walsh et al. (1999). The protein was purified by resuspension of IPTG-induced bacterial cells in binding buffer (20 mM Hepes, pH 8.0, 500 mM NaCl, 5% glycerol, 10 mM imidazole, and 10 mM β-mercaptoethanol). The cells were lysed by the addition of lysozyme to 1 mg/ml in the presence of a protease inhibitor cocktail (Sigma P8849) (0.25 ml/5 g cells) and sonication for 2–3 min. After clarification by centrifugation (30 min at 30,000g) and passage through a 0.2-μm filter (HT Tuffryn polysulfonate membrane; Pall Gelman), the lysate was applied to nitrilotriacetic acid (Ni-NTA) Superflow resin (Qiagen) and unbound proteins were removed by washing with 10 volumes of binding buffer. The protein was eluted from the column with 250 mM imidazole, and the fusion tag was cleaved with recombinant His-tagged tobacco etch virus (TEV) protease (Kapust et al., 2001). Native protein was purified from the His tag and undigested protein, persistent contaminant proteins from E. coli, and TEV protease (gift from Dr. D.S. Waugh) by application of the solution to a second Ni-NTA column. The purified protein was dialyzed in 20 mM Hepes, pH 7.5, 500 mM NaCl, and 2 mM DTT and concentrated using Centricon Plus-20 (Millipore).
2.2. Size-exclusion chromatography
The molecular weight of YXKO protein in solution was determined by size-exclusion chromatography on a Superdex-200 10/30 column (Pharmacia) calibrated by ribonuclease A (13.7 kDa), ovalbumin (43 kDa), albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), and thyroglobulin (669 kDa) as standards. The calibration curve of Kav versus log molecular weight was prepared using the equation Kav = Ve – Vo/Vt – Vo, where Ve is the elution volume for the protein, Vo is the column void volume, and Vt is the total bed volume.
2.3. Crystallization
Crystals of YXKO were grown using vapor diffusion in hanging drops containing equal volumes of protein at 25 mg/ml and reservoir containing 28% PEG 400, 0.1 M Hepes, pH 7.5, and 0.15 MgCl2. YXKO Se-Met derivative was crystallized under the same conditions. For data collection a single crystal of approximately 0:4 × 0:3 × 0:3 mm was flash frozen in liquid nitrogen directly from mother liquor.
2.4. Data collection
Diffraction data were collected at 100 °K at the 19ID beam line of the Structural Biology Center at the Advanced Photon Source, Argonne National Laboratory. A three-wavelength multiwavelength anomalous diffraction (MAD) dataset was collected at 2.0 Å resolution from a single Se-Met-labeled protein crystal using inverse-beam strategy (Table 1). Frames of 0.5° were collected at 100 °K using 4-s exposures and 150-mm crystal-to-detector distance. The total oscillation range covered 60° of unique reciprocal space, as predicted using the strategy module within the HKL2000 suite (Otwinowski and Minor, 1997). The space group was I422, with cell dimensions of a = b = 91.90 Å c = 170.506 Å. All data were processed and scaled with HKL2000 (Table 1) and show strong anomalous signal. The high-resolution native dataset (completeness 97.8%) was collected to 1.6 Å and was used for phase extension, automated model building, and refinement.
Table 1.
Summary of crystal and MAD data
| Crystal data |
||
|---|---|---|
| Unit cell | a = b = 91.90 Å, c = 170Å, α = β = γ = 90° | |
| Space group | I422 | |
| MW Da (residues) | 29,900 | |
| Mol (AU) | 1 | |
| Se-Met (AU) | 4 |
| MAD data |
||||
|---|---|---|---|---|
| Edge | Peak | Remote | High resolution | |
| Wavelength (Å) | 0.9794 | 0.9793 | 0.9538 | 1.0332 |
| Resolution range (Å) | 2.0 | 2.0 | 2.0 | 30–1.6 (1.60–1.60) |
| No. of unique reflections | 21,822 | 21,742 | 24,278 | 48,555 |
| Completeness (%) | 89.9 | 89.6 | 100.0 | 97.3 (86.0) |
| R merge (%) | 11.2 | 11.9 | 9.9 | 14.6 (54.6) |
2.5. Structure determination and refinement
The structure was determined by MAD phasing using a crystallography and nuclear magnetic resonance software system (CNS; Brunger et al., 1998) and was refined with CNS initially to 2.0 Å using the averaged peak data. The initial model was built automatically using ARP/wARP (Perrakis et al., 1999). The manual adjustment of the model was completed using QUANTA (Molecular Simulations, 2000). The model was further refined against native data to 1.6 Å. The final crystallographic R factor is 0.228 and the free R value is 0.232 with 1σ data (Table 2). Electron density calculated at 1.2σ is well connected for most of the main chain except at the N-terminal Met and at residues 19–25, which are disordered in the crystal structure (Fig. 2). The stereochemistry of the structure was verified with PROCHECK (Laskowski et al., 1996) and the Ramachandran plot. All main-chain torsion angles for all residues are in the allowed regions and in additional allowed regions.
Table 2.
Statistics of structure determination of refinement
| Phasing |
|||||||
|---|---|---|---|---|---|---|---|
| Centric |
Acentric |
All |
|||||
| Resolution range (Å) | FOM | Phasing power | FOM | Phasing power | No. | FOM | Phasing power |
| 50.0–2.0 | 0.6023 | 3.24 | 0.6745 | 2.83 | 46083 | 0.669 | 2.85 |
| Density modification | 0.88 | ||||||
| Refinement | |||||||
| Resolution range (Å) | 50–1.6 | ||||||
| No. of reflections | 92,275 | ||||||
| σ cutoff | 1.0 | ||||||
| R value (%) | 22.8 | ||||||
| Free R value (%) | 23.2 | ||||||
| Rms deviations from ideal geometry | |||||||
| Bond length (1-2) (Å) | 0.005 | ||||||
| Angle (°) | 1.40 | ||||||
| Dihedral (°) | 22.0 | ||||||
| Improper (°) | 0.80 | ||||||
| No. of atoms | |||||||
| Protien | 2044 | ||||||
| Water | 209 | ||||||
| Mean B factor (Å2) | |||||||
| All atoms | 22.9 | ||||||
| Protein atoms | |||||||
| Protein main chain | 22.1 | ||||||
| Protein side chain | 23.3 | ||||||
| Water | 38.0 | ||||||
| Ramachandran plot statistics (%) | |||||||
| Residues in most favored regions | 99.6 | ||||||
| Residues in additional allowed regions | 0.4 | ||||||
| Residues in disallowed region | 0.0 | ||||||
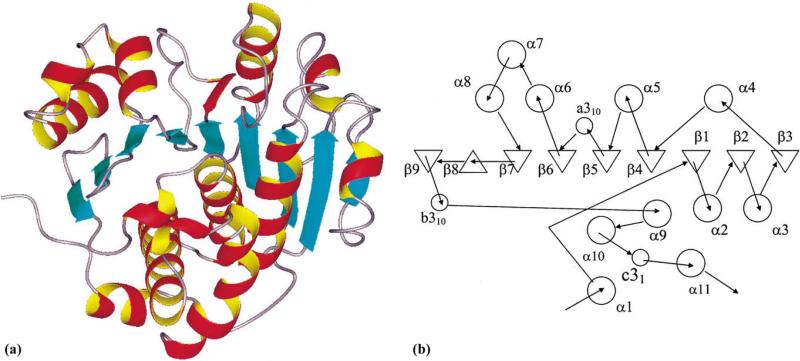
Fig. 2.
Ribbon drawing and topology of the subunit structure of YXKO. (a) The ribbon drawing of the YXKO subunit structure showing the overall fold. β strands are presented as blue arrows, α helices are depicted as red and yellow coils, and loops are shown in gray. (b) Topology diagram of the YXKO fold (α helices are shown as circles and β strands as triangles).
3. Results and discussion
3.1. Description of YXKO structure
The protein crystallized in centered tetragonal crystal form in an I422 space group with one subunit in the asymmetric unit. In the crystal the YXKO monomer has an α/β fold and shows structural similarity to the RK-like carbohydrate kinase superfamily (Fig. 2) (Murzin et al., 1995; Orengo et al., 1997). Each subunit of YXKO contains 276 amino acids, with approximately 42% of the amino acids in α helices, 12% in β sheets, and 3% in 310 helices. The remaining portion of the structure is composed of loops and coils. Each subunit contains a central, mainly parallel, nine-stranded β sheet; 11 helices; and 3 310 helices. The β sheet has the topology β3 ↓ β2 ↓ β1 ↓ β4 ↓ β5 ↓ β6 ↓ β7 ↓ β8 ↑ β9 ↓, with eight strands parallel to each other strand 8 antiparallel to all other strands. Two β hairpins involving strands 7, 8, and 9 form one edge of the central β sheet (Fig. 2b).
Helices α4α5α6α7α8 and the a310 helix cover the convex side of the β sheet, while helices α1α2α3α9α10α11 and the remaining two (b and c) 310 helices are on the concave side of the β sheet (Fig. 2). Helices α2α3α5α6α8α9 are approximately antiparallel to the strands of the β sheet, while the α10 helix is parallel to these strands and anti-parallel to strand 8, which approaches it most closely. Helices α2α9α10α11 form a four-helix bundle on the concave side of the β sheet (Fig. 2b).
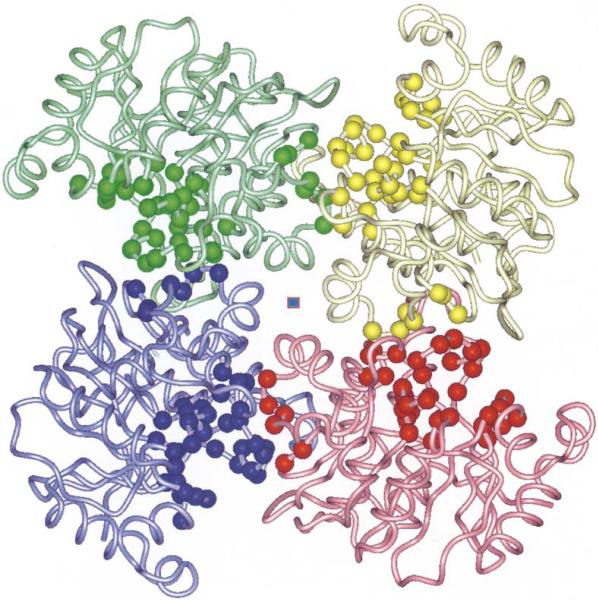
3.2. Yxko is a tetramer
YXKO monomers interact extensively with adjacent subunits, creating a tetramer with 4/1 symmetry (the monomers are related by a fourfold crystallographic axis). The overall shape of the tetramer resembles the blades of a fan with the approximate dimensions of 90 × 90 × 50 Å (Fig. 3) and a solvent channel in the middle. Size-exclusion chromatography showed protein migration consistent with a 130-kDa protein (data not shown), which indicates that YXKO is also a tetramer in solution (the molecular weight predicted from the gene sequence of the tetramer is 119.5 kDa).
Fig. 3.
The quaternary structure of the YXKO tetramer. The ribbon diagram is viewed along the fourfold axis (center square). The Cα atoms of residues forming a putative ATP- and substrate-binding pocket are indicated as spheres.
Subunits interact using loops and helices from both the N- and the C-terminal segments of the monomer. The interface is composed of a loop connecting α10 and α11, which involves the c310 helix; a loop between β9 and α9, including a second, b310, helix; a loop preceding the N-terminal residues of the α2 helix and the loop joining β2 and α3; the α3 helix; and the loop connecting α3 with β3 (blue spheres on the interface between blue and pink monomers, Fig. 3).
From the opposing monomer (red spheres on the interface between blue and pink monomers, Fig. 3), contacts involve α11; α3, a loop connecting α1 with β1, a loop connecting α2 with β2, and a loop between α2 and β2. The interface is stabilized by intersubunit polar interactions [salt bridges (E253/R270′ and E263/R52′)] (Fig. 3), hydrogen bonds involving main-chain and side-chain atoms (S255/P269′ and E276′, H257/P16′, R18/G54′, T258/R52′, H262/E76′, G207/R18′, A210/R18′, K211/M51′, G212/G56′, E37/R81′, D38/R81′, and Y76′), and hydrophobic interactions involving L45, P40, L69, and V73 from one subunit and P75, V71, Y79, P68, and P72 from the opposite subunit. Some of these residues are highly conserved in the RK-like family (V71, G212) (Fig. 1a). Others are conserved only among the YXKO subfamily (R18, R52, G54, G56, E76, G207, A210) (Fig. 1b).
3.3. Structural similarity of YXKO to ribokinase-like superfamily
The DALI (Holm and Sander, 1995) three-dimensional similarity search found five structural homologues:
Hydroxyethylthiazole kinase (ThiK) from B. subtilis (PDB code 1c3q) (Campobasso et al., 2000), with the highest score (Z = 20.6)
RK from E. coli (Z = 13.1) (PDB code 1rkd) (Sigrell et al., 1998)
4-Amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase (HMPP kinase) from Salmonella typhimurium (Z = 10.7) (PDB code 1jxh) (Cheng et al., 2002)
Human adenosine kinase (AK) (Z = 10:7) (PDB code 1bx4) (Mathews et al., 1998)
The adenosine diphosphate (ADP)-dependent glucokinase from Thermococcus litoralis (Z = 8.2) (PDB code 1gc5) (Ito et al., 2001)
All structural homologues of YXKO are members of a RK-like carbohydrate kinase superfamily (Murzin et al., 1995; Orengo et al., 1997), which is composed of three families: hydroxyethylthiazole kinase, RK-like, and ADP-dependent glucokinase (Gough and Chothia, 2002; Murzin et al., 1995). YXKO is a new member of this superfamily. All known proteins belonging to this superfamily have similar functions: they are phosphotransferases, they catalyze the phosphorylation of substrates containing hydroxymethyl groups, and they require magnesium and ATP (or ADP) for activity. The vicinity of the ATP- and substrate-binding sites is similar in a structural sense but different in terms of the amino acid framework within the members of this superfamily. A detailed comparison of RK-like superfamily members was recently published (Campobasso et al., 2000; Cheng et al., 2002).
Here we compare YXKO with ATP-dependent kinases of this superfamily. The overall fold of YXKO most closely resembles ThiK (hydroxyethylthiazole kinase family) and is similar to HMPP kinase, RK, and AK (RK-like family). In general, each of these proteins contains a central nine-stranded β-sheet (eight-stranded in HMPP kinase) flanked by several α helices. However, in contrast to ThiK and YXKO, the remaining members of the ATP-dependent RK-like family whose structures have been determined so far have an additional structural unit: a four-stranded β sheet in RK, a five-stranded β sheet and two helices in AK, and two β strands in HMPP kinase (corresponding to the second β-sheet of RK and AK) (Cheng et al., 2002).
The sequence alignment using Combinatorial Extension (Shindyalov and Bourne, 1998) shows that 215 amino acids of YXKO can be superimposed on structurally equivalent residues in ThiK with an RMSD of Cα 3.0 Å and 21% sequence identity. Similarly, the 227 amino acids of YXKO can be superimposed on RK from E. coli with an RMSD of 3.5 Å (14% sequence identity), 208 amino acids on HMPP kinase with an RMSD of 3.5 Å (15.4% sequence identity), 226 amino acids on human AK with an RMSD of 3.7 Å (12.5% sequence identity), and 191 amino acids on AK from Toxoplasma gondii with an RMSD of 3.3 Å (13% sequence identity).
These results point to the strongest similarity between YXKO and ThiK, but they also suggest that very different protein sequences can assume very similar 3D structures and engage in similar functions.
3.4. Putative ATP-binding site
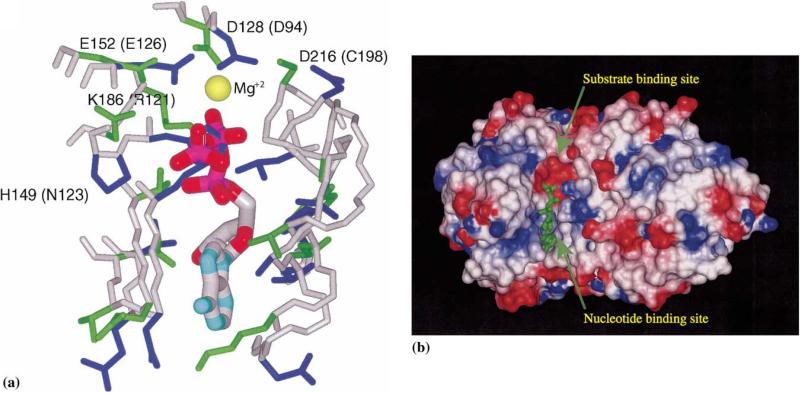
Several crystal structures of RK-like superfamily members in complex with nucleotides are available: ThiK with ATP (PDB code 1esq), RK with ADP (PDB codes 1rk2 and 1rkd), and AKs with ATP analogues (PDB codes 1dgy and 1bx4). Therefore, detailed structural comparisons with YXKO can be carried out. The nucleotide-binding site shows high structural similarity within all ATP-dependent members of the RK-like superfamily (Campobasso et al., 2000; Cheng et al., 2002). The putative ATP-binding site in YXKO was assigned as a result of the superimposition of YXKO on the structures of RK-like superfamily members. The structural superposition with ThiK is shown in Fig. 4. The appropriate space and environment for ATP binding in YXKO are similar to those in other members of the superfamily. The site most closely resembles the ATP-binding site of ThiK.
Fig. 4.
The putative ATP-binding site in YXKO. (a) The superposition of the YXKO structure onto ThiK, with ATP originating from ThiK; magnesium ion conserved within the family is indicated as a yellow sphere; the side chains of residues involved in the interactions with ATP are blue (YXKO) and green (ThiK); the names of ThiK residues corresponding to YXKO residues are in parentheses. (b) The putative ATP- and substrate-binding sites in YXKO, with van der Waals surfaces colored according to the molecular electrostatic potential on the residues: blue indicates positive potential, red indicates negative potential.
The putative ATP-binding site in YXKO is a shallow groove formed by strands β6, β7, and β8 along the C-terminal edge of the central β sheet and is open to the solvent. Docking studies performed with the LiganFit/Cerius2 program (Molecular Simulations, 2000) showed that the position of the nucleotide interacting with YXKO should be very similar to that found in the ThiK/ATP complex. No major steric hindrances were observed except for a clash between γ-phosphate and H149, suggesting that phosphates in ATP may assume somewhat different orientation or YXKO may undergo conformational changes during nucleotide binding, compared with ThiK (the corresponding residue in ThiK is N123).
The area surrounding the putative ATP-binding pocket in YXKO involves a loop connecting the β9 strand with the α9 helix, the beginning of the α9 helix, the loop connecting the β7 and β8 strands, and the loop connecting the β6 strand with the α6 helix, as well as residues L218 (from α9 helix) and H243 (from α10 helix) at the bottom of the pocket. Despite high structural similarity of the binding site, the residues forming the ATP-binding pocket are less rigorously conserved among the superfamily and even among its particular members from different sources (Campobasso et al., 2000; Cheng et al., 2002), with the exception of Arg/Lys residues involved in Mg2+/ATP binding that are strongly conserved within the hydroxyethylthiazole kinase family (K186). In ThiK R121is in an equivalent position and has also been implicated in catalysis. Thus, it is not surprising that only G187 of YXKO is strictly conserved in the members of the RK-like superfamily, while G205 of YXKO is conserved only within ThiK kinase family members (Campobasso et al., 2000). Lack of strong sequence conservation in the ATP-binding site suggests that the effective ligand binding can be accomplished by a surprisingly wide constellation of amino acid side chains.
3.5. Putative metal-binding site in YXKO
Almost all phosphate-transferring enzymes have been shown to bind divalent metal in their active sites, which interacts with both the β- and γ-phosphates to assist the reaction by orienting and stabilizing the γ-phosphate during transfer to the acceptor. Although the Mg2+ ion was not found in the structure, YXKO potentially utilizes the Mg2+ ion to interact with ATP similar to other members of RK-like superfamily.
One Mg2+ ion was found in RK and AK, while in ThiK there are two Mg2+ ions present (Campobasso et al., 2000). The Mg2+ ion in ThiK, which structurally corresponds to the Mg2+ ions in other members of the superfamily, interacts with the oxygen atoms of the β-phosphate of ATP and the phosphate of the product. It also interacts through water molecules with two highly conserved residues, D94 and E126, and with C198. The structurally equivalent residues in YXKO are D128, E152, and D216. Moreover, in RK and AK, the aspartic acid, which serves as the catalytic base, also coordinates the Mg2+ ion (Mathews et al., 1998). The corresponding residue in YXKO is D216. In ATP (or GTP)-binding proteins, an Asp or Glu residue coordinates the Mg2+ ion either directly or through a water molecule (Wild et al., 1997). This suggests that the magnesium ion in YXKO is positioned similar to that of the other members of the superfamily (Fig. 4).
3.6. Residues important for catalysis within the ribokinase-like superfamily
Several residues in the RK-like superfamily have been implicated in catalysis. In YXKO residues 213–216 (GTGD) are predicted to form the kinase anion (Mathews et al., 1998). This region is strictly conserved within the superfamily members (Fig. 1).
The most intriguing difference between the members of the RK-like superfamily is the residue playing the role of a catalytic base proposed to be involved in the deprotonation of a substrate hydroxyl group during phosphorylation. RK and AK utilize the aspartate residues (D255 and D300, respectively) for that function, while ThiK and HMPP have Cys residues (C198 and C213, respectively) at these positions. The mutagenesis studies of ThiK show that the role of the Cys residues in catalysis is not clear, as for instance the C198D mutant exhibits nine times higher activity than the wild-type form of the enzyme (Campobasso et al., 2000). The corresponding residue in YXKO is D216. We propose that this highly conserved residue is a key catalytic base.
3.7. Putative substrate-binding site
The common feature of the members of the RK-like superfamily is the phosphorylation of the primary alcohol functional group. Although for YXKO, the specific substrate has not been identified thus far, the location of the substrate-binding site in this protein may be deduced by analogy to the structures of other RK-like superfamily members. Superposition of the structures of the members of this superfamily revealed that although there are significant differences in the specific amino acid residues in this site—because different substrates are bound there—the overall geometry of the binding site is quite similar. ThiK catalyzes the phosphorylation reaction of a strongly hydrophobic substrate (4-methyl-5-b-hydroxyethylthiazole), while the substrate for HMPP kinase (4-amino-5-hydroxymethyl-2-methylpyrimidine) is slightly more polar (Campobasso et al., 2000; Cheng et al., 2002). In RK and AK, however, the 5′-hydroxyl group of ribose is phosphorylated and these enzymes have similar and strongly hydrophilic substrate-binding sites, because many hydrogen bonds are formed there (Mathews et al., 1998; Sigrell et al., 1998). The putative substrate-binding site in YXKO is significantly less polar compared with RK and AK; therefore, it appears that the substrate could be quite hydrophobic.
The binding sites in YXKO (a tetramer in solution) and ThiK (a trimer in solution) (Campobasso et al., 2000) have one substrate-binding site per subunit. The residues from two monomers are involved in the formation of the substrate-binding site in both proteins (Figs. 3 and 4). In ThiK the residues from the second subunit strand, β2, and helix α2 form a lid over the substrate-binding site, while YXKO engages the residues from β2 and the loop connecting α3 and β3 as well as the residues from the β3 strand of the second subunit (Fig. 3). It is interesting that α3, α3, and the loop between them correspond to an insertion between β2 and α4 in the ThiK structure.
Other members of the RK-like superfamily have additional structural elements within the same subunit that also function as a lid over the substrate-binding site (Cheng et al., 2002; Mathews et al., 1998; Sigrell et al., 1998). These additional structural elements functionally replace the adjacent subunit in ThiK and YXKO in shielding the substrate-binding site: in the HMPP kinase dimer the flap is formed by two additional short β strands and a connecting loop, in the RK dimer the lid is formed by four β strands from one subunit and one β strand from the twofold-related subunit, and in the AK monomer the lid is formed by five β strands and two α helices, which form the second domain).
3.8. Implications for evolution of ribokinase-like superfamily
The RK-like superfamily members have a common monomer fold, but they differ in quaternary structure and substrate specificity. The evolution pathway of RK-like superfamily members with known structure (excluding YXKO) has been proposed recently (Cheng et al., 2002). It was suggested that the divergence of substrate specificity and quaternary structure might be correlated with the evolution of the active-site lid that shields the substrate from the solvent. It seems that the evolution progressed from two members: (1) ThiK (and YXKO), which appear to utilize oligomerization of subunits to create the active site, and (2) RK and AK, which evolved a second domain—a flap β sheet—to shield the active site. HMPP kinase seems to be a transitional form between these two groups because it has two more strands between β2 and α2 than ThiK has. The structure of YXKO fits well into this hypothesis.
3.9. Functional implications
The sequence- and structure-similarity searches confirmed that the YXKO protein belongs to the RK-like superfamily and is more similar to ThiK kinase than to RK-like family members. Moreover, the ThiK kinase domain, which is present in the members of the ThiK kinase family, was identified in the YXKO sequence (for the sequence fragment of 100–250 amino acid residues), according to conserved-domain searches (Altschul et al., 1997).
On the other hand, all members of the RK-like family contain the pfkB domain, which was not found in YXKO. These findings suggest that YXKO is probably a member of the hydroxyethylthiazole kinase family.
A search using PSI-BLAST (Altschul et al., 1997) detected a strong relationship between YXKO and ThiK kinases from different sources. This search also revealed the relationship of YXKO to HMPP kinase, which is also involved in the thiamine biosynthetic pathway and is responsible for the sequential addition of two phosphate groups to the hydroxymethylpyrimidine. The PSI-BLAST searches for ThiK also revealed the relationship to HMPP kinase (Campobasso et al., 2000). This indicates that some enzymes within the thiamine biosynthetic pathway are related to each other; therefore, we cannot exclude the possibility that YXKO might be also involved in this pathway.
It is worth emphasizing that the residues proposed in YXKO as important for catalysis, binding of ATP and Mg2+ ion, and formation of a kinase anion hole are conserved in the members of the Pfam UPF0031 family. The same residues and several additional residues involved in the formation of the putative substrate and ATP-binding sites are strictly conserved in the YXKO sequence homologues—hypothetical proteins with unknown function—as well as in the predicted sugar kinases (Fig. 1b). This indicates that these proteins might have functions similar to those of the members of the RK-like carbohydrate kinase superfamily (Murzin et al., 1995; Orengo et al., 1997).
The Prosite signature sequence UPF0031_1corresponds to residues 98–109 in YXKO. Residues 103–107 are involved in forming the presumed substrate-binding pocket. The Prosite signature sequence UPF0031_2 corresponds to residues 212–222 in YXKO (loop before α9 and α9 helix). These residues form a presumable kinase anion hole in YXKO, with D216 predicted to be the catalytic base.
Although further biochemical and structural studies are necessary to reveal the detailed biochemical function of the YXKO protein, determination of the structure of this protein sheds light on possible functions of UPF0031 family members and other homologues of the YXKO protein.
Acknowledgments
We thank all the members of the Structural Biology Center at Argonne National Laboratory for their help in conducting experiments. This work was supported by National Institutes of Health Grant GM62414 and by the U.S. Department of Energy, Office of Biological and Environmental Research. Atomic coordinates have been deposited with the Protein Data Bank as 1KYH.
Footnotes
This article has been created by the University of Chicago as Operator of Argonne National Laboratory under Contract W-31-109-ENG-38 with the US Department of Energy. The US Government's right to retain a nonexclusive royalty-free license in and to the copyright covering this paper, for governmental purposes, is acknowledged.
Abbreviations used: AK, human adenosine kinase; HMPP, 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate; ORF, open reading frame; RK, ribokinase; ThiK, 4-methyl-5-β-hydroxyethylthiazole kinase; YXKO, putative kinase UPF0031 family; PDB, Protein Data Bank; TEV, tobacco etch virus; Ni-NTA, nitrilotriacetic acid; MAD, multiwavelength anomalous diffiraction; CNS, crystallography and nuclear magnetic resonance software system.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids. Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic. Acids. Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic. Acids. Res. 1999;27:260–262. doi: 10.1093/nar/27.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta. Crystallogr D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Campobasso N, Mathews II, Begley TP, Ealick SE. Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 Å resolution. Biochemistry. 2000;39:7868–7877. doi: 10.1021/bi0000061. [DOI] [PubMed] [Google Scholar]
- Cheng G, Bennett EM, Begley TP, Ealick SE. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure (Cambridge) 2002;10:225–235. doi: 10.1016/s0969-2126(02)00708-6. [DOI] [PubMed] [Google Scholar]
- Dieckman L, Gu M, Stols L, Donnelley MI, Collart FR. High throughput methods for gene cloning and expression. Protein Expression Purif. doi: 10.1006/prep.2001.1602. in press. [DOI] [PubMed] [Google Scholar]
- Gough J, Chothia C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic. Acids. Res. 2002;30:268–272. doi: 10.1093/nar/30.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Ito S, Fushinobu S, Yoshioka I, Koga S, Matsuzawa H, Wakagi T. Structural basis for the ADP-specificity of a novel glucokinase from a hyperthermophilic archaeon. Structure (Cambridge) 2001;9:205–214. doi: 10.1016/s0969-2126(01)00577-9. [DOI] [PubMed] [Google Scholar]
- Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;12:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Martinez-Cruz LA, Dreyer MK, Boisvert DC, Yokota H, Martinez-Chantar ML, Kim R, Kim SH. Crystal structure of MJ1247 protein from M. jannaschii at 2.0 Å resolution infers a molecular function of 3-hexulose-6-phosphate isomerase. Structure. 2002;10:195–204. doi: 10.1016/s0969-2126(02)00701-3. [DOI] [PubMed] [Google Scholar]
- Mathews II, Erion MD, Ealick SE. Structure of human adenosine kinase at 1.5 Å resolution. Biochemistry. 1998;37:15607–15620. doi: 10.1021/bi9815445. [DOI] [PubMed] [Google Scholar]
- Molecular Simulations Inc.; San Diego: 2000. [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Sigrell JA, Cameron AD, Jones TA, Mowbray SL. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 Å resolution: insights into a new family of kinase structures. Structure. 1998;6:183–193. doi: 10.1016/s0969-2126(98)00020-3. [DOI] [PubMed] [Google Scholar]
- Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelley MI. A new vector for high throughput, ligation independent cloning encoding a TEV protease cleavage site. Protein Expression Purif. doi: 10.1006/prep.2001.1603. in press. [DOI] [PubMed] [Google Scholar]
- Vitkup D, Melamud E, Moult J, Sander C. Completeness in structural genomics. Nat. Struct. Biol. 2001;8:559–566. doi: 10.1038/88640. [DOI] [PubMed] [Google Scholar]
- Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr D Biol. Crystallogr. 1999;55:1168–11673. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- Wild K, Bohner T, Folkers G, Schulz GE. The structures of thymidine kinase from herpes simplex virus type1 in complex with substrates and a substrate analogue. Protein Sci. 1997;6:2097–2106. doi: 10.1002/pro.5560061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Huang H, Arminski L, Castro-Alvear J, Chen Y, Hu ZZ, Ledley RS, Lewis KC, Mewes HW, Orcutt BC, Suzek BE, Tsugita A, Vinayaka CR, Yeh LS, Zhang J, Barker WC. The Protein Information Resource: an integrated public resource of functional annotation of proteins. Nucleic. Acids Res. 2002;30:35–37. doi: 10.1093/nar/30.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Shindo K, Sano H, Seki S, Fujimura M, Yanai N, Miwa Y, Fujita Y. Sequencing of a 65 kb region of the Bacillus subtilis genome containing the lic and cel loci, and creation of a 177 kb contig covering the gnt-sacXY region. Microbiology. 1996;142:3113–3123. doi: 10.1099/13500872-142-11-3113. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Hung LW, Mueller-Dieckmann HJ, Kim KK, Yokota H, Kim R, Kim SH. Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics. Proc. Natl. Acad. Sci. USA. 1998;95:15189–15193. doi: 10.1073/pnas.95.26.15189. [DOI] [PMC free article] [PubMed] [Google Scholar]