Abstract
Ionizing radiation induces chronic metabolic oxidative stress and a mutator phenotype in hamster fibroblasts that is mediated by H2O2, but the intracellular source of H2O2 is not well defined. To determine the role of mitochondria in the radiation-induced mutator phenotype, end points of mitochondrial function were determined in unstable (CS-9 and LS-12) and stable (114) hamster fibroblast cell lines derived from GM10115 cells exposed to 10 Gy X rays. Cell lines isolated after irradiation demonstrated a 20–40% loss of mitochondrial membrane potential and an increase in mitochondrial content compared to the parental cell line GM10115. Surprisingly, no differences were observed in steady-state levels of ATP (P > 0.05). Unstable clones demonstrated increased oxygen consumption (two- to threefold; CS-9) and/or increased mitochondrial electron transport chain (ETC) complex II activity (twofold; LS-12). Using Western blot analysis and Blue Native gel electrophoresis, a significant increase in complex II subunit B protein levels was observed in LS-12 cells. Furthermore, immunoprecipitation assays revealed evidence of abnormal complex II assembly in LS-12 cells. Treatment of LS-12 cells with an inhibitor of ETC complex II (thenoyltrifluoroacetone) resulted in significant decreases in the steady-state levels of H2O2 and a 50% reduction in mutation frequency as well as a 16% reduction in CAD gene amplification frequency. These data show that radiation-induced genomic instability was accompanied by evidence of mitochondrial dysfunction leading to increased steady-state levels of H2O2 that contributed to increased mutation frequency and gene amplification. These results support the hypothesis that mitochondrial dysfunction originating from complex II can contribute to radiation-induced genomic instability by increasing steady-state levels of reactive oxygen species.
INTRODUCTION
We showed previously that increased steady-state levels of H2O2 are observed in radiation-induced genomically unstable cells that are causally linked to a mutator phenotype (1). However, it is not clear which subcellular organelle(s) and component(s) are responsible for the increased H2O2 in the unstable cells. Since mitochondria are a well-known source of H2O2 production, defects in mitochondrial electron transport chains (ETCs) could contribute to the H2O2-induced mutator phenotype (1–6). In most cases, electron transport through the ETCs is a tightly controlled process, minimizing the possibility of H2O2 formation. However, when defects in electron transport chain proteins occur, respiratory complexes can become less efficient at transferring electrons, thereby increasing the residence time and/or accessibility of electrons to oxygen (2). Therefore, instead of flowing to complex IV where the 4-electron reduction of O2 to H2O occurs, there could be an increased probability of 1-electron reduction of O2 leading to the production of reactive oxygen species such as H2O2 (2). Thus defects in ETC proteins could contribute to increased steady-state levels of H2O2 and conditions of metabolic oxidative stress in irradiated cells (1–7).
It has been hypothesized that mitochondrial dysfunction originating at ETC complexes may contribute to radiation-induced genomic instability (2, 6). This hypothesis is strengthened by previous studies that have shown some evidence of dysfunctional mitochondria measured in terms of membrane potential, mitochondrial proteins, and mitochondria content in populations of cells consisting of a pool of various unstable clones obtained after exposure of GM10115 cells to 10 Gy X rays (3–6). There are also reports showing that cells expressing a known mutation in genes coding for ETC proteins show evidence of genomic instability and steady-state levels of O2•− and H2O2 (7). In addition, certain radiation-induced genomically unstable clones have decreased respiration and complex IV activity (3–6). Together these data suggest that mitochondrial defects may be critically linked to genomic instability.
In the current study, three subsets of Chinese hamster ovary fibroblasts were used: wild-type GM10115, genomically unstable cells (CS-9 and LS-12) derived after exposure of GM10115 cells to 10 Gy X rays, and genomically stable cells (114) also derived after exposure of GM10115 cells to 10 Gy X rays (8). Using these cell lines we showed that the unstable clones exhibited evidence of mitochondrial dysfunction as well as alterations in ETC complex II assembly that apparently contribute to increased steady-state levels of H2O2 and genomic instability based on studies using an ETC complex II inhibitor.
MATERIALS AND METHODS
Cell Culture
Cells of the Chinese hamster ovary parental cell line GM10115 were obtained from ATCC. Cells were grown in DMEM containing high glucose and 1 mM sodium pyruvate (CellGro, Herndon, VA), 10% FBS (Hyclone), 0.2 mM L-proline, and 1% L-glutamine (Gibco). Previously described stable (114) and unstable (CS-9, LS-12) clones isolated after radiation exposure were grown in the same medium (8).
Mitochondrial Membrane Potential using JC-1
Cells were seeded in 60-mm dishes at least 2 days before the experiment. On the day of the experiment, cells were trypsinized and resuspended in medium containing 10 μg/ml JC-1 (Invitrogen, Carlsbad, CA) with or without 50 μM carbonyl cyanide 3-chlorophenylhydrazone (CCCP), which was used as a positive control. Samples were incubated at 37°C for 15 min. Cells were then spun at 500 g to wash away unbound JC-1 and the pellet was resuspended in 500 μl of PBS. The samples were then filtered and analyzed using a Becton Dickinson FACS machine. Ten thousand cells were gated on forward scatter (FSC) and side scatter (SSC) to approximate live cells. These cells were then analyzed for fluorescence in the FL1 channel (λEx 488 nm, λEm 530 nm, 30 nm bandpass) and in the FL2 channel (λEx 488 nm, λEm 585 nm, 42 nm bandpass). The ratio of FL2/FL1 was plotted as a measure of mitochondrial membrane potential.
Mitochondrial Content Measurement using Mitotracker Green
One million cells were plated in 60-mm dishes 1 day before the assay. On the day of the assay, cells were washed with PBS and incubated with Mitotracker Green (80 nM) (Invitrogen) for 15 min at 37°C. The probe was then washed off, and cells were resuspended in 500 μl of PBS, filtered through a mesh, and analyzed using a Becton Dickinson FACS machine. Ten thousand cells were gated on FSC and SSC to approximate live cells. These cells were then analyzed for fluorescence in the FL1 channel (λEx 488 nm, λEm 530 nm, 30 nm bandpass).
ATP Assay
ATP levels were analyzed using a Sigma (St. Louis, MO) ATP-assay kit (Kit code FLASC) according to the manufacturer's instructions. Briefly, the sample ATP concentration was calculated based on a standard curve run in the 0.01–1-μM range. A master mix containing luciferin, firefly luciferase, DTT and sodium azide in a reaction buffer was added to both standards and samples, and luminescence was read immediately. Since the luciferase enzyme requires 1 molecule of ATP for each reaction, the luminescence produced by the reaction is directly proportional to the amount of ATP present in the cell.
Oxygen Consumption
Oxygen consumption was measured using a YSI oxygen monitor containing a Clark-type electrode. The cell pellet was resuspended at a density of 3 × 106 cells/ml in PBS containing 5 mM glucose. A 3-ml sample was placed in the electrode chamber and was allowed to equilibrate with air for 3 min. Oxygen consumption was then recorded for 14 min.
Isolation of Mitochondria
Mitochondria were isolated from cells as described (9) using density gradient centrifugation with a sucrose gradient in isolation buffer (0.25 M sucrose, 5 mM Hepes, 0.1 mM EDTA, pH 7.25). Briefly, exponentially growing cells were harvested and centrifuged at 500g for 5 min. The cell pellet was resuspended in mitochondria isolation buffer containing 1:1 w/v fatty acid free bovine serum albumin (BSA) and homogenized using a Dounce homogenizer. Unbroken cells and cell debris were removed by centrifugation at 1000g for 10 min. The supernatant from this step was collected and centrifuged at 10,000g for 10 min to obtain mitochondria. The mitochondrial pellet was resuspended in isolation buffer as described previously and used for further analysis (9).
Complex II Activity Assay
The complex II activity assay [i.e., mitochondrial membrane-associated succinate quinone oxidoreductase (SQR) activity] was done as described previously (10) using coenzyme Q (CoQ) as an electron acceptor and 2,6-dichloroindophenol (DCIP) as the chromophore. Oxidized DCIP is blue and becomes colorless upon acceptance of electrons from CoQ. Thus loss of DCIP absorbance at 600 nm correlates with complex II activity. Briefly, isolated whole mitochondria [resuspended in 20 mM potassium phosphate buffer (pH 7.5)] were lysed using repeated freezing-thawing. Each sample was divided into two parts. One was used as blank and contained mitochondria (100 μg), 68.5 mM potassium phosphate buffer with EDTA and neutral cyanide. The assay cuvette contained mitochondria (100 μg), 68.5 mM potassium phosphate buffer, neutral cyanide and 600 nM succinate. The blank and assay cuvettes for each sample were incubated in the spectrophotometer at 37°C for 10 min. Coenzyme Q and DCIP were then added, and recording of absorbance was started immediately. The rate of the blank was subtracted from each corresponding sample, and complex II activity was calculated using a DCIP standard curve.
Blue Native Gel Electrophoresis
Blue Native polyacrylamide gel electrophoresis (BN-PAGE) was done as described previously (11, 12). Briefly, mitochondrial protein (300 μg) was resuspended in isotonic buffer (20 mM Tris-HCl pH 7.5, 0.6 M sorbitol and protease inhibitors) and centrifuged at 10,000g for 5 min. Mitochondrial pellets were then resuspended in 2× solubilization buffer (40 mM Tris-HCl, pH 7.5, 0.2 mM EDTA, 100 mM NaCl, 20% glycerol and protease inhibitors). An equal volume of digitonin was added to the samples to achieve a protein (μg):digitonin (μg) ratio of 1:6. Samples were incubated on ice for 30 min and centrifuged at 240,000g for 30 min at 4°C. The supernatant was mixed with loading buffer and incubated on ice for 5 min. Samples (in duplicate) were analyzed on 4–13% gradient gels. The gel containing one set of samples was stained with Coomassie blue, and the other gel containing the same set of samples was analyzed by immunoblotting using SDHB antibodies (Invitrogen).
Western Blot for Complex II Assembly
The cell pellet was resuspended in PBS and lysed using three freeze-thaw cycles. Samples were then sonicated briefly using a bath sonicator for 30 s. Samples were then spun at 600g to remove the DNA as a pellet. The supernatant was then spun at 10,000g to pellet the membrane fraction. The supernatant from this spin was collected as the soluble fraction. The membrane fraction was resuspended in an appropriate volume of phosphate buffer. Protein was estimated for samples, and 50 μg protein was loaded on a 12% SDS gel. The proteins were then blotted on a nitrocellulose membrane, blocked for 1 h using 5% milk solution, and probed for SDHB by Western blotting.
Immunoprecipitation Assay
Immunoprecipitation assays were carried out on 150 μg of isolated mitochondria from wild-type, stable and unstable clones. Mitochondria were lysed using NP-40 lysis buffer and incubated with SDHA antibody (Invitrogen) for 2 h at 4°C on a rocker platform. Then the samples were incubated with Sepharose protein A beads (Zymed) for 4 h at 4°C to precipitate the immuno-complex. The immuno-complex was then eluted from the beads using SDS gel loading buffer and loaded on a 4–20% gradient gel. The gel was finally fixed and stained with Coomassie blue.
Extracellular Hydrogen Peroxide
The hydrogen peroxide assay was carried out as described previously (1, 13).
TTFA Treatment
Cells in complete medium were treated with 20 μMTTFA for 45 min prior to the H2O2 assay. For the mutation frequency assay, cells were treated with 20 μM TTFA at the same time as 6-thioguanine addition and remained in the cloning dishes for 14–21 days. For the CAD (carbamyl-P synthetase/aspartate transcarbamylase/dihydro-orotase) gene amplification assay, cells were treated with 20 μM TTFA at the same time as 100 μM PALA (N-phosphonacetyl-l-aspartate) addition and remained in the cloning dishes for 14 days. TTFA did not affect cloning efficiency in dishes without 6-thioguanine and PALA.
Mutation Frequency Assay
Three million cells from exponentially growing cultures were seeded onto 100-mm dishes in complete medium. 6-Thioguanine (Sigma) dissolved in DMSO was added at a final concentration of 40 μM. Tukey's post hoc analysis was The dishes were left undisturbed for 2–3 weeks until colonies with at least 50 cells appeared. The colonies were then fixed with 70% ethanol, stained with Coomassie blue, and counted. Data were normalized to the plating efficiency of cells treated identically in the absence of 6-thioguanine.
CAD Gene Amplification Assay
Different cell dilutions (1000, 1 × 104, 1 × 105 and 3 × 106) from exponentially growing cultures were seeded onto 100-mm dishes in complete medium. PALA (NSC 224131, obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment, National Cancer Institute, Bethesda, MD) dissolved in DMEM was added to produce a final concentration of 100 μM. The dishes were left undisturbed for 14 days until colonies with at least 50 cells appeared. The colonies were then fixed with 70% ethanol, stained with Coomassie blue and counted. Data were normalized to the plating efficiency of cells treated identically in the absence of PALA.
Statistics
One-way ANOVA for non-parametric measurements was performed for all experiments having N ≥ 3. Tukey's post hoc analysis was performed wherever appropriate. Differences were considered significantly different if P ≤ 0.05 unless otherwise stated. All statistical analysis was done using GraphPad Prism software.
RESULTS
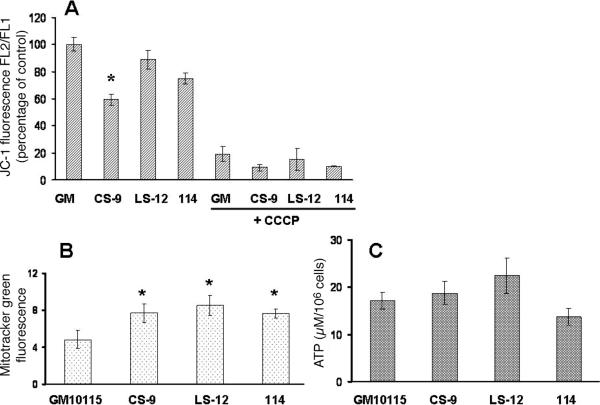
One of the parameters commonly used to describe mitochondrial function is membrane potential. The monomeric cationic dye JC-1 fluoresces in the green channel (FL1) and upon electrostatic entry into mitochondria forms large aggregates that fluoresce in the red channel (FL2) that can be monitored as an indicator of mitochondrial membrane potential using flow cytometry. The radiation-induced CS-9 unstable clone showed a significant decrease of 40% in mitochondrial membrane potential relative to the wild-type GM cells (Fig. 1A). These results clearly show that the radiation-induced genomically unstable CS-9 cells demonstrate significant reductions in mitochondrial membrane potential that could be indicative of some abnormality in mitochondrial electron transport chain function. Interestingly, neither the LS-12 unstable clone nor the stable 114 clone showed significant decreases in JC-1 labeling (Fig. 1A). When carbonyl cyanide 3-chlorophenylhydrazone (CCCP), a proton ionophore and an uncoupler of oxidative phosphorylation in mitochondria, was used as a positive control, the mitochondrial membrane potential did collapse (Fig. 1A).
FIG. 1.
Genomically unstable cells show a depolarized mitochondrial membrane potential, an increase in mitochondrial content and no difference in ATP levels. Panel A: Mitochondrial membrane potential as measured by JC-1. The error bars represent ±1 SD from three independent experiments. * P < 0.05 compared to wild-type cells. Panel B: Mitochondrial content as shown by Mitotracker green staining showed modest differences among clones. Error bars represent ±1 SD from three separate dishes. * P < 0.05 compared to wild-type cells. Panel C: ATP production measured by the luciferase assay. The error bars represent ±1 SD from three separate experiments. ATP levels were not significantly different.
To determine whether reductions in mitochondrial content could contribute to the observed reductions in membrane potential, the cells were labeled with Mitotracker green dye as a marker indicative of mitochondrial mass (Fig. 1B). Unexpectedly, all irradiated clones demonstrated 1.5- to twofold increases in mitochondrial mass (Fig. 1B), clearly demonstrating that the reductions in membrane potential shown in Fig. 1A are not likely to be the result of a decrease in the number of mitochondria. Despite apparent alterations in mitochondrial membrane potential and mitochondrial mass, there were no significant differences in the ATP levels of the cell lines (Fig. 1C) as measured by the luciferin-luciferase reaction. Since mitochondrial membrane potential appears to be lower (or the same) in irradiated clones (Fig. 1A) while the mitochondrial mass in irradiated cells appears to be increased (Fig. 1B), radiation exposure could have resulted in some persistent injury to mitochondrial function that caused the cells to increase mitochondrial mass to maintain adequate levels of ATP from oxidative phosphorylation (Fig. 1C). The specific mechanism(s) underlying this increased mitochondrial mass in irradiated cells remains obscure. However, previous authors suggested that increases in mitochondrial mass could be induced by increased steady-state levels of ROS caused by defective mitochondrial metabolism or could represent a compensatory response necessitated by increases in mitochondrial dysfunction (6, 14, 15).
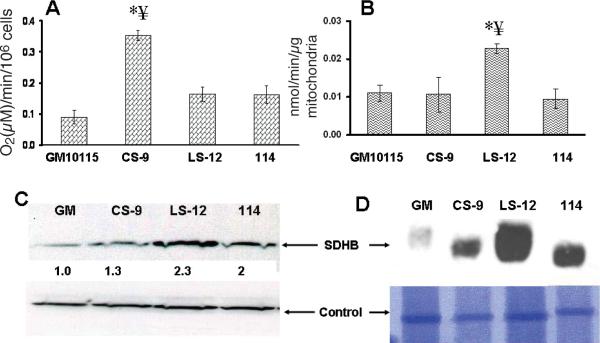
Interestingly, measurements of O2 consumption (Fig. 2A) showed that the unstable CS-9 cells demonstrated a significant increase in oxygen consumption relative to the wild-type and stable clones. This finding suggests that the unstable clone with a compromised membrane potential (CS-9; Fig. 1A) may have increased electron flux through ETCs to O2 to provide enough of a proton gradient to maintain ATP at control levels (Fig. 1C). Even though this experiment was conducted at an ambient O2 tension, we speculate that similar results might also be obtained in unstable clones in vivo.
FIG. 2.
Genomically unstable clones show high oxygen consumption and high complex II levels. Panel A: Oxygen consumption. The error bars represent ±1 SD from five independent experiments. CS-9 cells were significantly different from GM10115 cells and stable clone 114. * P < 0.001 compared to wild-type cells, ¥ P < 0.001 compared to 114 cells. Panel B: Complex II activity normalized to mitochondrial protein. * P < 0.05 compared to wild-type cells, ¥ P < 0.05 compared to 114 cells. Panel C: Western blots of whole cell lysate using 100 μg protein and SDHB antibody. Quantification shown by numerical data was achieved by normalizing SDHB density to actin density. Panel D: Coomassie blue staining of a Blue Native gel (lower panel) and a Western blot of a parallel gel probed with SDHB antibody.
Since defects in ETC complex II [a.k.a. succinate dehydrogenase (SDH)] have been shown to cause persistent metabolic oxidative stress and genomic instability (7), mitochondrial membrane-associated succinate quinone oxidoreductase (SQR) activity was measured in stable and unstable cell lines. Surprisingly, the unstable LS-12 cells (but not the CS-9 cells) demonstrated significant increases in SQR activity (Fig. 2B) that were also accompanied by a 2.3-fold increase in the SDHB protein level as measured by Western blot analysis done on whole cell lysates (Fig. 2C). Furthermore, Blue Native gel electrophoresis followed by Western blot analysis of isolated mitochondria (Fig. 2D) showed an increase in immunoreactive SDHB relative to wild-type cells. Thus both unstable clones demonstrated significant alterations in mitochondrial function, including alterations of the ETC complex II (LS-12).
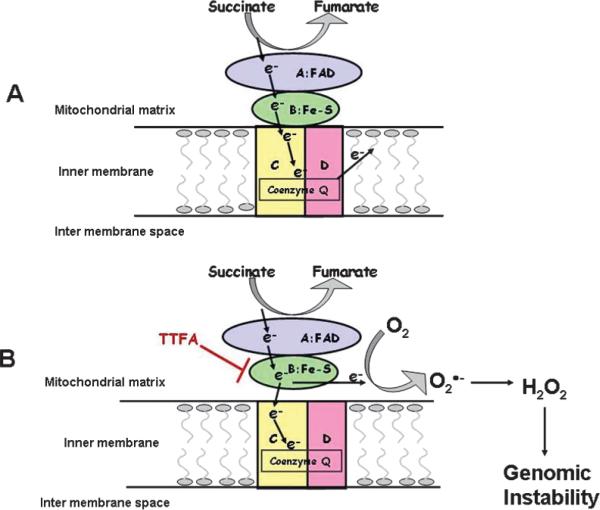
Complex II (succinate dehydrogenase) is not a proton pump. It removes electrons from succinate and transfers them (via FAD) to CoQ. Complex II contains four subunits (A, B, C and D). Subunits C and D are transmembrane integral proteins, whereas subunits A and B are tethered to C/D by hydrophobic and electrostatic interactions (16) (Fig. 3A). FAD in SDHA is the initial electron acceptor. FAD is reduced to FADH2 during oxidation of succinate to fumarate. FADH2 is then reoxidized by transfer of electrons through a series of three iron-sulfur centers (SDHB, SDHC, SDHD) to CoQ, yielding QH2. The QH2 product may then be reoxidized via complex III, providing a pathway for transfer of electrons from succinate into the respiratory chain. Thus, if complex II is properly assembled, all four subunits appear to be associated with the mitochondrial membrane fraction and electron transfer should proceed efficiently from SDHA to SDHD, avoiding univalent reductions of O2 to form superoxide or hydrogen peroxide. However, if the association between SDHA/SDHB and SDHC/SDHD is destabilized by radiation injury, then SDHC and SDHD would be expected to appear in the membrane fraction, whereas SDHA/SDHB would be expected to appear in the soluble fraction of the mitochondrial matrix (Fig. 3B). If this were to occur, then radiation injury could result in the assembly of a less stable form of complex II and the electron transfer from SDHB to SDHC could be disrupted, resulting in an increased probability of univalent reduction of oxygen to form superoxide that would dismute to form H2O2. In this way radiation-induced alterations in the assembly of complex II could contribute to increased steady-state levels of ROS, resulting in chronic metabolic oxidative stress (2).
FIG. 3.
Model by which disruption of complex II assembly may participate in radiation-induced genomic instability.
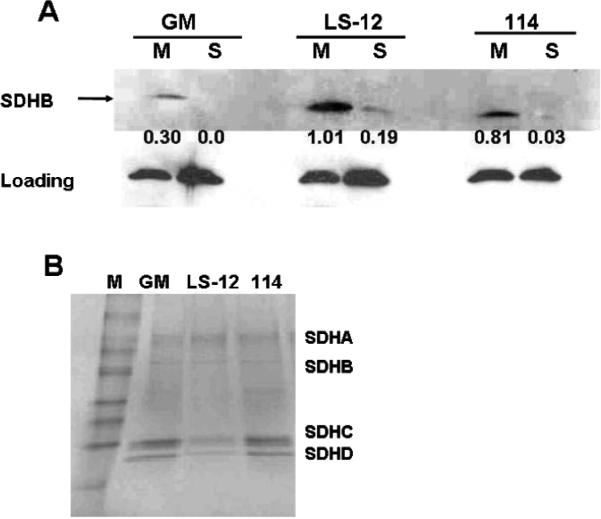
To further investigate complex II assembly, differential centrifugation was used to isolate subcellular fractions from sonicated and freeze-thawed whole cell homogenates in PBS. These subcellular fractions consisted of the 10,000g supernatants, which include cytosolic and mitochondrial matrix proteins (soluble fraction), and 10,000g pellets, which include the crude membrane fraction and membrane-bound proteins (insoluble fraction) from GM10115, LS-12 and 114 clones. The proteins from the soluble and insoluble fractions were separated using SDS PAGE followed by Western blot analysis using SDHB antibody. The results shown in Fig. 4A corroborate our findings (Fig. 2C and D) of increased membrane-bound SDHB in LS12 cells relative to wild-type and stable clones, in addition to showing that immunoreactive SDHB is detectable in the soluble fraction of LS-12 cells. Since LS-12 cells have a greater abundance of SDHB protein in the soluble fraction, these data suggest that misassembly or weak bonding between subunit B and the membrane-bound subunits SDHC and SDHD may contribute to complex II instability in the LS-12 cells.
FIG. 4.
Genomically unstable clone LS-12 shows improper assembly of complex II. Panel A: Western blot showing levels of immunoprecipitated SDHB after transfer on a nitrocellulose membrane. Unstable clone LS-12 shows the presence of SDHB in the soluble fraction. Panel B: Immunoprecipitation of SDHA proteins carried out on 150 μg isolated mitochondria using antibody against SDHA and Sepharose protein A beads. Samples were separated on a 4%–20% gradient gel, which was stained with Coomassie blue.
The assembly of complex II proteins in LS-12 cells was analyzed further by immunoprecipitation assays using antibodies against SDHA (Fig. 4B) using homogenates of isolated mitochondria. Figure 4B shows that in the case of LS-12 mitochondria, less SDHC and SDHD appeared to be immunoprecipitated with the SDHA antibody relative to the wild-type or stable cells as observed by Coomassie blue staining of the gel. Overall, the results in Fig. 4 support the hypothesis that LS-12 cells have increased immunoreactive SDHB protein, distributed between various states that are likely to reflect both proper and improper assembly into the inner mitochondrial membrane. Improper assembly may be characterized by less tightly bound states that appear in either the soluble fraction of the mitochondrial matrix or loosely tethered to SDHC or SDHD.
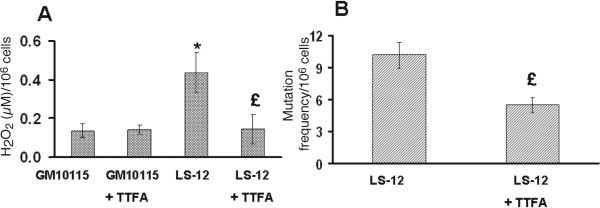
SDHB contains iron-sulfur clusters that are believed to pass electrons to CoQ, which binds to the interface of SDHC and SDHD embedded in the inner mitochondrial membrane. If SDHB is misassembled in LS-12 and either remains soluble in the mitochondrial matrix or has weak bonding to SDHC/SDHD, an increased probability of one-electron reduction of O2 to form reactive oxygen species (such as O2•− and H2O2) could occur. This could contribute to the increased steady-state levels of ROS and genomic instability observed in LS-12 cells. To examine this possibility, H2O2 levels were measured in wild-type and LS-12 cells treated with a complex II electron transport chain blocker [thenoyltrifuoroacetone (TTFA)] thought to bind to iron-sulfur clusters (17). Interestingly, LS-12 cells treated with TTFA (20 μM) demonstrated significantly reduced levels of H2O2 (Fig. 5A), but there was no measurable effect on the wild-type cells. These treatments were not found to affect the plating efficiency of either cell type (data not shown). In addition, we found that 20 μM TTFA was able to significantly reduce the mutation frequency in the LS-12 cells (Fig. 5B). To examine the possible involvement of complex II in chromosomal instability, the CAD gene amplification assay measuring PALA resistance was used in parental and LS-12 cell lines. The emergence of PALA resistance in a cell population is believed to be a good marker of chromosomal instability because it involves the amplification of CAD genes, which may also involve the formation of dicentrics, rings, isochromatid fusions and acentric fragments (8). The results in Table 1 clearly show that LS-12 cells demonstrated chromosomal instability as measured by the greater clonogenic survival after PALA exposure (approximately 400 times) relative to PALA-treated wild-type cells. Table 1 also shows that treatment of LS-12 cells with 20 μM TTFA caused a statistically significant reduction in PALA-resistant clones, suggesting that blocking ETC complex II suppressed chromosomal instability similarly to what was found with the HPRT point mutation assay. Taken together, these data provide evidence consistent with the hypothesis that defects in electron transport chain complex II contribute significantly to oxidative stress and genomic instability in LS-12 cells.
FIG. 5.
TTFA treatment reduces steady-state levels of H2O2 and the mutation frequency of LS-12 clone. Panel A: H2O2 per 106cells. Error bars are ±1 SD for three separate experiments (n = 4–9). * P < 0.05 compared to GM10115 cells, £ P < 0.05 compared to LS-12 cells. Panel B: HPRT mutation frequency. Error bars are ±1 SD for three separate experiments (n = 6–9). £ P < 0.05 compared to LS-12 cells.
TABLE 1.
CAD Gene Amplification in GM 10115 and LS-12 Cells Treated with or without 20 μM TTFA at the Same Time as PALA Treatment (100 μM)
| Clone | Gene amplification frequency/104 cells |
|---|---|
| GM 10115 | 19 ± 6 |
| LS-12 | 7840 ± 172a |
| LS-12 + TTFA | 6630 ± 376ab |
P < 0.001 compared to GM 10115 cells.
P < 0.001 compared to LS-12 cells.
DISCUSSION
Cellular metabolic processes provide reducing equivalents (i.e. NADPH, NADH, FADH2) and energy (i.e. ATP) to carry out the biosynthetic reactions necessary for maintaining cellular structure and function. Mitochondria are key organelles that provide the major site for Krebs cycle activity and ATP production in mammalian cells. These biochemical activities also lead to the production of mitochondrially derived ROS, which can act as signaling molecules that couple shifts in metabolism with signal transduction and gene expression pathways to coordinately regulate cellular homeostasis [reviewed in ref.(2)]. Furthermore, due to their complexity (~3000 proteins), mitochondrial structure and function can become disrupted, leading to increases in steady-state ROS levels that can contribute to pathological processes associated with genomic instability and degenerative diseases of aging (2, 18). Since radiation exposure can recapitulate many of the pathological changes seen with aging, we investigated the role of mitochondrial dysfunction in radiation-induced genomic instability. We found that genomically unstable clones demonstrated increased steady-state levels of intracellular H2O2 (1) as well as evidence of mitochondrial alterations, including changes in O2 consumption, alterations in complex II activity and alterations in mitochondrial membrane potential in the absence of any significant differences in steady-sate levels of ATP (Figs 1 and 2). Interestingly the increases in steady-state levels of H2O2 were noted in both radiation-induced unstable clones (LS-12 and CS-9) relative to controls (1), but the other changes in mitochondrial function noted (changes in O2 consumption, alterations in complex II activity, and alterations in membrane potential) were not common to these two unstable clones isolated from the irradiated population. Given the complexity of mitochondrial structure/function, this suggests that a multitude of persistent and heritable changes to mitochondrial metabolism could result in genomic instability as long as such changes promote increased steady-state levels of damaging reactive oxygen species.
In the current studies, changes in ETC complex II activity in LS-12 unstable cells were followed up with structural studies of complex II. Our data indicated that complex II subunit B immunoreactive protein (SDHB) was increased in LS-12 cells (Fig. 2). Increased levels of SDHB appeared to be in both the soluble fraction and the membrane-bound fraction (Fig. 4), and when SDHA was immunoprecipitated, less SDHC and SDHD were bound to SDHA relative to control and stable cell lines (Fig. 4). These findings suggest that compared to the normal assembly of complex II (Fig. 3A), where SDHA and SDHB are associated with the membrane-bound SDHC and SDHD, more SDHA and SDHB are disassociated (or loosely associated such that they dissociate during isolation) from the membrane-bound components (Fig. 3B) in genomically unstable cells such as LS-12. This could disrupt the flow of electrons to CoQ in complex II, leading to increased steady-state levels of O2•− and H2O2 (Fig. 3B).
To determine if the flow of electrons through complex II could be contributing to increases in H2O2 and genomic instability, LS-12 cells were treated with an ETC complex II inhibitor (TTFA) that binds iron-sulfur clusters (17) (Fig. 3B). Interestingly, treatment of LS-12 cells with TTFA significantly decreased steady-state levels of H2O2 as well as HPRT mutation frequency and CAD gene amplification (Fig. 5A and B and Table 1). These results provide strong support for the hypothesis that alterations in complex II assembly contribute significantly to oxidative stress and genomic instability in unstable cells isolated after irradiation. While it is uncertain whether these changes in complex II assembly involve mutations in genes coding for complex II proteins or simply disruptions in processes involved with the proper import and assembly of the complexes, our data support the idea that radiation can cause persistent and heritable changes in ETC function that can promote the onset of genomic instability.
Thus radiation can cause damage to mitochondria either directly or by damaging the genes that encode for ETC proteins. The combined result of these insults could lead to mitochondrial dysfunction, leading to increased metabolic oxidative stress and genomic instability. Our results emphasize that targets other than genes coding for repair of nuclear DNA could be important for the persistent effects of radiation on genomic instability. Other targets converging on mitochondrial function such as genes coding for mitochondrial proteins (including the mitochondrial genome) may therefore contribute significantly to the delayed effects of radiation, including genomic instability.
Based on previous (1, 4–6, 19) and current data, we propose a unifying hypothesis to explain one of the several possible mechanisms of radiation-induced genomic instability. It appears that, in addition to other targets, radiation damages mitochondrial function and assembly, creating a pro-oxidant environment in the cell via increased production of ROS (particularly H2O2). This along with other factors appears to contribute significantly to persistent radiation-induced genomic instability.
ACKNOWLEDGMENTS
We would like to thank Dr. Edouard I. Azzam (UMDNJ, Newark, NJ) for suggestions regarding Blue Native gel electrophoresis. We would also like to thank Dr. C. Michael Knudson (The University of Iowa) for suggestions regarding the JC-1 protocol. This work was supported by the U.S. Department of Energy grant DE-FG02-05ER64050 and the following NIH grants: R01CA100045, P42ES01366, P30CA086862, R01CA111365, T32-CA78586 and F32CA110611.
REFERENCES
- 1.Dayal D, Martin SM, Limoli CL, Spitz DR. Hydrogen peroxide mediates the radiation-induced mutator phenotype in mammalian cells. Biochem. J. 2008;413:185–191. doi: 10.1042/BJ20071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitz DR, Azzam EI, Li JJ, Gius D. Metabolic oxidation/reduction reactions and cellular responses to ionizing radiation: A unifying concept in stress response biology. Cancer Metastasis Rev. 2004;23:311–322. doi: 10.1023/B:CANC.0000031769.14728.bc. [DOI] [PubMed] [Google Scholar]
- 3.Kim GJ, Chandrasekaran K, Morgan WF. Mitochondrial dysfunction, persistently elevated levels of reactive oxygen species and radiation-induced genomic instability: A review. Mutagenesis. 2006;21:361–367. doi: 10.1093/mutage/gel048. [DOI] [PubMed] [Google Scholar]
- 4.Kim GJ, Fiskum GM, Morgan WF. A role for mitochondrial dysfunction in perpetuating radiation-induced genomic instability. Cancer Res. 2006;66:10377–10383. doi: 10.1158/0008-5472.CAN-05-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller JH, Jin S, Morgan WF, Yang A, Wan Y, Aypar U, Peters JS, Springer DL. Profiling mitochondrial proteins in radiation-induced genome-unstable cell lines with persistent oxidative stress by mass spectrometry. Radiat. Res. 2008;169:700–706. doi: 10.1667/RR1186.1. [DOI] [PubMed] [Google Scholar]
- 6.Limoli CL, Giedzinski E, Morgan WF, Swarts SG, Jones GD, Hyun W. Persistent oxidative stress in chromosomally unstable cells. Cancer Res. 2003;63:3107–3111. [PubMed] [Google Scholar]
- 7.Slane BG, Aykin-Burns N, Smith BJ, Kalen AL, Goswami PC, Domann FE, Spitz DR. Mutation of succinate dehydrogenase subunit C results in increased O2•−, oxidative stress, and genomic instability. Cancer Res. 2006;66:7615–7620. doi: 10.1158/0008-5472.CAN-06-0833. [DOI] [PubMed] [Google Scholar]
- 8.Limoli CL, Kaplan MI, Corcoran J, Meyers M, Boothman DA, Morgan WF. Chromosomal instability and its relationship to other end points of genomic instability. Cancer Res. 1997;57:5557–5563. [PubMed] [Google Scholar]
- 9.Kaschnitz RM, Hatefi Y, Pedersen PL, Morris HP. Isolation of mitochondria from Morris hepatomas. Methods Enzymol. 1979;55:79–88. doi: 10.1016/0076-6879(79)55011-3. [DOI] [PubMed] [Google Scholar]
- 10.Singer TP. Determination of the activity of succinate, NADH, choline, and alpha-glycerophosphate dehydrogenases. Methods Biochem. Anal. 1974;22:123–175. doi: 10.1002/9780470110423.ch3. [DOI] [PubMed] [Google Scholar]
- 11.Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 12.Amutha B, Gordon DM, Gu Y, Pain D. A novel role of Mgm1p, a dynamin-related GTPase, in ATP synthase assembly and cristae formation/maintenance. Biochem. J. 2004;381:19–23. doi: 10.1042/BJ20040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panus PC, Radi R, Chumley PH, Lillard RH, Freeman BA. Detection of H2O2 release from vascular endothelial cells. Free Radic. Biol. Med. 1993;14:217–223. doi: 10.1016/0891-5849(93)90013-k. [DOI] [PubMed] [Google Scholar]
- 14.Mancini M, Anderson BO, Caldwell E, Sedghinasab M, Paty PB, Hockenbery DM. Mitochondrial proliferation and paradoxical membrane depolarization during terminal differentiation and apoptosis in a human colon carcinoma cell line. J. Cell Biol. 1997;138:449–469. doi: 10.1083/jcb.138.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spodnik JH, Wozniak M, Budzko D, Teranishi MA, Karbowski M, Nishizawa Y, Usukura J, Wakabayashi T. Mechanism of leflunomide-induced proliferation of mitochondria in mammalian cells. Mitochondrion. 2002;2:163–179. doi: 10.1016/s1567-7249(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 16.Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 17.Spitz DR, Oberley LW. An assay for superoxide dismutase activity in mammalian tissue homogenates. Methods Enzymol. 1989;179:8–18. doi: 10.1016/0003-2697(89)90192-9. [DOI] [PubMed] [Google Scholar]
- 18.Johnson DT, Harris RA, Blair PV, Balaban RS. Functional consequences of mitochondrial proteome heterogeneity. Am. J. Physiol. Cell Physiol. 2007;292:C698–707. doi: 10.1152/ajpcell.00109.2006. [DOI] [PubMed] [Google Scholar]
- 19.Loeb LA. A mutator phenotype in cancer. Cancer Res. 2001;61:3230–3239. [PubMed] [Google Scholar]