Abstract

Macrophage scavenger receptor A (SR-A) is a multifunctional, multiligand pattern recognition receptor with roles in innate immunity, apoptotic cell clearance, and age-related degenerative pathologies, such as atherosclerosis and Alzheimer’s disease. Known endogenous SR-A ligands are polyanionic and include modified lipoproteins, advanced glycation end products, and extracellular matrix proteins. No native plasma ligands have been identified, but it is known that SR-A recognition of unidentified serum components mediates integrin-independent macrophage adhesion, which may drive chronic local inflammation. In this study, we used a high-throughput fractionation and screening method to identify novel endogenous SR-A ligands that may mediate macrophage adhesion. SR-A was found to recognize the exchangeable apolipoproteins A-I and E (apo A-I and apo E, respectively) in both lipid-free and lipid-associated form, suggesting the shared amphipathic α-helix as a potential recognition motif. Adhesion of RAW 264.7 macrophages to surfaces coated with apo A-I and apo E4 proved to be integrin-independent and could be blocked by anti-SR-A antibodies. The presence of apo A-I and apo E in pathological deposits, such as atherosclerotic lesions and neurotoxic Alzheimer’s plaques, suggests a possible contribution of SR-A-dependent adhesion of macrophages to an inflammatory microenvironment.
Scavenger receptor A (SR-A)1 is a multifunctional, multiligand receptor expressed mainly by myeloid cells, which plays a role both in innate immune defense and removal of modified or aged self and has been termed molecular flypaper for its low-affinity, broad specificity ligand binding capacities (1−5). Most known SR-A ligands are exogenous compounds discovered and defined by their ability to inhibit binding of receptor to the archetypal ligand acetylated LDL (2). The majority of endogenous SR-A ligands are connected to age-related degenerative diseases, oxidized lipoproteins being the driving force behind atherosclerosis, AGE-modified proteins resulting from diabetic glucose overload, and β-amyloid fibrils representing major components of neurotoxic Alzheimer’s plaques (6,7). A characteristic shared by all known SR-A ligands is their structurally defined, repetitive anionic charge distribution (2). Ligand binding and specificity are controlled by a positively charged stretch of lysines in the collagenous binding domain of the receptor (8,9), and receptor engagement is followed by endocytic uptake, dissociation of the receptor−ligand pair at acidic pH, and lysosomal degradation (10−12).
Macrophage retention within tissues relies on both metal ion-dependent and -independent mechanisms, the former including integrins and selectins and the latter scavenger receptors and immunoglobulins (13,14). Prolonged or pathological retention of macrophages may create an inflammatory microenvironment, which in many cases drives disease, as seen for atherosclerosis, neurodegeneration, or diabetes-induced nephropathy (15). Previous studies established a role for SR-A in integrin-independent adhesion of macrophages to an uncharacterised serum ligand (16). Subsequent adhesion studies have implicated SR-A in adhesion of macrophages to various extracellular matrix molecules, including glycated type IV collagen in diabetic patients, denatured type I and II collagens, and the proteoglycans biglycan and decorin (17−19).
To identify plasma-borne endogenous SR-A ligands that contribute to SR-A-mediated macrophage adhesion, we screened human plasma for candidate ligands and tested their ability to sustain macrophage adhesion. Identification of single molecules from a highly complex mixture such as plasma requires a combination of separation techniques to reduce complexity and a stringent large-scale screening method. As the whole-cell adhesion assays or standard ligand competition assays used to identify most known SR-A ligands are poorly adapted to multisample analyses, a rapid high-throughput screening assay for identifying novel bacterial and endogenous SR-A ligands was developed (20). In this ELISA-based assay, lysate from bone marrow-derived macrophages from WT and SR-A−/− mice is used in combination with a monoclonal anti-SR-A antibody to detect receptor−ligand interactions. This allowed an extensive and rapid screen of individual chromatography fractions. In addition, SR-A is mostly intracellular (21), rendering binding studies with whole cells suboptimal, while lysis increases receptor availability by releasing this intracellular receptor pool. Since human, murine, bovine, and rabbit SR-A share a high degree of homology and similar ligand affinities, SR-A from any available species can be used to screen human plasma (22). In particular, the basic residues in the collagenous domain responsible for ligand binding are conserved between human and murine SR-A (9). Our results represent a successful application of this novel ligand identification strategy. We propose that apolipoproteins A-I and E are novel ligands for SR-A, bind to the receptor via the known polyanion binding site, and, when immobilized, contribute to SR-A-mediated macrophage adhesion. This may have consequences for local macrophage-driven microinflammation at sites of apolipoprotein deposition, such as atherosclerotic lesions or Alzheimer’s disease plaques.
Experimental Procedures
Reagents
Human pooled outdated citrated plasma was obtained from HD Supplies (Aylesbury, U.K.). Recombinant human apo E and apo SAA were from Peprotech (London, U.K.), and purified human apo A-I and apo A-II were from Calbiochem (Nottingham, U.K.). MalBSA was prepared by reacting BSA with maleic anhydride (23). Fully oxidized LDL (oxLDL) was obtained by overnight incubation of freshly prepared LDL (1 mg/mL) with 5 μM CuSO4. To stop the reaction, 100 μM EDTA and 20 μM butylated hydroxytoluene (BHT) were added, and the extent of oxidation was controlled by SDS−PAGE. Chromatography size calibration standards were bovine IgG, factor B, and complement factor H (MRC Immunochemistry Unit) and BSA and soybean trypsin inhibitor (Sigma). Primary antibodies were the monoclonal anti-murine SR-A antibody (clone 2F8) (Serotec, Oxford, U.K.), monoclonal rat anti-CD52 (Campath, kindly provided by H. Waldman, Sir William Dunn School of Pathology) as an isotype control for anti-murine SR-A, and the sheep polyclonal anti-human apo A-I antibody (AH213) from Serotec; for secondary antibodies, HRP-coupled goat anti-rat and donkey anti-sheep polyclonal antibodies from Jackson laboratories (Stratech, Suffolk, U.K.) were used.
Preparation of Cell Lysates
Confluent monolayers of murine bone marrow-derived macrophages (BMMφ) from 129/ICR WT and SR-A−/− mice were washed four times in PBS, placed on ice, and incubated with 1.5 mL of NP-40 lysis buffer [150 mM NaCl, 10 mM EDTA, 10 mM NaN3, 10 mM Tris (pH 8.0), 1 mM PMSF, 5 mM iodoacetamide, and 1% Nonidet P-40] per dish. Cells were mechanically detached and lysates collected, pooled, and cleared of nuclear and cell debris. After the determination of the protein concentration by the BCA assay (Pierce, Perbio Science), postnuclear cell lysates were frozen and stored at −20 °C.
ELISA and Competition ELISA
An ELISA-based assay for the detection of scavenger receptor ligands using BMMφ lysates is described elsewhere (20). Briefly, High Bind EIA/RIA 96-well plates (Costar) were coated overnight at 4 °C with plasma fractions (in the respective chromatography elution buffer, up to 125 μg of protein per well) or purified ligands (positive control, MalBSA or acLDL; negative control, BSA, all coated at 5 μg/mL in PBS), blocked with PBS, 10 mg/mL BSA, and 5 mM EDTA (blocking buffer), and overlaid with either WT or SR-A−/− BMMφ lysate (∼60 μg of protein/mL) in blocking buffer. For competition studies, lysates were preincubated for 1.5 h at RT with excess concentrations of competitors before addition to wells. Bound SR-A was detected by incubation for 1 h at RT with the rat anti-mouse SR-A monoclonal antibody at 10 μg/mL or isotype-matched control antibody, followed by incubation for 45 min with HRP-coupled goat anti-rat polyclonal antibody at a dilution of 1:2000. All antibodies were diluted in blocking buffer. After color had developed for 5 min, plates were read at 450 nm. If required, SR-A−/− OD450 values were subtracted from WT OD450 values to yield SR-A-specific binding and normalized to the internal positive control for interassay comparability (binding expressed as a percentage of the positive control). For the apo A-I ELISA, plates were coated and blocked as described above and then incubated at RT for 1 h with the sheep polyclonal anti-human apo A-I antibody at a dilution of 1:40000, followed by a 45 min incubation at RT with the HRP-coupled donkey anti-sheep polyclonal antibody at a dilution of 1:2000. Detection was conducted as described above.
SDS−PAGE and Far-Western Blot
Proteins were separated on SDS−PAGE gels in Tris-glycine running buffer (24) and either stained with Coomassie Blue or transferred to HybondC nitrocellulose (GE Healthcare). Blots were blocked overnight in blocking buffer (PBS and 4% dried milk powder) and incubated for 1.5 h at RT with WT or SR-A−/− BMMφ lysate at a level of 0.6 mg of cell protein/mL in blotting buffer (PBS, 4% dried milk powder, and 5 mM EDTA). To detect bound receptor, blots were washed for 3 × 5 min with washing buffer (PBS and 0.1% Tween 20), incubated for 1.5 h at RT with the primary antibody (same as ELISA) at 10 μg/mL in blotting buffer, washed for 3 × 5 min in washing buffer, and incubated for 45 min at RT with the secondary antibody (same as ELISA) in blotting buffer. Finally, blots were washed for 3 × 5 min in washing buffer, developed using the enhanced chemiluminescence kit (ECLplus, GE Healthcare), and visualized on Kodak film. For sequential detection with different antibodies, membranes were stripped with Restore Western Blot Stripping Buffer (Pierce, Perbio Science) according to the manufacturer’s instructions.
In-Gel Tryptic Digest and Mass Spectrometry
Coomassie blue-stained bands of interest were cut out, diced, and transferred to low-adhesion grade Eppendorf tubes (Eppendorf, Cambridge, U.K.). Gel pieces were washed twice for 20 min in 100 μL of 100 mM ammonium bicarbonate and 50% acetonitrile, washed once for 10 min in 100 μL of 100% acetonitrile, then air-dried for 10 min and covered with ∼25 μL of sequencing grade modified trypsin solution (Promega) at 50 μg/mL in 25 mM ammonium bicarbonate, and incubated overnight at 37 °C. To stop the digest, formic acid was added to a final concentration of 2.5%, and the solution was transferred to a new tube. For extraction of peptides from the gel pieces, 100 μL of 50% acetonitrile in water was added for 1 h at RT, transferred to a new tube, and replaced with 100% acetonitrile for an additional 1 h at RT. Peptide-containing supernatant was collected again, and the collected peptides were dried down by vacuum centrifugation. Mass spectrometry was conducted on a Waters Q-TOF micro mass spectrometer (Waters, Manchester, U.K.) coupled to a CapLC device. The reference database used for mass spectrometric identification of candidate ligands was MSDB (Mass Spectrometry protein sequence DataBase, www.matrixscience.com), searching for human entries only. Acceptance criteria for protein hits were set as a minimum of two unique peptides per protein and a probability-based Mowse score of >45, where Mowse stands for Molecular Weight Search and allows assignment of a probability to each identified peptide of a given MW (25).
Chromatography
All chromatography runs were conducted on ÄKTA FPLC apparatus operated by Pharmacia Unicorn software. Ion exchange was conducted on a MonoQ HR 5/5 column or on a QSepharose High Resolution 16/10 Fast Flow column equilibrated in running buffer (10 mM Na2HPO4 and 15 mM NaCl, adjusted to pH 7.4). Fresh plasma was cleared of chylomicrons and precipitates by a 40 min centrifugation at 15000 rpm and 4 °C in a Beckman J2-HS centrifuge with a JA20 rotor, filtered through a 0.22 μm filter (Pall, VWR), and diluted ∼1:1 in running buffer before injection. Unbound material was washed off with running buffer, and bound material was eluted with a two-stage linear gradient of high-salt buffer (buffer B) [10 mM Na2HPO4 and 1 M NaCl (pH 7.4)] at a flow rate of 1 mL/min, and 1 mL fractions were collected. The gradient stages (for the MonoQ column) included a slow increase from 0 to 40% buffer B (20 mL) and then a steep increase from 40 to 100% B (5 mL), with a plateau at 100% B (2 mL) and re-equilibration to 0% B. Size exclusion was conducted on a Superose12 HR 10/30 column (ligand fractionation) or on two coupled Superose6 10/30 columns (lipoprotein fractionation) equilibrated in running buffer [PBS and 0.5 mM EDTA (pH 7.4)]. Samples were injected and eluted with 1.2 column volumes of running buffer at a flow rate of 1 mL/min. Fractions (1 mL) were collected throughout the run. Proteins of known size [bovine IgG (156 kDa), factor B (90 kDa), BSA (67 kDa), soybean trypsin inhibitor (21 kDa), and complement factor H (155 kDa)] were run separately to calibrate the column. For concentration of samples between chromatography runs, pooled fractions were centrifuged on Vivaspin 6 mL concentrators with a 30 kDa cutoff (Vivascience, VWR) according to the manufacturer’s instructions.
Depletion of Immunoglobulins and Albumin
Plasma was depleted of immunoglobulins on a fast flow protein G Sepharose column (GammaBind Plus Sepharose, GE Healthcare). Five milliliters of plasma was processed per run. Plasma was passed through the column with running buffer [PBS and 0.5 mM EDTA (pH 7.4)], and bound IgG was eluted with 0.2 M glycine-HCl at pH 2.2. The column was regenerated with running buffer. For albumin depletion, affinity-purified polyclonal rabbit anti-human serum albumin antibodies (MRC Immunochemistry Unit) were coupled to CNBr-activated Sepharose (Pharmacia, GE Healthcare) (26), to a final column capacity of 1 mg of albumin/mL of bed volume. The column (bed volume of 15 mL) was equilibrated in 1 column volume (CV) of running buffer (PBS and 0.5 mM EDTA), and then plasma was loaded at a flow rate of 1 CV/30 min, equivalent to 1 mL/2 min, and washed through with 1 CV of running buffer. Bound albumin was eluted with 1 CV of water, followed by 2 CV of 3 M MgCl2 (pH 6.8) and 1 CV of water. The column was re-equilibrated with 1 CV of running buffer. All runs were conducted at 4 °C.
Isolation of Total Plasma Lipoproteins (27)
Ultracentrifugations were conducted in a Beckman L8-80M ultracentrifuge, using a VTi80 vertical rotor (Beckman) and QuickSeal polyallomer tubes with a capacity of 5.1 mL (Beckman). For total lipoprotein preparation, the density of plasma (d = 1.006 g/mL) adjusted to 10 mM EDTA and 1 mM PMSF (protease inhibitor) was increased to 1.21 g/mL with solid KBr, and the mixture was centrifuged at 4 °C for 11.5 h at 55000 rpm. The top fraction containing total lipoproteins was recovered using a 19-gauge needle, dialyzed extensively against 150 mM NaCl and 0.5 mM sodium EDTA (pH 7.4), and stored at 4 °C. For size separation of total lipoproteins, preparations were processed within a week.
Delipidation of Plasma
PHM-L LIPOSORB resin (Calbiochem) was used to delipidate plasma, according to the batch procedure of the manufacturer’s instructions. Twenty milliliters of plasma was mixed with 15 mL of resin by vortexing for 1 min, and then the resin was pelleted by centrifugation at 3000 rpm for 10 min. The lipid-free supernatant was kept, and the resin was resuspended in 30 mL of PBS for washing. Bound lipid material was eluted when the resin was mixed once with 35 mL of 200 mM sodium deoxycholate and once with 35 mL of 100 mM NaOH. Eluted material was dialyzed against PBS (pH 7.4).
Adhesion Assay
RAW 264.7 cells were grown in DMEM supplemented with 10% fetal bovine serum, 10 mM Hepes, 2 mM glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin (all from Gibco/Invitrogen Corp.) and subcultured in bacteriologic plastic dishes for at least 24 h before an adhesion assay, to allow detachment with 5 mM EDTA alone; 96-well flat bottom tissue culture plates were coated with 30 μL of 2% (w/v) gelatin containing ligands at the indicated concentrations and left to dry at 37 °C. Cells were washed three times with PBS and then resuspended at a density of 3 × 106 cells/mL in DMEM and 1% BSA. Inhibitors were prepared in DMEM and 1% BSA as follows: 10 mM EDTA, 10 μg/mL rat anti-mouse SR-A monoclonal antibody, or a combination of both. For studies with apo A-I mimetic peptide 4F, the anti-SR-A antibody was replaced by 4F or control scrambled peptide scr4F at the indicated concentrations; 100 μL of inhibitors or medium alone was dispensed to the ligand-coated wells, followed by 100 μL of cell suspension. Plates were incubated at 4 °C for 30 min to allow binding of antibodies and then shifted to 37 °C for 1.5−2 h to let the cells adhere. We washed plates carefully by dispensing PBS into the wells and aspirating washes with a multichannel pipet. Adherent cells were fixed by incubation with ice-cold methanol for 20 min at 4 °C and then stained with a crystal violet solution. The stain was solubilized in 100 μL of 1% SDS per well and absorbance read on a plate reader at 550 nm.
Three-Dimensional Modeling
To generate a three-dimensional model of the collagenous and SRCR domains of human SR-AI (hSR-AI), the sequence of residues 273−341 (collagenous domain) of hSR-A (UniProt entry P21757) was entered into the FFAS server (28) searching against the PDB (Protein Data Bank) as of September 2007, and PDB entry 1y0f (type I collagen) was used as a starting model for the collagenous domain (structure by fiber diffraction; Cα atoms only). The human SR-A sequence was docked onto the Cα positions of the collagen structure using the SCWRL server (29), and main and side chain atoms were further modeled using MODELLER (30). The SRCR of SR-A (residues 350−450) was modeled onto the trimeric SRCR of MARCO, for which a crystal structure is known (PDB entry 2oya, using the AB dimer in the asymmetric unit and copy B in cell x−1,y,z). The model for hSR-AI was built in MODELLER by combining the model of the trimeric head of MARCO described above and the collagen 1 trimer.
The model hSR-AI structure and either the human apo A-I structure (PDB entry 2a01) or the human apo E structure (PDB entry 1ea8) were docked together using the suite of Haddock scripts (31), with constraints on docking residues of SR-A as follows: Arg325, Lys332, Lys335, and Lys338. After water refinement, the lowest-energy pairs of 200 structures were chosen. The MSDPisa server (32) was used to determine contact residues and interaction characteristics.
Data Analysis
All data presented were analyzed using GraphPad Prism, version 5.0 (GraphPad Software, San Diego, CA). Unless otherwise indicated, data are shown as means of triplicates with the standard deviation (SD). For comparison of grouped variables (ELISA, adhesion), two-way ANOVA with Bonferroni postcomparison tests were used to determine significance. One asterisk indicates P < 0.05; two asterisks indicate P < 0.01 and three asterisks P < 0.001.
Results
Purification of SR-A Candidate Ligands from Plasma
A combination of chromatographic separation techniques and an ELISA-based SR-A binding screen (20) paralleled by far-Western blot was used to identify candidate SR-A ligands from human plasma (see Figure 1 of the Supporting Information for details of the workflow). Although serum and plasma exhibit comparable ligand activity (data not shown), plasma was used for fractionation to prevent unwanted modifications induced by the proteases activated during the clotting cascade. Since all known SR-A ligands are polyanions, the complexity of the plasma was first reduced by anion exchange, followed by an ELISA screen with lysates from WT and SR-A−/− BMMφ on the eluted fractions. The resulting two-peak SR-A binding pattern is shown in Figure 1A. Ligand detection was selective for specific proteins as the SR-A binding trace did not simply reflect the protein elution profile. The two specific ligand peaks eluted at ∼300 mM NaCl (30% high-salt buffer) and after 1 M salt had been reached, suggesting either a very highly charged ligand or one that interacts with the column resin. Unbound material eluting before the salt gradient gave an equal signal in the presence and absence of detecting SR-A in ELISA (Figure 2A of the Supporting Information). The same was observed when an isotype control antibody instead of anti-SR-A was used (Figure 2B of the Supporting Information), but not when IgG-depleted plasma was fractionated, demonstrating that this signal was due to cross-reaction of the secondary antibody with immunoglobulins and not to specific SR-A ligands in these neutral or basic fractions. To remove nonspecific background from subsequent screens, either plasma was cleared of IgG prior to fractionation [which did not affect the two-peak SR-A binding pattern (Figure 1A)] or binding was expressed as the SR-A-specific signal (WT minus SR-A−/−). To allow interassay comparability, this value was normalized to an internal positive control (known SR-A ligand).
Figure 1.
Identification of novel endogenous SR-A ligands in human plasma. (A) One milliliter plasma devoid of precipitates and chylomicrons or 3.5 mL of the same plasma depleted of immunoglobulins was diluted with running buffer to a total volume of 5 mL, injected onto a MonoQ High Resolution 5/5 column, and eluted with a two-stage salt gradient from 0 to 1 M NaCl. Fractions (1 mL) were collected and assayed via an ELISA for SR-A binding activity as described in Experimental Procedures. The FPLC elution profile with absorbance at 280 nm (line) and conductance (millisiemens per centimeter) indicating salt strength (dotted line) is shown along with SR-A-specific binding (●), where specific binding is calculated as WT minus SR-A−/−. Arrows indicate peaks of SR-A binding; the bar indicates fractions pooled for further fractionation. (B) One milliliter of concentrated albumin-depleted active pool (fractions 30−35 from panel A, plasma, and IgG-depleted plasma) was injected onto a Superose12 HR 10/30 column. Fractions (1 mL) were collected and assessed for SR-A binding activity in ELISA (fractions 6−16). Binding (●), expressed as the percentage of binding to the internal positive control (5 μg/mL acLDL), is plotted vs absorbance (···). (C) Ten microliters of fractions 9−13 was boiled in nonreducing loading buffer, subjected to 8% SDS−PAGE, and either stained with Coomassie blue or transferred to a nitrocellulose membrane and subjected to far-Western analysis, using WT and SR-A−/− BMMφ lysates as described in Experimental Procedures. The strongest SR-A binding areas are indicated by boxes (around 160, 75−35, and 25 kDa).
While the first peak had a clearly detectable absorbance at 280 nm, suggesting a protein ligand, the second peak did not contain any detectable material at this wavelength, impeding further analysis with conventional protein biochemistry methods. Therefore, only ligand-containing fractions 30−35 (peak 1) from plasma and IgG-depleted plasma ion exchange were pooled, affinity-depleted of albumin, concentrated, and further fractionated by size exclusion on Superose12 (fractionation range up to approximately 300 kDa). Albumin depletion had no notable effect on SR-A reactivity, and the discarded albumin-enriched material showed no significant SR-A binding activity (not shown). Albumin-free material eluted in two contiguous peaks, with SR-A reactivity localizing to the second peak only (Figure 1B). A parallel chromatography with molecular mass standards allowed the ligand-containing peak (fractions 9−12) to be centered on 160 kDa. However, the activity peak was quite broad, suggesting that the ligand might not be homogeneous. Nonreducing SDS−PAGE and far-Western analysis (Figure 1C) of the corresponding fractions revealed several areas of SR-A reactivity, corresponding to apparent molecular masses of approximately 160, 75−35, and 25 kDa, suggestive of a ligand that would form a protein complex or aggregate in solution but (partially) dissociate into monomers under denaturing conditions.
Identification of Candidate Ligands by Mass Spectrometry
Four strong SR-A-reactive bands from fractions 9 and 10, at apparent molecular masses of <75, >50, 35, and 25 kDa (Figure 1C), were excised and subjected to in-gel tryptic digest, and peptides were analyzed by electrospray mass spectrometry. The first candidate list (Table 1 of the Supporting Information, first run) included a large number of complement breakdown products (C3), protease inhibitors (inter-α-trypsin inhibitor, C1 inhibitor), and numerous hits of apolipoprotein A-I. To increase the reliability of mass spectrometry hits, the described ligand purification (ion exchange, size exclusion) was repeated at a larger scale and seven putative SR-A-reactive bands were excised and subjected to mass spectrometry. The second candidate list (Table 1 of the Supporting Information, second run) confirmed initial hits and made it possible to select complement C3 (C3) and apolipoprotein A-I (apo A-I) as the two most promising candidate ligands. While both C3 and apo A-I were found in all four bands of the first run, C3 scored better than apo A-I (average scores of 558 and 211, respectively) and was identified by a higher average number of unique peptides (12 and 3.75, respectively). However, average sequence coverage was higher for apo A-I (16.25%) than for C3 (8%). As both the number of unique peptides and sequence coverage are somewhat dependent on the length of the protein, score was considered the most reliable rating criterion.
In a manner independent of score, however, several considerations argued in favor of apo A-I being the main candidate ligand. Since the constitutive low level of complement activation in plasma leads to covalent association of C3 with random plasma proteins, it is highly likely to find complement components in mass spectrometry searches. Similarly, high-abundance plasma proteins (albumin, immunoglobulins, antitrypsin, transferrin, and haptoglobin) are known to interfere with mass spectrometry sensitivity because they mask minor components. Apo A-I is the main protein constituent of high-density lipoprotein (HDL), which spans a size range from 155 to 416 kDa in healthy subjects (33). This agrees with the apparent molecular mass of the SR-A ligand determined by size exclusion chromatography (160 kDa). In addition, most table entries belonged to functional classes (proteinase inhibition, complement regulation, acute phase response, and lipid metabolism) recently found to be associated with HDL by shotgun proteomics (34). Therefore, apo A-I was retained as the main SR-A candidate ligand.
Novel SR-A Ligands Are Derived from HDL
To test whether the identified SR-A-reactive compound was carried on HDL, total plasma lipoproteins were isolated by ultracentrifugation, subjected to size exclusion chromatography, and assessed for SR-A reactivity via an ELISA (Figure 2A). Consistent with SR-A recognition of modified LDL, which occurs at basal levels in plasma, the LDL peak showed weak SR-A binding. Astonishingly, the HDL peak bound SR-A strongly, contradicting the established conviction that HDL is not an SR-A ligand. However, oxidation of HDL by plasma enzymes such as myeloperoxidase is a physiologically occurring modification and has been shown to target HDL to various scavenger receptors, including SR-A (35). Oxidation results in a ladderlike running pattern of HDL proteins on gels, mainly due to covalent aggregation of apo A-I and/or apo A-II. When HDL fractions were tested for SR-A reactivity by far-Western blot (Figure 2B), the observed banding pattern was similar to that in Figure 1C, with SR-A-reactive bands at <75, 50, and 25 kDa. A parallel apo A-I Western blot proved the presence of apo A-I in four different bands, at <75, 50, <35, and 25 kDa (filled arrowheads, Figure 2B). Note that the relative intensities of SR-A- and anti-apo A-I antibody-dependent recognition of higher-order apo A-I aggregates differ, suggesting preferential recognition of aggregated material by SR-A. The additional SR-A-reactive band at 14 kDa (empty arrowhead, Figure 2B) was not identified on the apo A-I blot and might correspond to apo A-II, the second-most abundant apolipoprotein of HDL and structurally similar to apo A-I (36). These findings point toward recognition of oxidized or aggregated rather than native apo A-I. To verify whether signatures of enzymatic oxidation could be found among the apo A-I peptides identified by mass spectrometry in our study, we reanalyzed all peptides with search criteria modified to include oxidative modifications. Myeloperoxidase specifically oxidizes selected methionine and tyrosine residues of apo A-I, including Met86 (37), which was indeed identified on several distinct peptides.
Figure 2.
Distribution of SR-A binding activity in plasma lipoproteins. (A) One milliliter of total plasma lipoproteins from a single donor was isolated by ultracentrifugation at a density of 1.25 g/mL and separated by size exclusion chromatography via FPLC (---), and fractions were analyzed for SR-A reactivity (●). Data are representative of three different donor profiles. (B) The HDL peak fractions (28−30) were separated by nonreducing 6 to 20% SDS−PAGE, transferred to a blotting membrane, and detected in the following order: far-Western blot with SR-A-containing cell lysate and isotype control, stripped for 15 min, far-Western blot with SR-A-containing lysate and anti-SR-A antibody, stripped for 10 min, and Western blot with anti-apo A-I antibody. Filled arrowheads indicate apo A-I bands at <75, 50, <35, and 25 kDa, and the empty arrowhead indicates the non-apo A-I, SR-A-reactive band at 14.3 kDa. Bands above 105 kDa on far-Western blots are due to cross-reacting secondary antibody.
Plasma Contains Lipid-Associated and Lipid-Free SR-A Ligands
To remove all lipid-associated SR-A ligands from plasma and test for remaining SR-A reactivity, plasma was delipidated using a lipophilic resin. Both the lipid-free plasma and the apolipoprotein-enriched eluates from the resin were then subjected to SDS−PAGE and far-Western blot (Figure 3A). On SDS−PAGE (Figure 3A, left panel), note the absence of strong bands in the apo A-I size region (25−30 kDa) in delipidated plasma and the resin wash, and corresponding strong bands in eluates, indicating efficient removal of apo A-I by the delipidation step and enrichment on the resin. On the far-Western blot (Figure 3A, right panel), four size areas reacted with SR-A, among which bands above 105 kDa were seen on both WT and SR-A−/− overlays, corresponding to cross-reaction of the secondary antibody with plasma immunoglobulins. A strong SR-A-specific band around 67 kDa (similar in size to the control SR-A ligand MalBSA) potentially corresponds to some form of modified albumin, as it is abundantly present in plasma and was unaffected by delipidation. Indeed, AGE-modified albumin occurs at low levels even in non-diabetic patients and has been proven to be a ligand for SR-A (38). Two lipid-associated SR-A-specific bands localized above 35 kDa (the size of apo E) and below 30 kDa, the latter likely corresponding to apo A-I. The far-Western assay used in this work may detect only ligands that are resistant to denaturation or that may renature at least partially on blotting. The ELISA, however, does not involve ligand denaturation and so might detect additional ligands that are more fragile. Therefore, delipidated plasma was fractionated by ion exchange, and all fractions were screened for the presence of apo A-I and SR-A reactivity via an ELISA (Figure 3B, bottom panel). Fractionation of delipidated plasma was compared to an identical fractionation of total plasma (Figure 3B, top panel). As in Figure 1A, ion exchange fractionation of plasma yielded two separate zones of SR-A binding activity, and again the second zone had very weak absorbance at 280 nm and eluted at a high salt strength. In plasma fractions, both zones contained apo A-I, with close overlap between SR-A binding and apo A-I content in zone 1. In delipidated plasma fractions, the SR-A-reactive, apo A-I-containing zone 1 was absent, confirming the hypothesis of lipid-associated apo A-I as a major SR-A ligand. However, binding and apo A-I traces in zone 2 were unaffected by delipidation, suggesting that zone 2 corresponds to small amounts of intact or fragmented lipid-free apo A-I and potentially additional ligands. Filtration on centrifugal columns with different molecular mass cutoffs showed that ligand activity in zone 2 was associated with a molecular entity above 10 kDa. Traces of apo A-I could be proven by Western blot, and SDS−PAGE showed additional bands between 10 and 14 kDa potentially corresponding to apo A-II (not shown). Similar to the oxidative hypothesis raised above, this suggested that fragmentation and/or aggregation generates SR-A ligands from apo A-I.
Figure 3.
Plasma contains lipid-associated and lipid-free SR-A binding activity. (A) Plasma (20 mL) was delipidated on 15 mL of PMH-L Liposorb resin slurry as described by the manufacturer. Eluates with sodium deoxycholate (eluate 1) and sodium hydroxide (eluate 2) were dialyzed against PBS and adjusted to the starting volume. Aliquots of delipidation steps (plasma, P; delipidated plasma, DP; PBS wash, w; eluates predialysis e1, e2; eluates postdialysis E1, E2) were run on a nonreducing 6 to 20% SDS−PAGE gradient, along with 5 μg of positive and negative controls (MalBSA, MB; BSA, B) per lane and 5 μg of purified apo A-I (AI) per lane. Identical gels were blotted on nitrocellulose for far-Western blots with WT and SR-A−/− lysates. The black arrowhead on the SDS−PAGE gel indicates the position of apo A-I, and white arrowheads on the blot indicate SR-A ligands in plasma, corresponding to proteins with apparent molecular masses of >25, >35, and ∼67 kDa. (B) One milliliter each of total plasma and Liposorb-delipidated plasma were fractionated on a QSepharose column [elution profile by absorbance at 280 nm (···)], and every second 1 mL fraction was tested for SR-A reactivity (●) and apo A-I content (—) via an ELISA. Zone 1 indicates delipidation-sensitive and zone 2 delipidation-resistant SR-A binding activity.
Confirmation of Novel SR-A Ligands
Ligand identification so far suggested apo A-I and potentially also apo A-II and apo E as novel SR-A ligands. Further, receptor binding seemed to be induced or enhanced by oxidative modifications, aggregation or degradation. To test whether native apolipoproteins are novel ligands for SR-A and to prove specificity for select apolipoproteins, various purified human apolipoproteins, including apo A-I and A-II, all three human apo E isoforms (E2, E3, and E4), and serum amyloid A (apo SAA), which replaces apo A-I in HDL during the acute phase response, were tested for SR-A reactivity. When WT was compared to the SR-A−/− signal via an ELISA (Figure 4A), significant binding was detected for apo A-I, HDL (containing both apo A-I and apo A-II), and all apo E isoforms, but not apo SAA. Comparison of WT and SR-A−/− far-Western blots (Figure 4B) confirmed this recognition pattern, with a strong SR-A-specific signal for all apo E isoforms and apo A-I, but not for apo A-II or monomeric apo SAA. The band above 50 kDa in the apo SAA lane most likely corresponds to aggregated or multimeric apo SAA, in agreement with known SR-A recognition of amyloid aggregates (39). An additional proof of receptor specificity is the ability to compete with established ligands. Therefore, the ELISA was repeated in the presence of competitors. OxLDL and, to some extent, acLDL blocked recognition of all novel SR-A ligands (Figure 4C), suggesting that the novel ligands share the same receptor binding site as known ligands. Taken together, these experiments suggested that SR-A recognition of apolipoproteins is lipid-independent and selective and does not seem to depend on oxidative modification. However, both ELISA and far-Western blot assays depend on immobilization or unfolding of the protein ligand and do not prove that native, soluble proteins will interact with the receptor.
Figure 4.
Apo A-I and apo E isoforms are novel ligands for SR-A. (A) Recombinant human apo E2, E3, E4, and SAA and purified human apo A-I (all at 500 nM), purified HDL (250 nM), and control ligands MalBSA (positive) and BSA (negative) (150 nM) were coated on ELISA plates, and SR-A reactivity was detected by comparison of WT (black bars) and SR-A−/− (white bars) lysates as described in Experimental Procedures. Three asterisks indicate P < 0.001 for WT vs SR-A−/− compared by two-way ANOVA and Bonferroni post-tests. Results are representative of at least three similar experiments. (B) Five micrograms of control ligands MalBSA (positive) and BSA (negative) or candidate ligands apo E2, E3, E4, and apo SAA and HDL containing apo A-I and apo A-II were separated by SDS−PAGE, transferred to a nitrocellulose membrane, and assessed for SR-A reactivity by far-Western blot as described in Experimental Procedures. Localization of proteins was visualized by Ponceau Red staining of the membrane prior to far-Western blot. (C) Apo A-I (250 nM) and apo E isoforms (125 nM) (total volume of 50 μL) were coated on ELISA plates and detected with WT or SR-A−/− lysates as described. For competition, lysates were preincubated with acLDL or oxLDL at a concentration of 50 μg of apolipoprotein/mL for 30 min at RT. Results are expressed as the percentage of residual SR-A-specific binding. One asterisk indicates P < 0.05, and three asterisks indicate P < 0.001 for competitor vs unblocked compared by two-way ANOVA and Bonferroni post-tests.
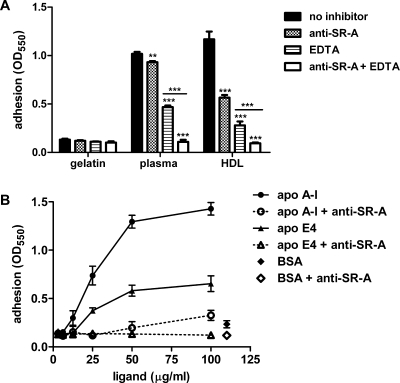
This study set off with the hypothesis that plasma contains endogenous ligands capable of mediating SR-A-dependent macrophage adhesion. Therefore, the novel candidate ligands were tested in an SR-A-specific adhesion assay. First, adhesion of RAW 264.7 macrophages was assessed on a matrix of gelatin, which does not sustain SR-A-dependent adhesion, or on a mix of gelatin matrix and plasma or HDL (Figure 5A). In the latter setting, macrophage adhesion is expected to occur through integrins, which can be inhibited by divalent metal ion chelation with EDTA, and through SR-A, which can be abolished by a monoclonal blocking antibody against SR-A, while a combination of both completely abrogates adhesion (16). Macrophages adhered to plasma and HDL in an SR-A-dependent manner, proving that plasma contains an adhesion ligand that copurifies with HDL. Next, protein candidate ligands apo A-I and apo E were tested for their capacity to mediate SR-A-specific macrophage adhesion. Apo A-I is found in atherosclerotic vascular plaques and apo E in neurodegenerative lesions of Alzheimer’s disease, both macrophage-driven chronic inflammatory pathologies characterized by amyloid-associated deposits of denatured, unfolded, or truncated proteins (40−42). Among the apo E isoforms, apo E4 was deemed the most physiologically relevant since the APOE4 allele is associated with late-onset Alzheimer’s disease (43). Integrin-independent adhesion of macrophages to immobilized apo A-I or apo E4 was dose-dependent, completely blocked by anti-SR-A antibodies, and specific for the novel ligands, since no adhesion was observed with gelatin and BSA (Figure 5B). Although RAW 264.7 macrophages secrete apo E at a rate comparable to that of immature monocytes (44), the lack of adhesion to BSA-coated wells proved this endogenous apo E did not contribute to adhesion in our assay. To exclude any additive effects from nonquantified endogenous apo E in future studies, it would be of experimental benefit to use J774 macrophages, which do not express apo E.
Figure 5.
Novel SR-A ligands apo A-I and apo E4 mediate macrophage adhesion. (A) Adhesion of RAW 264.7 macrophages to ligand-coated tissue culture plastic plates [coated with 2% (w/v) gelatin, nonadhesive negative control; plasma 1:10 or HDL at a concentration of 50 μg of apolipoprotein/mL diluted in 2% (w/v) gelatin] was assessed in the absence of inhibitors or in the presence of mouse monoclonal anti-SR-A blocking antibody, EDTA (for integrin-independent adhesion), or a combination of both. Two asterisks indicate P < 0.01 and three asterisks P < 0.001 comparing conditions with inhibitors to unblocked control by two-way ANOVA and Bonferroni post-tests. Results are representative of three similar experiments. (B) Dose-dependent adhesion of RAW 264.7 macrophages to immobilized apo A-I and apo E4 was tested as described for panel A. For the sake of clarity, only adhesion in the presence of EDTA (—) or EDTA and anti-SR-A (···) is shown. BSA was used as a negative control ligand. Results are representative of two similar experiments.
The Shared Amphipathic α-Helix of Apo A-I and Apo E Is a Minimal Binding Motif for SR-A
All known polyanionic SR-A ligands interact with a stretch of positively charged lysines within the receptor’s collagenous ligand binding domain (9,45). Competition between known SR-A ligands (acLDL and oxLDL) and the novel ligands identified here (Figure 4C) already established that both groups likely bound to overlapping or identical sites on the receptor. It would thus seem plausible that recognition relied on interaction between positive receptor residues and negative ligand residues, either present in the native state or induced by modification. Since purified and recombinant apolipoproteins were able to bind the receptor (Figure 4A,B), modification did not seem an absolute prerequisite, and native negative charge was favored as a hypothesis. Apo A-I and apo E belong to an evolutionarily conserved family of exchangeable apolipoproteins, which are characterized by amphipathic α-helical repeats (46). Superposition of electron density maps of lipid-free apo A-I and apo E has revealed that both molecules exhibit a large negative surface, which results from parallel packaging of α-helices and remains solvent-exposed even after rearrangement of helices around lipid globules (47). 4F is an apo A-I mimetic peptide 18 amino acids in length, which reproduces the features of apo A-I amphipathic α-helices based on a hydrophobic, lipid-associating and a negatively charged, solvent-exposed side without having any sequence similarity, and displays manifold anti-atherogenic and broadly anti-inflammatory properties (48−52). If the shared amphipathic α-helix was a minimal binding motif, then 4F should be able to compete with known and novel SR-A ligands alike, since both seem to bind the same receptor site. In an ELISA-based competition assay, soluble 4F, but not a scrambled control peptide (scr4F), blocked binding of SR-A to known ligands acLDL and oxLDL, as well as to novel ligands apo A-I, E2, E3, and E4, while nonligands BSA and apo SAA were unaffected (Figure 6A). In the whole-cell adhesion assay, the apo A-I mimetic peptide dose-dependently inhibited integrin-independent adhesion of RAW 264.7 macrophages to apo A-I and apo E4 (Figure 6B). We therefore assumed short amphipathic α-helices containing a negatively charged side represented a minimal binding motif as the basis of selective apolipoprotein recognition by SR-A.
Figure 6.
Identification of a minimal binding motif on novel apolipoprotein ligands. (A) Apo A-I mimetic peptide (4F, sequence DWFKAFYDKVAEKFKEAF) or scrambled control peptide (scr4F, sequence DWFAKDYFKKAFVEEFAK) was tested in a competition ELISA against known and novel SR-A ligands (5 μg of acLDL protein/mL, 20 μg of oxLDL protein/mL, 10 μg/mL BSA, or 250 nM apo A-I, E2, E3, E4, or SAA). Lysates were preincubated for 30 min at RT with competitors at 80 μg/mL. Binding is shown as the WT minus SR-A−/− signal. (B) Adhesion of RAW 264.7 macrophages to immobilized apo A-I and apo E4 was tested in the presence of increasing concentrations of 4F or control scr4F (single value shown, highest dose), and IC50 values were determined using a nonlinear regression model with a variable slope. Error bars represent the standard error of the mean.
Proposed Interaction Model of the Collagenous Ligand Binding Domain from Human SR-A with Apolipoproteins A-I and E
The data presented above are consistent with an interaction model in which both newly identified and known ligands share the same binding site on SR-A. No structure is available for the complex trimeric transmembrane receptor SR-A; however, its collagenous ligand binding domain closely resembles collagen I, and the SRCR domains of class A scavenger receptors SR-A and MARCO are highly conserved, allowing three-dimensional modeling of the binding and head regions (Figure 7A, residues involved in binding highlighted in red). The three-dimensional structures of apo A-I and apo E have been determined at 2.4 and 1.95 Å, respectively (PDB entries 2a01 and 1ea8, respectively) and show considerable overlap in structure. Most importantly, when bundled in a lipid-free state, they present a large solvent-accessible negative patch, which unfolds into smaller negative stretches upon association with lipid bilayers or disks (47). The four-helix domain common to both apo A-I and apo E (Figure 7B), corresponding to amino acids 1−190 of human apo A-I or amino acids 23−162 of human apo E3, was used to model the interaction with SR-A, as it contains the negative residues potentially involved in SR-A binding. Both apo A-I and apo E3 helix domains were separately docked with the model for the SR-A receptor; in both docking calculations, the only restraint imposed was that one of the known contact residues on the receptor side would contact the apolipoprotein. Of 200 possible receptor−ligand pairs in both docking calculations, the energetically most favorable pairs with maximal interaction surfaces were selected. All involved the apolipoprotein docking to the receptor binding domain in a parallel manner, with consecutive negative residues (Asp or Glu) on ligand helices locking one or more positive residues in the receptor binding domain (Arg317, Arg325, or Lys338) (Figure 7C, SR-A:apo AI, and Figure 7D, SR-A:apo E). The energetically most favorable models used a single helix for interaction, a finding compatible with lipid-free apolipoprotein (helix bundle), HDL (unfolded helices), or short lipid-free α-helical peptides binding to SR-A. Modeling with the murine collagenous receptor domain produced comparable interactions (not shown). The models are therefore consistent with the hypothesis of an interaction pair involving negative residues of a ligand α-helix contacting the known binding site on SR-A.
Figure 7.
Proposed model of interaction of human SR-A with apolipoproteins A-I and E3. (A) The collagenous ligand binding domain of human SR-A, modeled onto the NMR diffraction structure of collagen I (PDB entry 1y0f), and the SRCR domain modeled according to the SRCR of class A scavenger receptor MARCO (PDB entry 2oya) were assembled using MODELLER. Residues implicated in ligand binding are colored red and labeled for one chain only. (B) Overlay of apo A-I residues 1−183 (PDB entry 2a01, blue) and apo E3 residues 23−162 (PDB entry 1ea8, turquoise) showing the similarity of three-dimensional features. (C) SR-A collagenous domain and the first four helices of apo A-I docked together using Haddock. The inset shows a crucial receptor binding residue, Arg317, clamped between two negative residues on apo A-I, Asp24 and Asp28. (D) Similar docking with apo E3. The inset shows two conserved receptor binding residues, Arg325 and Lys338, clamped between Asp50 and Asp59 and between Asp66 and Asp70, respectively. The numbering of apolipoprotein residues corresponds to the mature protein (precursor numbering = mature protein numbering + 24 for apo A-I and + 18 for apo E3).
Discussion
Our aim was to investigate SR-A-mediated adhesion by identifying endogenous ligands in plasma. We propose that exchangeable apolipoproteins A-I and E represent a novel class of endogenous SR-A ligands, compete with known ligands acLDL and oxLDL for the known binding site on the receptor, and use their shared amphipathic α-helix as a minimal recognition motif for binding to SR-A. Immobilized apo A-I and apo E sustain integrin-independent, SR-A-mediated macrophage adhesion, which may represent a mechanism through which SR-A contributes to the trapping of macrophages at sites of pathological apolipoprotein deposition.
Structural Considerations
Exchangeable apolipoproteins are structurally conserved from insect apolipophorins to human apolipoproteins (53,54). Their lipid-free conformation is based on a helix bundle with hydrophobic domains shielded from the solvent and a large solvent-exposed negative domain. During lipid association, the amphipathic helix bundle unfolds and concatenates its helices to wrap around lipid disks or globules (47). This implies that SR-A could recognize exchangeable apolipoproteins via their solvent-exposed negative domains in both lipid-free (bundled) or lipid-associated (unfolded, concatenated) conformations. In addition, recognition of a small peptide containing as few as four negative residues supports the concept of a short, robust minimal binding motif, potentially found in unrelated molecules. However, not all apolipoproteins tested in this study bound to SR-A. Apo SAA presents only limited α-helical content; apo A-II, despite its amphipathic α-helical structure, is considerably shorter than apo A-I or apo E, and its charge distribution may differ. Interestingly, known SR-A ligands acLDL and oxLDL differed slightly in their ability to compete with the novel ligands (Figure 4C), and reciprocally, the apo A-I mimetic peptide differentially inhibited SR-A recognition of acLDL and oxLDL (Figure 6A). While this may simply reflect the differential receptor binding residue usage underlying the nonreciprocal cross-competition of oxLDL and acLDL (9,55), it could also indicate that exchangeable apolipoproteins recapitulate an oxidation-specific epitope present exclusively on oxLDL.
Novel SR-A Ligands Are Derived from High-Density Lipoprotein
A shotgun proteomics study comparing HDL from healthy subjects and coronary artery disease patients identified proteins involved in the acute phase response, proteinase inhibition, and complement regulation in addition to lipid metabolism (34). Overlapping hits between this and our study include acute phase proteins kininogen-1 and serum albumin, protease inhibitors inter-α-trypsin inhibitor heavy chain H4 and α-1-antitrypsin, complement components C3 and C4, and HDL-associated antioxidant enzyme paraoxonase, validating the accuracy of mass spectrometry-based identification of candidate ligands. To ascertain that apo A-I was the only HDL protein involved in SR-A binding, several high-scoring hits were tested for binding to SR-A. α-1-antitrypsin and α-1-antichymotrypsin both tested negative. Complement iC3b is known to promote macrophage adhesion via complement receptor CR3 (56), a mechanism that is divalent metal ion-dependent and would thus not account for the EDTA-resistant adhesion mechanism with respect to serum-coated surfaces at the base of this study.
Initial receptor binding studies described here involved immobilization of the ligands, which might have induced conformational changes or aggregation, thereby deriving SR-A ligands from HDL. It cannot immediately be concluded that apolipoproteins are sufficient for receptor interaction with HDL, even though apo A-I mimetic peptides in solution were able to compete with known ligands for receptor usage. Rather, SR-A is defined by its ability to scavenge obsolete material, including oxidized, denatured, and aggregated molecules, and is not known to bind native lipoproteins or HDL in particular. Indeed, preliminary cell association assays with HDL protected from oxidation by the addition of the antioxidant probucol showed no difference between WT and SR-A−/− BMMφ, in line with the lack of detectable HDL phenotype in SR-A−/− mice. This may indicate that additional factors, like oxidative modification, are needed to increase negative charge or that aggregation or unfolding on surfaces is necessary to confer SR-A binding capacity to apolipoproteins. In vivo enzymatic oxidation of HDL specifically targets tyrosine and methionine residues of apo A-I, which results in impaired acceptance of cholesterol from ABCA1 and SR-BI (37). SR-A has been implicated in the metabolism of myeloperoxidase-modified HDL (35) and may constitute a scavenging mechanism for the removal of this nonfunctional entity from the organism. Whether this relies partially or at all on recognition of apo A-I remains to be determined.
Similarly, whether SR-A-dependent adhesion to apo A-I and apo E is a phenomenon of frustrated endocytosis or a distinct mechanism remains to be investigated. Different cytoplasmic domains of SR-A are involved in adhesion and receptor internalization (57), and cell spreading after adhesion has been shown to induce distinct signaling pathways from ligand-activated receptor internalization (58,59). Bound SR-A ligands usually undergo receptor-mediated endocytosis followed by lysosomal delivery (12), but since HDL does not seem to be an endocytic ligand, it is likely that apolipoprotein recognition will solely elicit adhesion pathways.
Pathophysiological Implications and Therapeutic Potential
Age-related chronic inflammatory pathologies such as atherosclerosis and Alzheimer’s disease are characterized by combined accumulation of activated macrophages and microglia, which express high levels of SR-A (60,61), and pathological deposition of apolipoproteins. Macrophage adhesion through scavenger receptors is well-characterized, including binding of SR-A to extracellular matrix proteins, biglycan and decorin (19), glycated collagen IV (17), and β-amyloid (39), as well as adhesion of CD36 to myeloperoxidase-modified LDL (oxLDL) (62). Surprisingly, ablation of both receptors in atherosclerosis-prone apo E−/− or C57BL/6 mice does not decrease overall lesion area, suggesting that additional adhesion mechanisms share the responsibility of lesional macrophage retention (63,64). However, engagement of both receptors activates cell responses that contribute to the progression of disease: engagement of SR-A or CD36 with immobilized ligands induces a spread morphology with formation of filopodia and focal adhesion complexes, through a common signaling pathway (Src nonreceptor protein tyrosine kinases, MAPK serine/threonine kinases) (65−68), and blocks macrophage chemotaxis (39,62,69,70), suggesting both receptors are involved in macrophage accumulation in lesions. SR-A- and CD36-mediated adhesion to amyloid substrata or immobilized but not soluble oxLDL induces ROS production in microglia and macrophages (39,62,69,71), increasing the oxidative stress on their immediate environment. Our findings suggest that the novel SR-A ligands, which are found as insoluble deposits at sites of chronic inflammation, contribute to retention of macrophages and induce a pro-inflammatory, oxidative state, which in turn would drive accumulation of pro-atherogenic oxidized lipoproteins in the arterial wall or induce neurological damage in the brain. Indeed, several studies have proven the presence of apo A-I within atherosclerotic lesions (40,72), along with apo A-II and apo B (73−75). In many cases, apolipoproteins colocalize with extracellular matrix proteoglycans, many of which are scavenger receptor ligands in their own right (19,75,76). Similarly, senile Alzheimer’s plaques contain a plethora of apolipoproteins, including apo A-I and apo E (41). Apo A-I is present throughout all stages of lesion development, suggesting that SR-A- and apo A-I-dependent macrophage retention could occur from an early stage of disease. Early immunohistochemistry studies have shown colocalization of macrophages with apo A-I and/or apo E in human coronary artery lesions (73,76), but the direct functional relationship between SR-A and apo A-I in tissues remains to be proven. Interestingly, the novel ligands identified here are in part derived from the same macrophages they retain. Repopulation of irradiated apo E−/− mice with either WT or apo E−/− bone marrow indicates that macrophage secretion alone is able to reconstitute plasma apo E levels to approximately 10% (∼130 μg/dL) (77), sufficient to maintain a normal cholesterol profile in the absence of metabolic challenge. These studies further show that most lesion apo E is derived from resident macrophages, as opposed to infiltrated plasma lipoproteins. Macrophage apo E secretion is stimulated by cholesterol loading and can represent up to 16% of the total secreted protein (78,79). It would thus appear that apo E deposition, SR-A- and apo E-mediated macrophage retention, foam cell formation, and subsequent apo E production would form a self-propagating loop, exacerbating lesion progression. While it is technically challenging to determine the exact lesional concentration of apolipoproteins in vivo, several considerations justify the physiological relevance of apolipoprotein concentrations used in our in vitro assays. Secreted apo E is retained and enriched on the surface of macrophages through interaction with cell surface and extracellular matrix components, and constitutive apo E re-uptake and degradation is slowed by the lysosomal blockage that occurs in foamy macrophages saturated with nondegradable oxidized lipids (80). Hence, the apo E concentration in the extracellular space surrounding macrophages might reach levels higher than those corresponding to secretion.
The hypothesis of a pathological role for apo E proposed above is in contradiction with the atheroprotective role of plasma apo E established in hypercholesterolemic apo E−/− mice (81). However, it is important to distinguish between peripheral (macrophage-derived) and hepatic apo E. Indeed, hepatic apo E plays an essential role in lipoprotein catabolism through its interaction with hepatic lipoprotein receptors (81), and overexpression studies suggest that hepatic apo E, rather than macrophage apo E, is essential in lesion regression through reverse cholesterol transport (82,83). In contrast, macrophage-specific deletion of apo E in an atherosclerosis-prone background (C57Bl/6) and under metabolic challenge inhibits rather than enhances atherosclerosis development, consistent with a pro-atherogenic role of macrophage-derived, local apo E (84).
Along with CD36 and LOX-1, SR-A is one of the major receptors involved in cholesterol accumulation in macrophages. Our finding that 4F is able to compete efficiently with modified LDL suggests it may prevent foam cell formation by inhibiting oxLDL uptake via SR-A. The capacity of the small peptide 4F to abrogate the pro-atherogenic SR-A-mediated processes of oxLDL binding and adhesion of macrophages to lesional substrata may add therapeutic potential to this molecule, which is already on the way to medical approval (85,86). In addition, 4F provides a useful tool for potentially targeting one class A scavenger receptor family member specifically, as it is not recognized by the class A member MARCO (S. Mukhopadhyay, personal communication).
In summary, the identification of exchangeable apolipoproteins as novel endogenous ligands for scavenger receptor A described here raises a number of implications for macrophage adhesion, lipid homeostasis, and innate immunity.
Acknowledgments
We greatly appreciate the gift of apo A-I mimetic peptide 4F and control from Prof. Alan Fogelman (David Geffen School of Medicine, UCLA, Los Angeles, CA) and of HDL and HDL subclasses from Samuel Wright (Merck) and Marcielle de Beer (University of Kentucky), respectively. We thank Dr. David Greaves and Prof. Monty Krieger for helpful discussions and Dr. Susan Lea for access to computing resources.
Supporting Information Available
Outline of the ligand isolation strategy (Figure 1), plasma fractionated by ion exchange that contains two SR-A binding peaks (Figure 2), and mass spectrometry identification of candidate ligands in SR-A binding bands from two similar purification runs (Table 1). This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by the Wellcome Trust (Grant 076256/Z/04/Z to C.N.), the MRC (A.P., P.R., R.B.S., and S.G.), National Institutes of Health Grant P01HL086670 (D.R.v.d.W.), and the Ministry for Culture, Research and Higher Education of Luxembourg (C.N.).
Funding Statement
National Institutes of Health, United States
Footnotes
Abbreviations: SR-A, scavenger receptor A; BMMφ, bone marrow-derived macrophage; HDL, high-density lipoprotein; LDL, low-density lipoprotein; acLDL, acetylated LDL; oxLDL, oxidized LDL; apo, apolipoprotein; BSA, bovine serum albumin; MalBSA, maleylated BSA; AGE, advanced glycation end product; FCS, fetal calf serum; PBS, phosphate-buffered saline; RT, room temperature; IgG, immunoglobulin G; PMSF, phenylmethanesulfonyl fluoride; SDS−PAGE, sodium dodecyl sulfate−polyacrylamide gel electrophoresis; WB, Western blot.
Supplementary Material
References
- Goldstein J. L.; Ho Y. K.; Basu S. K.; Brown M. S. (1979) Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc. Natl. Acad. Sci. U.S.A. 76, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S.; Basu S. K.; Falck J. R.; Ho Y. K.; Goldstein J. L. (1980) The scavenger cell pathway for lipoprotein degradation: Specificity of the binding site that mediates the uptake of negatively-charged LDL by macrophages. J. Supramol. Struct. 13, 67–81. [DOI] [PubMed] [Google Scholar]
- Kodama T.; Reddy P.; Kishimoto C.; Krieger M. (1988) Purification and characterization of a bovine acetyl low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 85, 9238–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M.; Kodama T.; Matsumoto A.; Doi T.; Takahashi K. (1991) Tissue distribution, intracellular localization, and in vitro expression of bovine macrophage scavenger receptors. Am. J. Pathol. 139, 1411–1423. [PMC free article] [PubMed] [Google Scholar]
- Krieger M.; Acton S.; Ashkenas J.; Pearson A.; Penman M.; Resnick D. (1993) Molecular flypaper, host defense, and atherosclerosis. Structure, binding properties, and functions of macrophage scavenger receptors. J. Biol. Chem. 268, 4569–4572. [PubMed] [Google Scholar]
- Horiuchi S.; Sakamoto Y.; Sakai M. (2003) Scavenger receptors for oxidized and glycated proteins. Amino Acids 25, 283–292. [DOI] [PubMed] [Google Scholar]
- Husemann J.; Loike J. D.; Anankov R.; Febbraio M.; Silverstein S. C. (2002) Scavenger receptors in neurobiology and neuropathology: Their role on microglia and other cells of the nervous system. Glia 40, 195–205. [DOI] [PubMed] [Google Scholar]
- Acton S.; Resnick D.; Freeman M.; Ekkel Y.; Ashkenas J.; Krieger M. (1993) The collagenous domains of macrophage scavenger receptors and complement component C1q mediate their similar, but not identical, binding specificities for polyanionic ligands. J. Biol. Chem. 268, 3530–3537. [PubMed] [Google Scholar]
- Doi T.; Higashino K.; Kurihara Y.; Wada Y.; Miyazaki T.; Nakamura H.; Uesugi S.; Imanishi T.; Kawabe Y.; Itakura H.; et al. (1993) Charged collagen structure mediates the recognition of negatively charged macromolecules by macrophage scavenger receptors. J. Biol. Chem. 268, 2126–2133. [PubMed] [Google Scholar]
- Doi T.; Kurasawa M.; Higashino K.; Imanishi T.; Mori T.; Naito M.; Takahashi K.; Kawabe Y.; Wada Y.; Matsumoto A.; et al. (1994) The histidine interruption of an α-helical coiled coil allosterically mediates a pH-dependent ligand dissociation from macrophage scavenger receptors. J. Biol. Chem. 269, 25598–25604. [PubMed] [Google Scholar]
- Mommaas-Kienhuis A. M.; van der Schroeff J. G.; Wijsman M. C.; Daems W. T.; Vermeer B. J. (1985) Conjugates of colloidal gold with native and acetylated low density lipoproteins for ultrastructural investigations on receptor-mediated endocytosis by cultured human monocyte-derived macrophages. Histochemistry 83, 29–35. [DOI] [PubMed] [Google Scholar]
- Araki N.; Higashi T.; Mori T.; Shibayama R.; Kawabe Y.; Kodama T.; Takahashi K.; Shichiri M.; Horiuchi S. (1995) Macrophage scavenger receptor mediates the endocytic uptake and degradation of advanced glycation end products of the Maillard reaction. Eur. J. Biochem. 230, 408–415. [DOI] [PubMed] [Google Scholar]
- Yong K.; Khwaja A. (1990) Leucocyte cellular adhesion molecules. Blood Rev. 4, 211–225. [DOI] [PubMed] [Google Scholar]
- Plüddemann A.; Neyen C.; Gordon S. (2007) Macrophage scavenger receptors and host-derived ligands. Methods 43, 207–217. [DOI] [PubMed] [Google Scholar]
- Usui H. K.; Shikata K.; Sasaki M.; Okada S.; Matsuda M.; Shikata Y.; Ogawa D.; Kido Y.; Nagase R.; Yozai K.; Ohga S.; Tone A.; Wada J.; Takeya M.; Horiuchi S.; Kodama T.; Makino H. (2007) Macrophage scavenger receptor-a-deficient mice are resistant against diabetic nephropathy through amelioration of microinflammation. Diabetes 56, 363–372. [DOI] [PubMed] [Google Scholar]
- Fraser I.; Hughes D.; Gordon S. (1993) Divalent cation-independent macrophage adhesion inhibited by monoclonal antibody to murine scavenger receptor. Nature 364, 343–346. [DOI] [PubMed] [Google Scholar]
- El Khoury J.; Thomas C. A.; Loike J. D.; Hickman S. E.; Cao L.; Silverstein S. C. (1994) Macrophages adhere to glucose-modified basement membrane collagen IV via their scavenger receptors. J. Biol. Chem. 269, 10197–10200. [PubMed] [Google Scholar]
- Gowen B. B.; Borg T. K.; Ghaffar A.; Mayer E. P. (2000) Selective adhesion of macrophages to denatured forms of type I collagen is mediated by scavenger receptors. Matrix Biol. 19, 61–71. [DOI] [PubMed] [Google Scholar]
- Santiago-Garcia J.; Kodama T.; Pitas R. E. (2003) The class A scavenger receptor binds to proteoglycans and mediates adhesion of macrophages to the extracellular matrix. J. Biol. Chem. 278, 6942–6946. [DOI] [PubMed] [Google Scholar]
- Plüddemann A.; Neyen C.; Gordon S.; Peiser L. (2008) A sensitive solid-phase assay for identification of class A macrophage scavenger receptor ligands using cell lysate. J. Immunol. Methods 329, 167–175. [DOI] [PubMed] [Google Scholar]
- de Villiers W. J.; Fraser I. P.; Hughes D. A.; Doyle A. G.; Gordon S. (1994) Macrophage-colony-stimulating factor selectively enhances macrophage scavenger receptor expression and function. J. Exp. Med. 180, 705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenas J.; Penman M.; Vasile E.; Acton S.; Freeman M.; Krieger M. (1993) Structures and high and low affinity ligand binding properties of murine type I and type II macrophage scavenger receptors. J. Lipid Res. 34, 983–1000. [PubMed] [Google Scholar]
- Butler P. J.; Harris J. I.; Hartley B. S.; Lebeman R. (1969) The use of maleic anhydride for the reversible blocking of amino groups in polypeptide chains. Biochem. J. 112, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Pappin D. J.; Hojrup P.; Bleasby A. J. (1993) Rapid identification of proteins by peptide-mass fingerprinting. Curr. Biol. 3, 327–332. [DOI] [PubMed] [Google Scholar]
- Sim R. B.; Day A. J.; Moffatt B. E.; Fontaine M. (1993) Complement factor I and cofactors in control of complement system convertase enzymes. Methods Enzymol. 223, 13–35. [DOI] [PubMed] [Google Scholar]
- Poumay Y.; Ronveaux-Dupal M. F. (1985) Rapid preparative isolation of concentrated low density lipoproteins and of lipoprotein-deficient serum using vertical rotor gradient ultracentrifugation. J. Lipid Res. 26, 1476–1480. [PubMed] [Google Scholar]
- Jaroszewski L.; Rychlewski L.; Li Z.; Li W.; Godzik A. (2005) FFAS03: A server for profile−profile sequence alignments. Nucleic Acids Res. 33, W284–W288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canutescu A. A.; Shelenkov A. A.; Dunbrack R. L. Jr. (2003) A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12, 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., and Sali A. (2007) Comparative protein structure modeling using MODELLER. Current Protocols in Protein Science, Chapter 2, Unit 2, p 9, Wiley, New York. [DOI] [PubMed] [Google Scholar]
- Dominguez C.; Boelens R.; Bonvin A. M. (2003) HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737. [DOI] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Kontush A.; de Faria E. C.; Chantepie S.; Chapman M. J. (2005) A normotriglyceridemic, low HDL-cholesterol phenotype is characterised by elevated oxidative stress and HDL particles with attenuated antioxidative activity. Atherosclerosis 182, 277–285. [DOI] [PubMed] [Google Scholar]
- Vaisar T.; Pennathur S.; Green P. S.; Gharib S. A.; Hoofnagle A. N.; Cheung M. C.; Byun J.; Vuletic S.; Kassim S.; Singh P.; Chea H.; Knopp R. H.; Brunzell J.; Geary R.; Chait A.; Zhao X. Q.; Elkon K.; Marcovina S.; Ridker P.; Oram J. F.; Heinecke J. W. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117, 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suc I.; Brunet S.; Mitchell G.; Rivard G. E.; Levy E. (2003) Oxidative tyrosylation of high density lipoproteins impairs cholesterol efflux from mouse J774 macrophages: Role of scavenger receptors, classes A and B. J. Cell Sci. 116, 89–99. [DOI] [PubMed] [Google Scholar]
- Bolanos-Garcia V. M.; Miguel R. N. (2003) On the structure and function of apolipoproteins: More than a family of lipid-binding proteins. Prog. Biophys. Mol. Biol. 83, 47–68. [DOI] [PubMed] [Google Scholar]
- Shao B.; Oda M. N.; Bergt C.; Fu X.; Green P. S.; Brot N.; Oram J. F.; Heinecke J. W. (2006) Myeloperoxidase impairs ABCA1-dependent cholesterol efflux through methionine oxidation and site-specific tyrosine chlorination of apolipoprotein A-I. J. Biol. Chem. 281, 9001–9004. [DOI] [PubMed] [Google Scholar]
- Nagai R.; Matsumoto K.; Ling X.; Suzuki H.; Araki T.; Horiuchi S. (2000) Glycolaldehyde, a reactive intermediate for advanced glycation end products, plays an important role in the generation of an active ligand for the macrophage scavenger receptor. Diabetes 49, 1714–1723. [DOI] [PubMed] [Google Scholar]
- El Khoury J.; Hickman S. E.; Thomas C. A.; Cao L.; Silverstein S. C.; Loike J. D. (1996) Scavenger receptor-mediated adhesion of microglia to β-amyloid fibrils. Nature 382, 716–719. [DOI] [PubMed] [Google Scholar]
- Mucchiano G. I.; Jonasson L.; Haggqvist B.; Einarsson E.; Westermark P. (2001) Apolipoprotein A-I-derived amyloid in atherosclerosis. Its association with plasma levels of apolipoprotein A-I and cholesterol. Am. J. Clin. Pathol. 115, 298–303. [DOI] [PubMed] [Google Scholar]
- Harr S. D.; Uint L.; Hollister R.; Hyman B. T.; Mendez A. J. (1996) Brain expression of apolipoproteins E, J, and A-I in Alzheimer′s disease. J. Neurochem. 66, 2429–2435. [DOI] [PubMed] [Google Scholar]
- Wisniewski T.; Golabek A. A.; Kida E.; Wisniewski K. E.; Frangione B. (1995) Conformational mimicry in Alzheimer's disease. Role of apolipoproteins in amyloidogenesis. Am. J. Pathol. 147, 238–244. [PMC free article] [PubMed] [Google Scholar]
- Schmechel D. E.; Saunders A. M.; Strittmatter W. J.; Crain B. J.; Hulette C. M.; Joo S. H.; Pericak-Vance M. A.; Goldgaber D.; Roses A. D. (1993) Increased amyloid β-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 90, 9649–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werb Z.; Chin J. R. (1983) Onset of apoprotein E secretion during differentiation of mouse bone marrow-derived mononuclear phagocytes. J. Cell Biol. 97, 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K.; Nishimura N.; Doi T.; Imanishi T.; Kodama T.; Suzuki K.; Tanaka T. (1997) The lysine cluster in the collagen-like domain of the scavenger receptor provides for its ligand binding and ligand specificity. FEBS Lett. 414, 182–186. [DOI] [PubMed] [Google Scholar]
- Narayanaswami V.; Ryan R. O. (2000) Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta 1483, 15–36. [DOI] [PubMed] [Google Scholar]
- Ajees A. A.; Anantharamaiah G. M.; Mishra V. K.; Hussain M. M.; Murthy H. M. (2006) Crystal structure of human apolipoprotein A-I: Insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. U.S.A. 103, 2126–2131. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Navab M.; Anantharamaiah G. M.; Reddy S. T.; Hama S.; Hough G.; Grijalva V. R.; Wagner A. C.; Frank J. S.; Datta G.; Garber D.; Fogelman A. M. (2004) Oral D-4F causes formation of pre-β high-density lipoprotein and improves high-density lipoprotein-mediated cholesterol efflux and reverse cholesterol transport from macrophages in apolipoprotein E-null mice. Circulation 109, 3215–3220. [DOI] [PubMed] [Google Scholar]
- Navab M.; Anantharamaiah G. M.; Hama S.; Hough G.; Reddy S. T.; Frank J. S.; Garber D. W.; Handattu S.; Fogelman A. M. (2005) D-4F and statins synergize to render HDL antiinflammatory in mice and monkeys and cause lesion regression in old apolipoprotein E-null mice. Arterioscler. Thromb. Vasc. Biol. 25, 1426–1432. [DOI] [PubMed] [Google Scholar]
- Navab M.; Anantharamaiah G. M.; Hama S.; Garber D. W.; Chaddha M.; Hough G.; Lallone R.; Fogelman A. M. (2002) Oral administration of an Apo A-I mimetic Peptide synthesized from d-amino acids dramatically reduces atherosclerosis in mice independent of plasma cholesterol. Circulation 105, 290–292. [DOI] [PubMed] [Google Scholar]
- Van Lenten B. J.; Wagner A. C.; Navab M.; Anantharamaiah G. M.; Hui E. K.; Nayak D. P.; Fogelman A. M. (2004) D-4F, an apolipoprotein A-I mimetic peptide, inhibits the inflammatory response induced by influenza A infection of human type II pneumocytes. Circulation 110, 3252–3258. [DOI] [PubMed] [Google Scholar]
- Navab M.; Anantharamaiah G. M.; Reddy S. T.; Hama S.; Hough G.; Grijalva V. R.; Yu N.; Ansell B. J.; Datta G.; Garber D. W.; Fogelman A. M. (2005) Apolipoprotein A-I mimetic peptides. Arterioscler. Thromb. Vasc. Biol. 25, 1325–1331. [DOI] [PubMed] [Google Scholar]
- Soulages J. L.; Arrese E. L. (2000) Dynamics and hydration of the α-helices of apolipophorin III. J. Biol. Chem. 275, 17501–17509. [DOI] [PubMed] [Google Scholar]
- Wang J.; Sykes B. D.; Ryan R. O. (2002) Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc. Natl. Acad. Sci. U.S.A. 99, 1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M.; Ekkel Y.; Rohrer L.; Penman M.; Freedman N. J.; Chisolm G. M.; Krieger M. (1991) Expression of type I and type II bovine scavenger receptors in Chinese hamster ovary cells: Lipid droplet accumulation and nonreciprocal cross competition by acetylated and oxidized low density lipoprotein. Proc. Natl. Acad. Sci. U.S.A. 88, 4931–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. D.; Reddy P. A.; Jong M. T.; Erickson B. W. (1987) C3bi receptor (complement receptor type 3) recognizes a region of complement protein C3 containing the sequence Arg-Gly-Asp. Proc. Natl. Acad. Sci. U.S.A. 84, 1965–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosswig N.; Rice S.; Daugherty A.; Post S. R. (2003) Class A scavenger receptor-mediated adhesion and internalization require distinct cytoplasmic domains. J. Biol. Chem. 278, 34219–34225. [DOI] [PubMed] [Google Scholar]
- Whitman S. C.; Daugherty A.; Post S. R. (2000) Regulation of acetylated low density lipoprotein uptake in macrophages by pertussis toxin-sensitive G proteins. J. Lipid Res. 41, 807–813. [PubMed] [Google Scholar]
- Post S. R.; Gass C.; Rice S.; Nikolic D.; Crump H.; Post G. R. (2002) Class A scavenger receptors mediate cell adhesion via activation of G(i/o) and formation of focal adhesion complexes. J. Lipid Res. 43, 1829–1836. [DOI] [PubMed] [Google Scholar]
- Gough P. J.; Greaves D. R.; Suzuki H.; Hakkinen T.; Hiltunen M. O.; Turunen M.; Herttuala S. Y.; Kodama T.; Gordon S. (1999) Analysis of macrophage scavenger receptor (SR-A) expression in human aortic atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 19, 461–471. [DOI] [PubMed] [Google Scholar]
- Alarcon R.; Fuenzalida C.; Santibanez M.; von Bernhardi R. (2005) Expression of scavenger receptors in glial cells. Comparing the adhesion of astrocytes and microglia from neonatal rats to surface-bound β-amyloid. J. Biol. Chem. 280, 30406–30415. [DOI] [PubMed] [Google Scholar]
- Park Y. M.; Febbraio M.; Silverstein R. L. (2009) CD36 modulates migration of mouse and human macrophages in response to oxidized LDL and may contribute to macrophage trapping in the arterial intima. J. Clin. Invest. 119, 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning-Tobin J. J.; Moore K. J.; Seimon T. A.; Bell S. A.; Sharuk M.; Alvarez-Leite J. I.; de Winther M. P.; Tabas I.; Freeman M. W. (2009) Loss of SR-A and CD36 activity reduces atherosclerotic lesion complexity without abrogating foam cell formation in hyperlipidemic mice. Arterioscler. Thromb. Vasc. Biol. 29, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. J.; Kunjathoor V. V.; Koehn S. L.; Manning J. J.; Tseng A. A.; Silver J. M.; McKee M.; Freeman M. W. (2005) Loss of receptor-mediated lipid uptake via scavenger receptor A or CD36 pathways does not ameliorate atherosclerosis in hyperlipidemic mice. J. Clin. Invest. 115, 2192–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic D. M.; Cholewa J.; Gass C.; Gong M. C.; Post S. R. (2007) Class A scavenger receptor-mediated cell adhesion requires the sequential activation of Lyn and PI3-kinase. Am. J. Physiol. 292, C1450–C1458. [DOI] [PubMed] [Google Scholar]
- Nikolic D. M.; Gong M. C.; Turk J.; Post S. R. (2007) Class A scavenger receptor-mediated macrophage adhesion requires coupling of calcium-independent phospholipase A2 and 12/15-lipoxygenase to Rac and Cdc42 activation. J. Biol. Chem. 282, 33405–33411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K. J.; El Khoury J.; Medeiros L. A.; Terada K.; Geula C.; Luster A. D.; Freeman M. W. (2002) A CD36-initiated signaling cascade mediates inflammatory effects of β-amyloid. J. Biol. Chem. 277, 47373–47379. [DOI] [PubMed] [Google Scholar]
- Medeiros L. A.; Khan T.; El Khoury J. B.; Pham C. L.; Hatters D. M.; Howlett G. J.; Lopez R.; O'Brien K. D.; Moore K. J. (2004) Fibrillar amyloid protein present in atheroma activates CD36 signal transduction. J. Biol. Chem. 279, 10643–10648. [DOI] [PubMed] [Google Scholar]
- Maxeiner H.; Husemann J.; Thomas C. A.; Loike J. D.; El Khoury J.; Silverstein S. C. (1998) Complementary roles for scavenger receptor A and CD36 of human monocyte-derived macrophages in adhesion to surfaces coated with oxidized low-density lipoproteins and in secretion of H2O2. J. Exp. Med. 188, 2257–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H.; Kurihara Y.; Takeya M.; Kamada N.; Kataoka M.; Jishage K.; Ueda O.; Sakaguchi H.; Higashi T.; Suzuki T.; Takashima Y.; Kawabe Y.; Cynshi O.; Wada Y.; Honda M.; Kurihara H.; Aburatani H.; Doi T.; Matsumoto A.; Azuma S.; Noda T.; Toyoda Y.; Itakura H.; Yazaki Y.; Kodama T.; et al. (1997) A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature 386, 292–296. [DOI] [PubMed] [Google Scholar]
- Coraci I. S.; Husemann J.; Berman J. W.; Hulette C.; Dufour J. H.; Campanella G. K.; Luster A. D.; Silverstein S. C.; El-Khoury J. B. (2002) CD36, a class B scavenger receptor, is expressed on microglia in Alzheimer's disease brains and can mediate production of reactive oxygen species in response to β-amyloid fibrils. Am. J. Pathol. 160, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucchiano G. I.; Haggqvist B.; Sletten K.; Westermark P. (2001) Apolipoprotein A-1-derived amyloid in atherosclerotic plaques of the human aorta. J. Pathol. 193, 270–275. [DOI] [PubMed] [Google Scholar]
- Vollmer E.; Brust J.; Roessner A.; Bosse A.; Burwikel F.; Kaesberg B.; Harrach B.; Robenek H.; Bocker W. (1991) Distribution patterns of apolipoproteins A1, A2, and B in the wall of atherosclerotic vessels. Virchows Arch. A: Pathol. Anat. Histopathol. 419, 79–88. [DOI] [PubMed] [Google Scholar]
- Carter R. S.; Siegel R. J.; Chai A. U.; Fishbein M. C. (1987) Immunohistochemical localization of apolipoproteins A-I and B in human carotid arteries. J. Pathol. 153, 31–36. [DOI] [PubMed] [Google Scholar]
- Kunjathoor V. V.; Chiu D. S.; O'Brien K. D.; LeBoeuf R. C. (2002) Accumulation of biglycan and perlecan, but not versican, in lesions of murine models of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 22, 462–468. [DOI] [PubMed] [Google Scholar]
- O'Brien K. D.; Olin K. L.; Alpers C. E.; Chiu W.; Ferguson M.; Hudkins K.; Wight T. N.; Chait A. (1998) Comparison of apolipoprotein and proteoglycan deposits in human coronary atherosclerotic plaques: Colocalization of biglycan with apolipoproteins. Circulation 98, 519–527. [DOI] [PubMed] [Google Scholar]
- Van Eck M.; Herijgers N.; Van Dijk K. W.; Havekes L. M.; Hofker M. H.; Groot P. H.; Van Berkel T. J. (2000) Effect of macrophage-derived mouse ApoE, human ApoE3-Leiden, and human ApoE2 (Arg158→Cys) on cholesterol levels and atherosclerosis in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 20, 119–127. [DOI] [PubMed] [Google Scholar]
- Basu S. K.; Brown M. S.; Ho Y. K.; Havel R. J.; Goldstein J. L. (1981) Mouse macrophages synthesize and secrete a protein resembling apolipoprotein E. Proc. Natl. Acad. Sci. U.S.A. 78, 7545–7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayden H. J.; Maschio F.; Traber M. G. (1985) The secretion of apolipoprotein E by human monocyte-derived macrophages. Arch. Biochem. Biophys. 239, 388–395. [DOI] [PubMed] [Google Scholar]
- Kockx M.; Jessup W.; Kritharides L. (2008) Regulation of endogenous apolipoprotein E secretion by macrophages. Arterioscler. Thromb. Vasc. Biol. 28, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Curtiss L. K.; Boisvert W. A. (2000) Apolipoprotein E and atherosclerosis. Curr. Opin. Lipidol. 11, 243–251. [DOI] [PubMed] [Google Scholar]
- Harris J. D.; Graham I. R.; Schepelmann S.; Stannard A. K.; Roberts M. L.; Hodges B. L.; Hill V.; Amalfitano A.; Hassall D. G.; Owen J. S.; Dickson G. (2002) Acute regression of advanced and retardation of early aortic atheroma in immunocompetent apolipoprotein-E (apoE) deficient mice by administration of a second generation [E1(-), E3(-), polymerase(-)] adenovirus vector expressing human apoE. Hum. Mol. Genet. 11, 43–58. [DOI] [PubMed] [Google Scholar]
- Shi W.; Wang X.; Wang N. J.; McBride W. H.; Lusis A. J. (2000) Effect of macrophage-derived apolipoprotein E on established atherosclerosis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 20, 2261–2266. [DOI] [PubMed] [Google Scholar]
- Boisvert W. A.; Curtiss L. K. (1999) Elimination of macrophage-specific apolipoprotein E reduces diet-induced atherosclerosis in C57BL/6J male mice. J. Lipid Res. 40, 806–813. [PubMed] [Google Scholar]
- Navab M.; Anantharamaiah G. M.; Reddy S. T.; Fogelman A. M. (2006) Apolipoprotein A-I mimetic peptides and their role in atherosclerosis prevention. Nat. Clin. Pract. Cardiovasc. Med. 3, 540–547. [DOI] [PubMed] [Google Scholar]
- Bloedon L.; Dunbar R.; Duffy D.; Pinell-Salles P.; Norris R.; Degroot B. J.; Movva R.; Navab M.; Fogelman A. M.; Rader D. J. (2008) Safety, pharmacokinetics and pharmacodynamics of a single dose of oral apolipoprotein A-I mimetic peptide D-4F in high-risk cardiovascular patients. J. Lipid Res. 49, 1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.