Abstract
The ATP-dependent Lon protease of Saccharomyces cerevisiae mitochondria is required for selective proteolysis in the matrix, maintenance of mitochondrial DNA, and respiration-dependent growth. Lon may also possess a chaperone-like function that facilitates protein degradation and protein-complex assembly. To understand the influence of Lon’s ATPase and protease activities on these functions, we examined several Lon mutants for their ability to complement defects of Lon-deleted yeast cells. We also developed a rapid procedure for purifying yeast Lon to homogeneity to study the enzyme’s activities and oligomeric state. A point mutation in either the ATPase or the protease site strongly inhibited the corresponding activity of the pure protein but did not alter the protein’s oligomerization; when expressed at normal low levels neither of these mutant enzymes supported respiration-dependent growth of Lon-deleted cells. When the ATPase- or the protease-containing regions of Lon were expressed as separate truncated proteins, neither could support respiration-dependent growth of Lon-deleted cells; however, coexpression of these two separated regions sustained wild-type growth. These results suggest that yeast Lon contains two catalytic domains that can interact with one another even as separate proteins, and that both are essential for the different functions of Lon.
The ATP-dependent protease Lon (La) is the most highly conserved member of energy-dependent proteases present in archaea, bacteria, as well as mitochondria (1–3). Lon-deficient Escherichia coli mutants are viable but exhibit defects in the degradation of abnormal and regulatory proteins (4–6). LON-related genes have been identified in humans (7), yeast (8, 9), and plants. Deletion of LON in yeast blocks ATP-dependent degradation of mitochondrial matrix proteins, prevents growth on a nonfermentable carbon source, and causes deletions in the mitochondrial genome as well as accumulation of electron-dense inclusion bodies within the matrix (8–10). In addition, we have proposed a chaperone-like function of Lon that serves to facilitate protein degradation and possibly the assembly of protein complexes (2, 11, 12). Therefore, Lon appears to have diverse functions in vivo.
To better understand the role of Lon in mitochondrial homeostasis, we compared the effect of mutations within the ATPase and protease sites on the activity and the oligomeric state of the pure Lon protein and on the function of Lon in vivo. Although mutation of the ATPase or protease site strongly inhibited the corresponding enzymatic activity of the purified protein, the oligomerization of Lon was not altered significantly. Thus, the different in vivo function of the Lon mutants is most likely a consequence of altered catalytic activities. In addition, we found that proteolysis and respiration-dependent growth of yeast cells could be maintained by coexpressing the ATPase and protease regions of Lon as separate proteins; neither region alone could support these functions. Our results suggest that yeast Lon consists of two catalytic domains that can interact functionally even as separate proteins and that both domains are required for the different roles of Lon within mitochondria.
EXPERIMENTAL PROCEDURES
Media, Yeast Strains, and Plasmids.
The yeast media yeast extract/peptone/dextrose, yeast extract/peptone/ethanol/glycerol, yeast extract/peptone/galactose, and synthetic medium supplemented with dextrose or galactose have been described (13). Where required, synthetic medium supplemented with dextrose or galactose were supplemented with Casamino acids (0.5%), histidine (0.002%), leucine (0.003%), tryptophan (0.002%), or uracil (0.002%). Synthethic medium supplemented with galactose was supplemented with 0.2% glucose to facilitate growth of respiratory-deficient strains.
The strains used in this study were the a, α, or a/α versions of JK93d (leu2-3,112, ura3-52, trp1, his4, rme1, HMLa) (14). The diploid strain JCJK1 (LON/Δlon) was constructed by replacing a 2,948-bp BamHI-SnaBI fragment of one LON gene with a bacterial kanamycin-resistance gene (15). The Δlon strain lacks sequences encoding the ATP-binding motifs, the charged region, and the proteolytic active site; therefore, the plasmid-borne genes encoding LonK638N, LonS1015A, or LonΔcharge cannot undergo recombination to restore a wild-type LON gene.
The centromere-based plasmid pSEYc68 (14) was used for the GAL1-directed expression of wild-type and mutant LON genes (11), whereas the 2 μ-based plasmid YEplac181 was used for the overexpression of the mutant LON gene encoding the truncated ATPase domain of Lon under the control of the authentic LON promoter.
To test whether the respiration-dependent growth defect of Δlon cells could be complemented by wild-type Lon or any of the Lon variants, the diploid JCJK1 first was transformed with the respective plasmids and sporulated. Tetrads were dissected on yeast extract/peptone/dextrose and yeast extract/peptone/galactose, and the spores were grown for approximately 4–6 days at 30°C and replica-plated onto yeast extract/peptone/ethanol/glycerol. The Δlon haploid cells expressing wild-type or mutant Lon were grown in galactose to an OD600 of 0.5, and cyanide-sensitive oxygen consumption was tested directly in a Clark-type oxygen electrode.
Molecular Genetic Techniques.
Procedures for DNA purification, restriction enzyme digestion, ligation, agarose gel electrophoresis, and Southern hybridization were performed as described (16). Standard protocols for yeast transformations, mating, sporulation, and tetrad dissection were used. PCRs were carried out with Vent DNA polymerase (New England Biolabs) or Taq DNA polymerase (Boehringer Mannheim).
Site-directed mutagenesis by PCR was carried out as described (17). The construction of mutant LON genes encoding LonK638N and LonS1015A has been described (11). The LonΔcharge variant lacks 54 aa from lysine 839 to serine 894. The primer used to produce this variant was 5′-CATCGAGAAAATATACCGTAAAGCAGCCCTACAAGTGGTA/ACCTCGAAAAGATCAATGTTTCCATATCCCAG-3′; the site of deletion is indicated by the slash; threonine-893 was replaced with isoleucine because of an error in the PCR. The primer used to produce the Lon ATPase domain was 5′-GTTTCGTATACGTACTATGTAGTGTAGACTGGAGGCCCAACGT-3′; it replaces codon 918 by a stop codon. Complementary primers used for producing the protease and proteaseΔcharge domains were 5′-GATCCAAGGTACATGGAACACGGA-3′ and 5′-CTAGTCCGTGTTCCATGTACCTTG-3′. Mutant LON genes were created by replacing restriction fragments of the wild-type LON gene cloned in pSEYc68 by the corresponding fragments with site-specific mutations or by annealed primers (11).
Purification of Hexahistidine-Tagged Lon Proteins and NH2-Terminal Sequencing.
Haploid Δlon cells that had been transformed with plasmid pSEYc68 carrying the genes encoding wild-type Lon, Lon K638N, LonS1015A, or LonΔcharge were grown to midlogarithmic phase in synthethic medium supplemented with galactose supplemented with Casamino acids, glucose, adenine, and tryptophan. Mitochondria were isolated (18) and solubilized at 10 mg/ml in solubilization buffer [20 mM Hepes, pH 8.0/150 mM NaCl/20% glycerol (wt/vol)] containing 1.6 mg of Lubrol per mg of mitochondrial protein. After a 15-min incubation on ice, the lysate was centrifuged at 200,000 × g for 5 min at 4°C, and the supernatant was applied to a 4- to 7-ml Ni2+-agarose column. The column was washed with 5 vol of solubilization buffer containing 40 mM imidazole. Bound proteins were eluted with solubilization buffer containing 0.2 M imidazole and concentrated on a Centricon 100 filter, and their concentration was determined by the bicinchoninic acid assay (Pierce). The concentrated Lon protein (4–8 μg) was subjected to NH2-terminal sequencing as described (19).
Gel Filtration.
Hexahistidine-tagged Lon proteins (60–90 μg) were isolated as described above and loaded in a volume of 200 μl onto a Superose 6 column (Pharmacia) equilibrated with solubilization buffer at 4°C. The absorbance at 280 nm was monitored, and fractions of 0.5 ml were collected (unless otherwise indicated) and analyzed by SDS/PAGE. The gels were analyzed either by Coomassie blue staining or immunoblotting as described below. Standards used for size calibration were Blue dextran, 2,000 kDa (eluted at a volume of 6.75 ml); thyroglobulin, 669 kDa (12.2 ml); apoferritin, 443 kDa (14 ml); alcohol dehydrogenase, 150 kDa (15.6 ml); and BSA, 66 kDa (16.3 ml).
In Vitro ATPase and Protease Assays.
The ATPase activity of Lon was assayed as described (20). A final volume of 390 μl containing 50 mM Hepes at pH 8.0, 10 mM MgCl2, 1 mM [γ-32P]ATP (0.04 Ci/mmol), and 13 μg of Lon was incubated at 37°C. Aliquots of 30 μl were withdrawn in duplicate at 2-min intervals over a 10- to 15-min period, quenched with 210 μl of 1 M perchloric acid/1 mM NaPi, and mixed with 480 μl of 20 mM ammonium molybdate. The resulting phosphomolybdate complexes were extracted into 480 μl of isopropyl acetate, of which 120 μl was then counted in a liquid scintillation counter. All ATPase values were calculated from initial rates during which the hydrolysis of ATP was linear with time. Background counts from reactions lacking Lon proteins were less than 1% of the total and represented 5% of the maximal measured values.
The in vitro protease activity of Lon was measured by the degradation of 14C-labeled casein to acid-soluble fragments. The reaction mixture (800 μl) contained 20 mM Tris at pH 8.0, 10 mM MgCl2, 5 mM ATP, 200 μg of casein (245 × 106 cpm/mmol), and 2 μg of Lon. Aliquots of 200 μl were quenched at 10-min intervals over a 30-min period with 40 μl of 10% BSA and 220 μl of 10% trichloroacetic acid. The mixture was held on ice for 10 min and spun in a microcentrifuge for 5 min, and 300 μl of the supernatant was counted in a liquid scintillation counter. Background counts from reactions lacking Lon protein were less than 1% of the total and represented 25% of the highest values measured.
Chemical Crosslinking.
Pure Lon, 30 μg or 10 μg, isolated on Ni2+-agarose, was incubated on ice with 0.1% glutaraldehyde (Sigma) for 0.5 or 30 min, respectively; 5 μg of Lon protein was used in the untreated control (0 min). Crosslinking was terminated by the addition of reducing sample buffer containing 1 M urea. The samples were run on SDS/3.3% PAGE tube gels as described (21).
Immunoblot Analysis.
Proteins were extracted from 2 OD600 units of yeast cells (22) and immunoblotted with monospecific antibodies recognizing the mitochondrial ribosomal protein Mrp20p (a kind gift from T. Mason, Univ. of Massachusetts, Amherst); blots were developed with 125I-protein A and autoradiography.
RESULTS
The Lon Mutants.
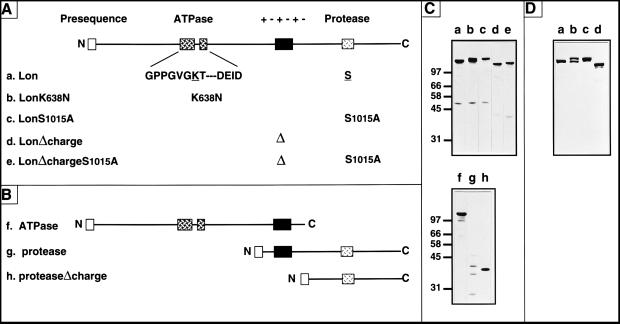
Two distinguishing hallmarks of all known Lon proteases are a highly conserved ATP-binding site composed of box A and box B motifs and a conserved region surrounding a serine residue essential for proteolytic activity; Lon from Saccharomyces cerevisiae is unique in that these two catalytic sites are separated by a highly charged stretch of approximately 50 aa (Fig. 1). The following Lon mutants were constructed (Fig. 1 A and B): the conserved lysine 638 within the box A motif for ATP binding was replaced by an asparagine (LonK638N); the conserved serine-1015 in the proteolytic site was replaced by an alanine (LonS1015A); the highly charged region between the ATPase and protease site was deleted (LonΔcharge and LonΔcharge S1015A); the proteolytic region was deleted (ATPase); and the ATPase region was deleted such that the mitochondrial-targeting sequence was joined either directly before the charged region (protease) or after it (proteaseΔcharge).
Figure 1.
(A and B) The Lon mutants used in this study. (A) Full-length Lon (a) was mutated in the following ways: LonK638N (b), the ATPase site (Boxes A and B are shown) was mutated by replacing the highly conserved lysine-638 with asparagine; LonS1015A (c), the protease site was inactivated by replacing serine-1015 with alanine; LonΔcharge (d), the charged region of 54 aa (+−+−+−) was deleted; LonΔchargeS1015A (e), deletion of the charged region was combined with replacement of serine-1015 with alanine. (B) Truncated variants of Lon: ATPase (f), a termination codon was introduced after threonine-917; protease (g), the first 75 aa, which include the targeting presequence, were fused in front of glycine-793; proteaseΔcharge (h), the first 75 aa of Lon were fused to glycine-793, thereby deleting the charged region. All truncated constructs carry the mitochondrial-targeting presequence of the authentic Lon precursor. (C) Total cellular proteins extracted from Δlon yeast cells overproducing wild-type Lon (a) or the Lon mutant proteins (b–h) were immunoblotted with antisera recognizing yeast Lon. (D) Coomassie blue-stained gel of purified wild-type Lon, LonK638N, LonS1015A, and LonΔcharge (a–d).
As we were interested in correlating the in vivo and in vitro functions of Lon, we needed to purify the wild-type and mutant Lon proteins. To do so, we overexpressed wild-type Lon and its variants in yeast cells lacking their chromosomal copy of LON (Δlon cells). When wild-type Lon, LonK638N, LonS1015A, and LonΔcharge were overexpressed from plasmid-borne genes in Δlon cells, they were all produced at similar high levels and were stable as determined by immunoblotting of total protein extracts; the same was true for the truncated ATPase and proteaseΔcharge variants, whereas the larger protease variant was less stable (Fig. 1C).
The different mobilities of wild-type and mutant Lon proteins were seen both by immunoblotting of total cell extracts (Fig. 1C) and by Coomassie blue staining of the pure proteins (Fig. 1D). Purified wild-type Lon had a higher mobility as compared with LonS1015A, whereas LonK638N migrated as two species, one comigrating with wild-type Lon, and the other with the more slowly migrating band of LonS1015A. The NH2 terminus of LonS1015A and wild-type Lon corresponded to alanine-38 and lysine-99, respectively, of the predicted Lon precursor. The precursor thus is cleaved twice: first, before alanine-38 by the matrix processing peptidase removing the targeting presequence, and then before lysine-99 through autocatalysis, converting the transient proform to the mature enzyme. These findings agree with those reported recently by Wagner et al. (23). Inactivation of the protease site by replacing serine-1015 with alanine prevented the second maturation step (Fig. 1C, lane c and e). LonK638N, which was partially active as a protease in vivo (see Fig. 2A), was present as both the proform and the mature form (Figs. 1D, lane b and 3D).
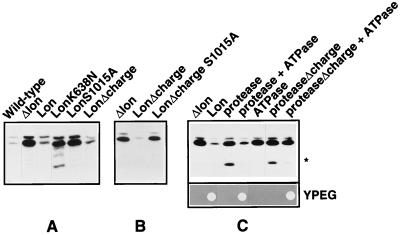
Figure 2.
The ATPase and protease regions of Lon are required for proteolysis and respiration-dependent growth. Proteolytic activity was monitored by the steady-state levels of Mrp20p, a protein of mitochondrial ribosomes; total cell extracts were immunoblotted with a mAb recognizing Mrp20p. (A and B) Wild-type, cells carrying an intact chromosomal copy of the LON gene and the “empty” plasmid pSEYc68 but lacking mtDNA; Δlon, cells whose LON gene had been deleted and that carry the “empty” plasmid pSEYc68; Lon, LonK638N, LonS1015A, LonΔcharge, and LonΔcharge S1015A, Δlon cells carrying plasmid pSEYc68, which contains the corresponding LON gene under the control of the GAL1 promoter. Transformants were grown in selective liquid medium containing Casamino acids and galactose at 30°C for approximately three generations. ∗, A 15-kDa degradation product of Mrp20p. (C) Coexpression of the separated ATPase and protease domains of Lon restores proteolytic activity and complements respiration-dependent growth of Δlon cells. (Upper) The protease, proteaseΔcharge, or ATPase regions of Lon were expressed in Δlon cells from plasmid-borne genes controlled by the GAL1 promoter except for the ATPase region, which was expressed under the authentic LON promoter on a 2-μ plasmid. Transformants were grown as in A and B, except that the selective liquid medium contained a mixture of nonessential amino acids (minus leucine) for plasmid maintenance. (Lower) A diploid yeast strain lacking one chromosomal copy of LON was transformed with two plasmids: a centromere-based plasmid (pSEYc68) carrying the gene for one of the two protease domain constructs of Lon under the control of the GAL1 promoter, and a 2-μ plasmid carrying the gene for the ATPase domain under the control of the authentic LON promoter. Transformants were sporulated and tetrads were dissected on galactose-containing rich medium. The resulting haploid segregants producing the indicated proteins were tested for respiration-dependent growth on yeast extract/peptone/ethanol/glycerol (YPEG).
Coexpression of the ATPase and the Protease Domains Supports Proteolysis and Respiration-Dependent Growth of Δlon Cells.
We tested whether the Lon variants could support full mitochondrial function in Δlon cells (see Experimental Procedures). When overproduced, either wild-type Lon or LonK638N sustained respiration-dependent growth; overproduction of LonS1015A did not (Table 1). We have confirmed that neither the ATPase nor protease mutant of yeast Lon could maintain respiration-dependent growth when expressed at low levels from the endogenous LON promoter (23). Interestingly, overexpression of LonΔcharge did not support respiration-dependent growth of Δlon cells (Table 1), even though it restored protein degradation in vivo, and was proteolytically active in vitro (Fig. 2 A and B and Table 2). However, when expressed at low levels from the endogenous LON promoter, LonΔcharge could sustain respiration-dependent growth (see Discussion).
Table 1.
Complementation of the respiration defect of Δlon cells
| Overexpressed plasmid-borne gene(s) | Respiration |
|---|---|
| LON | + |
| LON K638N | + |
| ATPase + protease | + |
| ATPase + proteaseΔcharge | + |
| LON S1015A | − |
| LONΔcharge | − |
| ATPase | − |
| Protease | − |
| ProteaseΔcharge | − |
The diploid strain JCJK1 (LON/Δlon) was transformed with a plasmid carrying the indicated LON gene and sporulated, and the tetrads were dissected on YPGa1. The spores were grown for ≈4 days at 30°C, replica-plated onto YPEG, and grown for 4–6 days. Wild-type Lon or its variants were expressed under the control of the GAL1 promoter from a centromere-based plasmid, with the exception of the ATPase domain, which was expressed from either the GAL1 promoter on a centromere-based plasmid or from the LON promoter from a 2-μ plasmid. The Δlon hapaloid cells expressing wild-type or mutant Lon proteins also were tested directly for cyanide-sensitive oxygen consumption.
Table 2.
Protease and ATPase activities of pure Lon proteins
| Protein | Protease activity
|
ATPase activity
|
||
|---|---|---|---|---|
| pmol/pmol per min | % | pmol/pmol per min | % | |
| Lon | 0.290 ± 0.014 | (100) | 37.1 ± 7.3 | (100) |
| LonK638N | 0.009 ± 0.005 | 3 | 4.4 ± 1.3 | 12 |
| LonS1015A | 0.007 ± 0.002 | 2 | 15.1 ± 3.2 | 41 |
| LonΔcharge | 0.162 ± 0.065 | 56 | 17.7 ± 4.7 | 48 |
Protease activity: pmol of casein hydrolyzed per pmol of monomer per min at 37°C in the presence of 5 mM ATP/20 mM Tris, pH 8.0/10 mM MgCl2/50 μg of casein/0.5 μg of Lon in 200 μl per time point. In the absence of ATP, less than 3% of wild-type activity was observed for Lon and all of the Lon variants. ATPase activity: pmol of ATP hydrolyzed per pmol of Lon monomer per min at 37°C in the presence of 1 mM ATP/20 mM Tris, pH 8.0/10 mM MgCl2/0.5 or 1 μg of Lon in 30 μl per time point.
We also expressed the ATPase and the protease regions of Lon as separate polypeptide chains in Δlon cells to test whether these regions could function independently of one another. When overproduced, neither region alone complemented the Δlon growth defect (Table 1). However, complementation was observed when both regions were coexpressed (Table 1 and Fig. 2C). Coexpression did not affect the stability or the solubility of these truncated proteins (data not shown). This suggested that the two separated Lon regions can interact functionally and perhaps physically in the mitochondrial matrix, thereby restoring sufficient Lon activity for complementing the growth defect of Δlon cells. This result also suggests that these two regions represent separate domains of Lon.
The in vivo proteolytic activity of the overproduced Lon variants was monitored by assessing the levels of Mrp20p, a subunit of the mitochondrial ribosome (24). Approximately 70 nuclear-encoded mitochondrial ribosomal proteins are synthesized in the cytosol, imported into the mitochondrial matrix, and assembled with the mitochondrially encoded protein Var1p and the two mitochondrially encoded rRNAs. In wild-type cells lacking mtDNA, mitochondrial ribosome assembly is defective and most imported ribosomal proteins are degraded. The degradation is Lon-mediated because deletion of the LON gene stabilizes the unassembled ribosomal proteins (T. Mason and S. Leonhardt, personal communication).
Cells expressing wild-type Lon but lacking mtDNA had barely detectable steady-state levels of Mrp20p (Fig. 2A). Δlon cells accumulated Mrp20p but could degrade it when overexpressing wild-type Lon or LonΔcharge (Fig. 2A). Mutation of serine 1015 in full-length Lon or LonΔcharge blocked Mrp20p degradation (LonS1015A, LonΔchargeS1015A, Fig. 2 A and B). LonK638N was partially active and cleaved Mrp20p to a 15-kDa degradation intermediate (Fig. 2A).
Proteolytic activity similar to that of LonK638N was observed upon coexpressing either the isolated protease or proteaseΔcharge domain, whereas the isolated ATPase domain had no detectable proteolytic activity (Fig. 2C). However, full degradation of Mrp20p was restored when the protease domain was coexpressed with the isolated ATPase domain (Fig. 2C). The proteaseΔcharge domain failed to interact with the ATPase domain as tested by the degradation of Mrp20p, but still gave functional complementation as tested by respiration-dependent growth (Fig. 2C). This result suggests that growth is a more sensitive indicator of domain interaction in vivo than is restoration of proteolysis.
The ATPase and Protease Activities of Yeast Lon Are Tightly Coupled.
To determine the effects of mutations in Lon on the activity of the pure proteins, we developed a procedure to rapidly purify Lon to homogeneity. Wild-type Lon, LonK638N, LonS1015A, and LonΔcharge were each tagged with six histidines at their carboxyl terminus and overproduced in yeast from plasmid-borne genes; Lon was purified from mitochondrial extracts by binding to Ni2+-agarose beads (Fig. 1C and Experimental Procedures).
Proteolysis was measured by the conversion of 14C-labeled casein to acid-soluble peptides in the presence or absence of ATP (Table 2). Wild-type Lon degraded 0.290 pmol casein/pmol monomer per min in the presence of ATP and less than 3% of this amount in the absence of ATP. LonΔcharge had 56% of wild-type activity, whereas LonK638N and LonS1015A were essentially inactive. The ATPase activity of the purified proteins was measured by the release of 32Pi from [γ-32P]ATP. Wild-type Lon hydrolyzed 37.1 ± 7.3 pmol ATP/pmol monomer per min. LonK638N had only 12% of wild-type activity, confirming that mutation of the ATP-binding site strongly inhibited ATPase activity. LonΔcharge and LonS1015A had 48% and 41% of wild-type ATPase activity, respectively (Table 2). The ATPase activity of Lon and LonΔcharge was stimulated 2- to 3-fold by the protein substrate casein, whereas no such stimulation was seen with LonS1015A. The addition of BSA, which is not a substrate, had no effect on ATP hydrolysis of wild-type Lon.
Oligomerization of Pure Yeast Lon Does Not Depend on Its ATPase or Protease Activities.
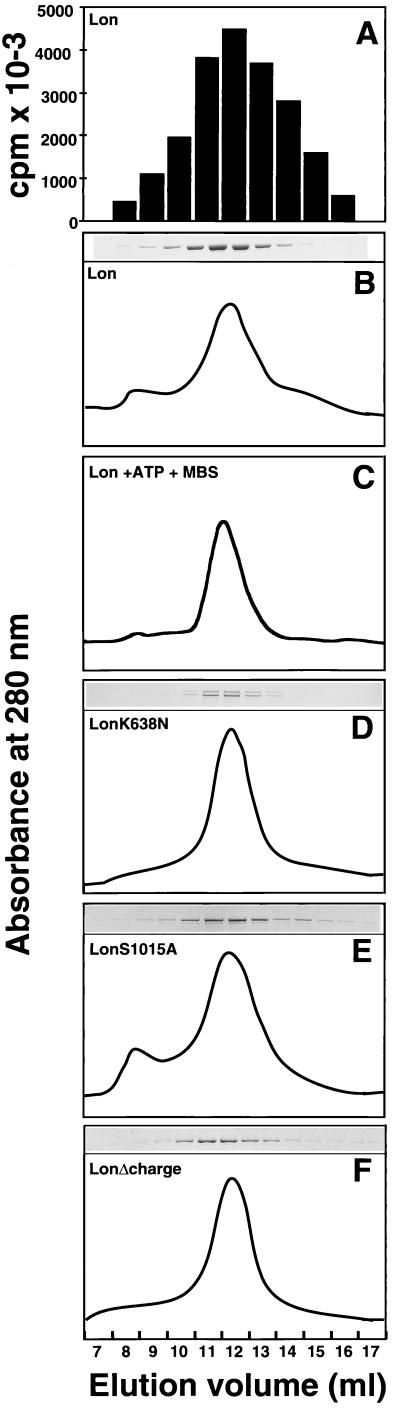
To assess whether the different in vivo and in vitro activities of the full-length Lon mutants reflected different oligomeric states, we analyzed the pure proteins by gel filtration. Previous work has shown that yeast Lon migrates as a hexamer (25). In agreement with this, we found that wild-type Lon eluted as a single 750- to 850-kDa species as measured by protein or ATP-dependent degradation (Fig. 3 A and B). The same result was found whether or not ATP was present during the purification (data not shown); furthermore, no difference in migration was observed when the enzyme was purified in the presence of ATP and then immediately crosslinked with m-maleimidobenzoyl-N-hydroxysuccinimide ester before gel filtration (Fig. 3C). The same apparent molecular mass was found for LonK638N, LonS1015A, and LonΔcharge (Fig. 3 D–F). With LonK638N, the mature and the proform of the subunits cofractionated (Fig. 3D), either because they formed mixed oligomers or could not be separated from one another in this system.
Figure 3.
Oligomeric state of wild-type Lon and its variants determined by gel filtration. Lon, LonK638N, LonS1015A, and LonΔcharge were isolated by binding to Ni2+-agarose, and 60–90 μg of pure protein was analyzed by gel filtration. (A) Purified wild-type Lon from the corresponding fractions shown in B was assayed for ATP-dependent protease activity. (B–F) Absorbance at 280 nm and Coomassie blue-stained protein of eluted fractions. (C) Wild-type Lon was purified as in A, except that 1 mM ATP was added during solubilization and purification on Ni2+-agarose. The isolated protein was crosslinked with 1 mM m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS) for 60 min on ice, and 60 μg of protein was analyzed by gel filtration.
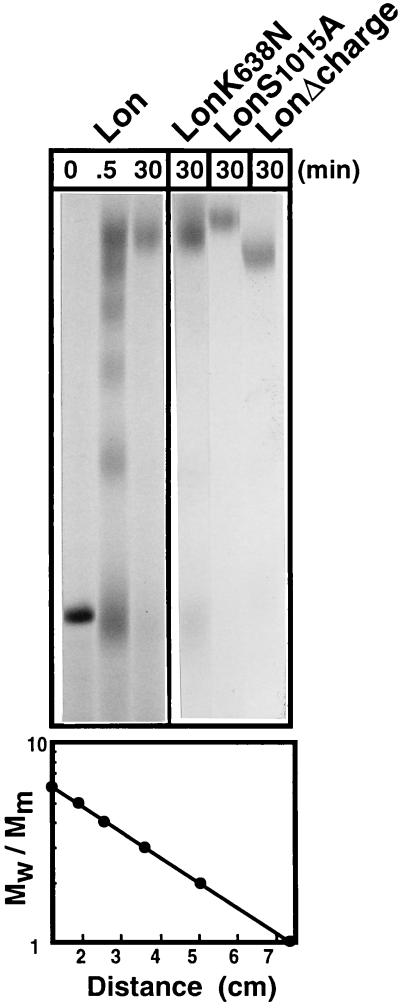
To determine the number of subunits in the Lon holoenzyme, we treated wild-type Lon with glutaraldehyde and analyzed the resulting crosslinked intermediates (Fig. 4) (21). Lon purified on Ni2+-agarose was treated with 0.1% glutaraldehyde on ice for 0.5 min and then analyzed by SDS/PAGE. We observed six distinct species that we interpreted as monomers, dimers, trimers, tetramers, pentamers, and hexamers because their electrophoretic mobility was linearly related to the logarithm of subunit number (Mw/Mm) and the distance each oligomer had migrated from the origin (Fig. 4 Lower). Incubation periods of 30 min or longer yielded primarily crosslinked hexamers although some crosslinked monomers were occasionally apparent. The same pattern of intermediates was observed after 0.5 min for the mutants LonK638N, LonS1015A, and LonΔcharge (data not shown), and only the species corresponding to hexamers was observed at 30 min (Fig. 4). Taken together we conclude that the mutations do not significantly affect the oligomeric state of the Lon complex.
Figure 4.
Intramolecular crosslinking of wild-type Lon and Lon variants. (Upper) Lon protein was isolated on Ni2+-agarose, treated with 0.1% glutaraldehyde for the indicated times, and analyzed on SDS/3.3% polyacrylamide tube gels stained with Coomassie blue. Five micrograms of Lon protein was used for the 0- and 30-min time points, whereas 35 μg of Lon was used for the 0.5-min time point. (Lower) Electrophoretic mobility of the products from the 0.5-min time point of crosslinking wild-type Lon (Upper) was plotted against the logarithm of Mw/Mm, where Mw is the relative molecular weight of the crosslinked species and Mm is the molecular mass of the mature Lon subunit (117 kDa).
DISCUSSION
Little is known about how the ATPase and protease activities of eukaryotic Lon influence one another in the holoenzyme and how they contribute to the different phenotypic effects of Lon in vivo. To begin to address this question we have tested several mutants of yeast Lon in which one of the two catalytic activities was inactivated by a point mutation or in which each activity was expressed as a separate polypeptide chain. Some of these engineered mutants have counterparts in nature. Lon-related proteins encoded by the sms genes (26) of Synechocystys sp., Listeria monocytogenes, or Bacillus subtilis resemble the LonS1015A mutant in that the serine at the putative proteolytic active site is replaced by alanine. Also, Lon-related proteins from Bacillus subtilis, Haemophilus influenzae, and Methanococcus jannaschii resemble the truncated protease domain mutants in that they lack sequences at the ATP-binding site (J.M.v.D. and C.K.S., unpublished observations). The physiological function of these Lon-like proteins has not been described, and whether they exhibit ATPase or protease activity remains to be tested. However, the existence of these natural Lon variants supports our present finding that the ATPase and protease domains may be able to act independently from one another in vivo.
Proteolysis and respiration-dependent growth require both the ATPase and protease activity of Lon. We have confirmed the report (23) that, when expressed at normal low levels, neither the ATPase nor the protease point mutant of Lon can maintain respiration-dependent growth in Δlon cells. However, when overexpressed, the ATPase mutant can substitute for the wild-type enzyme, presumably because it still has partial proteolytic activity in vivo (Table 1 and Fig. 2A). The opposite result was observed for LonΔcharge; it complemented the Δlon growth defect when expressed at lower levels (J.M.v.D. and C.K.S., unpublished observations) but failed to do so when overexpressed (Table 1). Overexpression of this mutant appears to be toxic: it blocked respiration-dependent growth even in wild-type cells, most probably because of an altered proteolytic activity or an altered substrate specificity (J.M.v.D. and C.K.S., unpublished observations).
The requirement of the ATPase and protease activities of Lon for full Lon function also can be demonstrated when each activity is provided by a separate polypeptide chain (Table 1 and Fig. 2C). Although each of the overexpressed protease domains resembled LonK638N in showing partial proteolytic activity in vivo, only LonK638N can support respiration-dependent growth. This result suggests that the region containing the ATP-binding site may be important for functions other than proteolysis. As we have excluded the possibility that the coexpressed ATPase domain merely stabilizes the protease domain, we favor the possibility that the two domains interact physically in vivo, although this has not yet been shown directly.
Proteolysis in vitro is coupled tightly with ATP hydrolysis. LonK638N has only low ATPase and protease activities. The same is true for bacterial Lon (27–30): an analogous mutation in the conserved lysine 362 of E. coli Lon blocks ATP hydrolysis by more than 90% and severely inhibits protease activity (27). By contrast, control of the ATPase activity by the protease activity of Lon is less stringent: purified wild-type Lon has significant ATPase activity in the absence of a protein substrate; the purified LonS1015A mutant still has 41% of wild-type ATPase activity (Table 2); and an analogous mutation in the proteolytic site of E. coli Lon did not affect the ATPase activity at all (31). However, casein degradation stimulates the ATPase activity of wild-type yeast Lon or of the LonΔcharge mutant 2- to 3-fold, whereas no such stimulation is observed with the proteolytically inactive LonS1015A mutant. Thus, the ATPase activity of Lon does not require proteolysis, but is stimulated by it.
The different activities of the Lon point mutants cannot be explained by differences in oligomerization. The effects of these mutations in vivo thus appear to reflect differences in Lon’s catalytic activities. We have shown directly that the 750- to 850-kDa complex isolated by gel filtration is enzymatically active and that the mass of this complex is unaffected by the presence or absence of either ATP or crosslinker. We do not exclude the possibility that this complex can associate with itself or other proteins under certain conditions (23), but the 750–850 kDa is the smallest unit capable of performing ATP-dependent proteolysis; therefore, we define it as the Lon holoenzyme.
How the binding and hydrolysis of ATP by yeast Lon contribute to ATP-dependent proteolysis still remains unclear. They may act to induce a conformational change in the holoenzyme activating the protease (32), maintain protein substrates in an unfolded state facilitating proteolysis (2, 6, 33), or transport the protein substrate from a recognition site to the proteolytic site (34). The availability of pure yeast Lon and of pure Lon mutants lacking either ATPase or protease activity, in addition to the functional interaction of the isolated ATPase and protease Lon domains, provides a basis for the rigorous molecular characterization of this enzyme and for understanding how it performs its diverse functions in vivo.
Acknowledgments
We thank S. A. Leonhardt and T. L. Mason for sharing unpublished data and for providing antibodies recognizing Mrp20p; P. Jenö and T. Mini for protein sequencing; A. Azem and other members of the Schatz laboratory for helpful discussions; and A. Engel for critical review of this manuscript. This study was supported by the Swiss National Science Foundation (to G.S. and E.K.); by the Human Capital and Mobility Programme of the European Union (to G.S.); by the European Molecular Biology Organization (to J.M.v.D.); by the Slovak Grant Agency (to E.K.); and by the Damon Runyon–Walter Winchell Cancer Research Foundation (to C.K.S.).
References
- 1.Desautels M, Goldberg A L. J Biol Chem. 1982;257:11673–11679. [PubMed] [Google Scholar]
- 2.Suzuki C K, Rep M, van Dijl J M, Suda K, Grivell L A, Schatz G. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 3.Swamy K H S, Goldberg A L. Nature (London) 1981;292:652–654. doi: 10.1038/292652a0. [DOI] [PubMed] [Google Scholar]
- 4.Goldberg A L, Moerschell R P, Chung C H, Maurizi M R. Methods Enzymol. 1994;244:350–375. doi: 10.1016/0076-6879(94)44027-1. [DOI] [PubMed] [Google Scholar]
- 5.Gottesman S, Maurizi M R. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurizi M R. Experentia. 1992;48:178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- 7.Wang N, Gottesman S, Willingham M C, Gottesman M M, Maurizi M R. Proc Natl Acad Sci USA. 1993;90:11247–11251. doi: 10.1073/pnas.90.23.11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki C K, Suda K, Wang N, Schatz G. Science. 1994;264:273–276. doi: 10.1126/science.8146662. , 891. [DOI] [PubMed] [Google Scholar]
- 9.Van Dyck L, Pearce D A, Sherman F. J Biol Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- 10.Wagner I, Arlt H, van Dyck L, Langer T, Neupert W. EMBO J. 1994;13:5135–5145. doi: 10.1002/j.1460-2075.1994.tb06843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rep M, van Dijl J M, Suda K, Schatz G, Grivell L A, Suzuki C K. Science. 1996;274:103–106. doi: 10.1126/science.274.5284.103. [DOI] [PubMed] [Google Scholar]
- 12.Rep M, van Dijl J M, Suda K, Schatz G, Grivell L A, Suzuki C K. Science. 1996;275:741. doi: 10.1126/science.274.5284.103. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G R. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic; 1991. [Google Scholar]
- 14.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 15.Wach A, Brachat A, Pöhlmann R, Philippsen P. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.van Dijl J M, de Jong A, Venema G, Bron S. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- 18.Glick B S, Pon L A. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 19.Jenö P, Mini T, Moes S, Hintermann E, Horst M. Anal Biochem. 1995;224:75–82. doi: 10.1006/abio.1995.1010. [DOI] [PubMed] [Google Scholar]
- 20.Viitanen P V, Lubben T H, Reed J, Goloubinoff P, O’Keefe D P, Lorimer G H. Biochemistry. 1990;29:5665–5671. doi: 10.1021/bi00476a003. [DOI] [PubMed] [Google Scholar]
- 21.Azem A, Weiss C, Goloubinoff P. Methods Enzymol. 1998;290:253–268. doi: 10.1016/s0076-6879(98)90024-6. [DOI] [PubMed] [Google Scholar]
- 22.Yaffe M P, Schatz G. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner I, van Dyck L, Savel’ev A S, Neupert W, Langer T. EMBO J. 1997;16:7317–7326. doi: 10.1093/emboj/16.24.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fearon K, Mason T L. J Biol Chem. 1992;267:5162–5170. [PubMed] [Google Scholar]
- 25.Kutejová E, Durcová G, Surovková E, Kužela Š. FEBS Lett. 1993;329:47–50. doi: 10.1016/0014-5793(93)80190-6. [DOI] [PubMed] [Google Scholar]
- 26.Neuwald A F, Berg D E, Stauffer G V. Gene. 1992;120:1–9. doi: 10.1016/0378-1119(92)90002-7. [DOI] [PubMed] [Google Scholar]
- 27.Fisher H, Glockshuber R. FEBS Lett. 1994;356:101–103. doi: 10.1016/0014-5793(94)01244-x. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg A L. Eur J Biochem. 1992;203:9–23. doi: 10.1111/j.1432-1033.1992.tb19822.x. [DOI] [PubMed] [Google Scholar]
- 29.Menon A S, Waxman L, Goldberg A L. J Biol Chem. 1987;262:722–726. [PubMed] [Google Scholar]
- 30.Waxman L, Goldberg A L. Proc Natl Acad Sci USA. 1982;79:4883–4887. doi: 10.1073/pnas.79.16.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher H, Glockshuber R. J Biol Chem. 1993;268:22502–22507. [PubMed] [Google Scholar]
- 32.Waxman L, Goldberg A L. Science. 1986;232:500–503. doi: 10.1126/science.2938257. [DOI] [PubMed] [Google Scholar]
- 33.Gottesman S, Maurizi M R, Wickner S. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 34.Rothman J E, Kornberg R D. Nature (London) 1986;322:209–210. doi: 10.1038/322209a0. [DOI] [PubMed] [Google Scholar]