Abstract
The rat 3Y1 derivative cell lines, EId10 and EId23, established by introducing the adenovirus E1A12S, Id-1H, and Id-2H cDNAs linked to the hormone-inducible promoter, express these proteins upon treatment with dexamethasone and elicit apoptosis, although these cell lines express mutated p53. The E1A mutants containing a deletion in either the N terminus or the conserved region 1 were unable to induce apoptosis in cooperation with Ids. Western blot analysis of the immunoprecipitates prepared from the dexamethasone-treated EId10 cell extract showed that Id-2H preferentially binds to E1A and E2A (E12/E47) helix–loop–helix transcription factors in vivo, but scarcely to the retinoblastoma protein. After induction of E1A and Ids, EId10 and EId23 cells began to accumulate in S phase and undergo apoptosis before entering G2 phase, suggesting that abnormal synthesis of DNA induced by coexpression of E1A, Id-1H, and Id-2H results in the induction of apoptosis. Apoptosis also is induced in mouse A40 (p53−/−) cells by E1A alone or E1A plus Ids after transient transfection of the expression vectors. The induction of apoptosis is stimulated by coexpression with wild-type p53; however, apoptosis induced by E1A alone was suppressed completely by coexpression with mutated p53, whereas apoptosis induced by E1A plus Ids was stimulated by the mutated p53 as done by wild-type p53. These results suggest that the suppressive function of mutated p53 is overcome by Ids.
The adenovirus E1A gene products (E1A) have functions to induce cell proliferation and death and to inhibit cell differentiation in a variety of cell types. The E1A gene of human adenovirus types 2 and 5 generates two major species of mRNA with the sizes of 13S and 12S that encode proteins of 289 aa (E1A13S) and 243 aa (E1A12S), respectively. E1A13S contains conserved regions CR1, CR2, and CR3, but E1A12S lacks CR3 (1). The N terminus of E1A 5′ to CR1 interacts with basic helix–loop–helix (bHLH) transcription factors and inhibits terminal differentiation of muscle cells (2). As reported here, Id (inhibitor of differentiation or inhibitor of DNA binding) proteins (3), a family of HLH proteins lacking the basic domain, also binds to the N terminus of E1A. The E1A domain required for induction of p53-dependent apoptosis recently has been mapped in the N terminus and the CR1 regions (4, 5), which overlap with the p300 binding domain (1).
bHLH proteins, such as MyoD and E2A (E12/E47), bind to DNA efficiently when these proteins heterodimerize (3). Id proteins also heterodimerize with bHLH proteins through the HLH domain, suppressing their DNA binding activity (3). Expression levels of Id proteins decrease in a variety of undifferentiated cells, upon induction of differentiation (3). The cDNA clones of four members of the mouse and human Id families have been isolated. Id-1H and Id-2H isolated by us from a human cDNA library (6) correspond to mouse Id-1 and Id-2, respectively. Id-1H and Id-2H are induced in quiescent human diploid fibroblasts by serum factors biphasically in early and late G1 phase, and their expression is essential for the G1 to S phase transition (6).
In the present work, to study the effects of coexpression of Id-1H and Id-2H with E1A on cell proliferation, rat 3Y1 derivative cell lines EId10 and EId23 carrying p53 mutation were established by introducing the E1A12S, Id-1H, and Id-2H cDNAs fused to the hormone inducible promoter. Unexpectedly, after expression of E1A and Ids in response to dexamethasone (dex), these cell lines undergo apoptosis after accumulation in S phase. Cotransfection of E1A and/or Id expression vectors into mouse A40 cells (p53−/−) also induced apoptosis, which was enhanced by simultaneous expression of wild-type (wt) p53. However, simultaneous expression of mutated p53 completely suppressed apoptosis induced by E1A alone, but enhanced apoptosis induced by E1A and Ids as done by wt p53. These results suggest that suppression of E1A-induced apoptosis by mutated p53 is overcome by Ids.
MATERIALS AND METHODS
Cell Lines.
The 3Y1–B cell line, clone 1–6, is a clonal cell line of Fischer rat embryo fibroblasts (7) and has been shown to carry p53 mutations (8). A 3Y1 derivative cell line, g12, was established by introducing 3Y1 cells with pMA12SG and expresses E1A12S in response to dex. EId10 and EId23 cell lines were established by introducing g12 cells with pMTV-Id-1H and pMTV-Id-2H and express E1A12S, Id-1H, and Id-2H in response to dex. YId6 and YId24 cell lines were established by introducing 3Y1 cells with pMTV-Id-1H and pMTV-Id-2H. ΔEId23 and ΔEId2 cell lines were established by introducing 3Y1 cells with pMTV-Id-1H, pMTV-Id-2H, and either pMTV-E1A12SΔ17–23 or pMTV-E1A12SΔ54–69, respectively (Fig. 1A). The A40 cell line was established from the cerebellum of a p53 null mouse. These cells were cultivated at 37°C in DMEM with 10% fetal calf serum (FCS). For cultivation of EId10 and EId23 cells, dialyzed FCS was used.
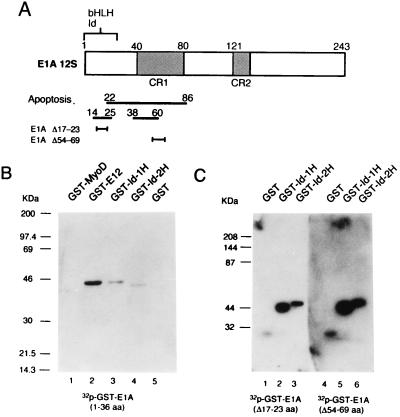
Figure 1.
The N terminus of E1A binds to Id-1H and Id-2H. (A) Functional domains and deletion mutants of E1A12S. The N-terminal domain required for binding to some bHLH transcription factors and Id proteins is shown by a caret. The domain required for induction of wt p53-dependent apoptosis and E1A12S mutants having a deletion in this domain are shown below. (B) GST fusion proteins were made by inserting the cDNAs into the multicloning site of pGEX-2TK in which the nucleotide sequence corresponding to amino acid sequence RRASV for the catalytic subunit of A kinase was placed between GST and the cloning site. The binding of these proteins to E1A was analyzed by Western blotting with 32P-labeled GST-N-terminal-E1A fragment (1–36 amino acids) after denaturation and renaturation of the proteins in buffer containing 6 and 3 M guanidinium hydrochloride, respectively. (C) GST-Id-1H and GST-Id-2H were similarly probed with 32P-labeled GST-E1AΔ17–23aa and GST-E1AΔ54–69aa.
Construction of E1A12S, Id-1H, and Id-2H Expression Vectors.
An expression vector, pMA12SG, was constructed by placing the adenovirus type 2 E1A12S cDNA downstream of the mouse mammary tumor virus (MMTV)-long terminal repeat (LTR). The E1A12SΔ17–23 cDNA lacking codons 17–23 was constructed by shortening the cDNA in pBRE1A12S after cleavage with PvuII between codons 22 and 23. The E1A12SΔ54–69 cDNA was constructed by using site-specific mutagenesis (9) with M13E1A12S. These cDNAs were inserted between HindIII and BglII sites of pMTV-dhfr (10), displacing the dhfr cDNA to generate pMTV-E1A12SΔ17–23 and pMTV-E1A12SΔ54–69 (Fig. 1A).
For construction of MMTV-LTR-linked Id-1H and Id-2H cDNAs, the DNA fragments containing Id-1H and Id-2H cDNAs were isolated from pBKS-Id-1H and pBKS-Id-2H (6) and inserted between the HindIII and BglII sites of pMTV-dhfr to generate pMTV-Id-1H and pMTV-Id-2H. The expression vectors pHβAPr-E1A12S, pCMV-Id-1H, and pCMV-Id-2H were constructed by inserting the E1A 12S cDNA between the HindIII and BamHI sites of pHβAPr-1 (11) and the Id-1H and Id-2H cDNAs between the HindIII and XbaI sites of pCMV1.
Reverse Transcriptase (RT)-Linked PCR.
Total cellular RNA (4 μg) was annealed with the downstream antisense primer (1 μM) of either E1A (nucleotide positions 576–595), Id-1H (nucleotide positions 124–141), or Id-2H (nucleotide positions 356–374) at 65°C for 5 min, and cDNA was synthesized with Moloney murine leukemia virus RT. The DNA/RNA hybrid was denatured, and the upstream sense primer (1 μM) containing the MMTV-LTR sequence (5′-AGTGACCCTTGTCTT TATTTC-3′) and each of the above antisense primers (0.2 μM) then were added. PCR was performed for 30 cycles at 94°C for 1.5 min, 55°C for 1.5 min, and 72°C for 3 min.
Flow Cytometric Analysis.
Cells were treated with dex, and both floating and attached cells were harvested at the times indicated. The cells were stained with propidium iodide (PI) containing 0.1% Triton X-100 and treated with 1,300 Kunitz unit⋅ml−1 RNase A. When the cells synthesizing DNA were analyzed, the cells were labeled with 100 μM 5-bromo-2′-deoxyuridine (BrdU) for 2 h before the harvest. The cells were fixed in 4% formaldehyde and stained with PI. The cells were incubated with 10 μg⋅ml−1 mouse anti-BrdU antibody (2B1, MBL) and then with 2 μg⋅ml−1 fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Cappel). The cells were analyzed for fluorescence intensity of FITC and PI.
When apoptosis was analyzed in transiently transfected cells, A40 cells (p53−/−) in 9-cm dishes were transfected with combinations of the expression vectors (each 0.8 μg) pHβAPr-E1A12S, pCMV-Id-1H, pCMV-Id-2H, pMoLVp53His175, and pMoLVp53Cys205 (12) together with 0.8 μg of pCMVCD20 (13) by lipofection. The cells were stained with FITC-labeled anti-CD20 antibody (B-B6, BioSource International, Camarillo, CA) and fixed with 4% formaldehyde. A set of cells was transfected with control empty vector pCMV-1 alone and similarly stained with anti-CD20 antibody to determine the background. The cell population that showed the fluorescence intensity over this background was gated and analyzed for cell cycle distribution.
Immunoprecipitation and Western Blotting.
The EId10 cell extract (200 μg of protein) was mixed with 1 μg of either anti-Id2 rabbit polyclonal antibody (C-20, Santa Cruz Biotechnology), anti-E1A rabbit polyclonal antibody (13S-5, Santa Cruz Biotechnology), anti-E2A (E12/E47) mouse mAb (Yae, Santa Cruz Biotechnology) or anti-pRB rabbit polyclonal antibody, and 20 μl of protein A-Sepharose beads [50% (vol/vol)] was added. The mixture was incubated at 4°C for 1 h, and the immune complex was washed with immunoprecipitation washing buffer (20 mM Hepes-KOH, pH 7.9, 100 mM KCl, 6 mM MgCl2, 1 mM EDTA, 1 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, and 10 μg⋅ml−1 each of aprotinin, antipain, and leupeptin) and treated with 20 μl of 0.8% deoxycholate (14). After addition of 5 μl of 6% Nonidet P-40, the mixture was centrifuged, and the supernatant was electrophoresed on 15% polyacrylamide gels. The proteins transferred to nitrocellulose membranes were reacted with anti-Id2 antibody at a dilution of 1:2,000 then with horseradish peroxidase-linked anti-rabbit IgG antibody, and detected by ECL (Amersham).
RESULTS
The N Terminus of E1A Binds Directly to Id-1H and Id-2H.
To see whether Id proteins interact with the N terminus of E1A as do bHLH proteins (2), glutathione S-transferase (GST)-Id-1H and GST-Id-2H were electrophoresed along with GST-MyoD, GST-E12, and GST as controls. After transfer to the filters, the proteins were denatured and renatured and reacted with 32P-labeled GST-N-terminal E1A fragment (Fig. 1B). The E1A fragment bound to Id-1H, Id-2H, and the ubiquitous bHLH protein E12, but not to the tissue-specific bHLH protein MyoD. Both N-terminal and CR1 deletion mutants, E1AΔ17–23 and E1AΔ54–69, also bound to Id-1H and Id-2H (Fig. 1C), suggesting that the sequence required for induction of p53-dependent apoptosis differs from that required for binding to Id-1H and Id-2H.
Establishment of 3Y1 Derivative Cell Lines that Express E1A and Ids in Response to Dex.
To see the effect of Id proteins on the E1A-induced cell cycle progression, g12 cells that express E1A12S in response to dex were doubly transfected with pMTV-Id-1H and pMTV-Id-2H together with pSV2neo. G418-resistant colonies developed were cultivated in two 24-well tissue culture plates in the presence and absence of 1 μM dex for 8 h to estimate expression levels of Id-1H, Id-2H, and E1A mRNAs by RNA dot hybridization. Among 96 clones examined, 12 clones expressed a significant level of Id-1H mRNA, and nine clones expressed Id-2H mRNA in response to dex. Two clones, designated EId10 and EId23, expressed both Id-1H and Id-2H mRNAs at significant levels in addition to E1A mRNA. Unexpectedly, microscopic examination revealed that some clones showed morphological changes in response to dex after prolonged incubation. The changes induced were most marked in clones EId10 and EId23, whereas the changes induced in other clones that expressed either Id-1H or Id-2H in addition to E1A were weak. A minor proportion of EId10 and EId23 cells also underwent morphological changes during cultivation even in the absence of dex.
As controls, 3Y1 cells were similarly transfected with three expression vectors pMTV-Id-1H, pMTV-Id-2H, and either pMTV-E1AΔ17–23 or pMTV-E1AΔ54–69. The clones ΔEId23 and ΔEId2 thus established expressed these three mRNAs in response to dex. The cell lines YId6 and YId24 cells were established by transfection of 3Y1 cells with pMTV-Id-1H and pMTV-Id-2H and expressed both Id mRNAs in response to dex. No morphological changes were observed in these cell lines after prolonged cultivation in the presence of dex.
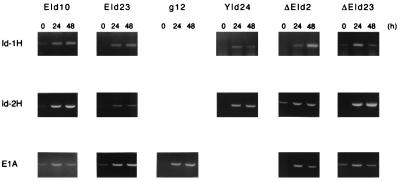
To quantify E1A12S, Id-1H, and Id-2H mRNAs synthesized from the MMTV-LTR of the exogenously introduced cDNA expression vectors, and to detect a trace amount of mRNA, the sequences between the MMTV LTR and the coding region of E1A, Id-1H, and Id-2H were amplified by RT-PCR. Total cellular RNAs (4 μg) were used as the templates. Under the conditions, the amount of each cDNA fragment synthesized was roughly proportional to the amount of input RNA within the range of 0.5 to 10 μg. The E1A, Id-1H, and Id-2H cDNA fragments with the expected lengths were synthesized quantitatively as shown in Fig. 2. EId10 and EId23 cells expressed very low levels of E1A, Id-1H, and Id-2H mRNAs in the absence of dex (0 h), but the levels increased sharply in response to dex. g12 cells expressed no detectable level of E1A mRNA in the absence of dex, but expressed a high level of E1A mRNA in response to dex. The levels of Id-1H and Id-2H mRNAs expressed in YId24 cells in the absence of dex were almost undetectable, but increased sharply after dex treatment. ΔEId2 and ΔEId23 cells expressed low levels of Id-2H and mutant E1A mRNAs, respectively, in the absence of dex, but all of the mRNAs were expressed at high levels after dex treatment.
Figure 2.
Expression of exogenously introduced E1A, Id-1H, and Id-2H cDNAs in response to dex. Total cellular RNA was prepared from the cell lines indicated before (0 h) and after treatment with dex (24 and 48 h). The exogenous mRNA sequences between the MMTV-LTR and the coding sequences of E1A, Id-1H, and Id-2H were amplified by RT-PCR and electrophoresed on 1.5% agarose gels. DNA was stained with ethidium bromide.
Induction of Apoptosis in Rat 3Y1 Cells After Coexpression of E1A and Ids.
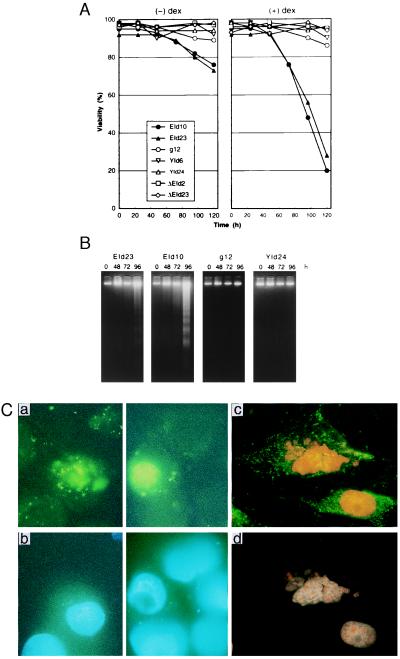
The correlation between the expression of E1A, Id-1H, and Id-2H and the changes in cell viability was analyzed in the above cell lines after treatment with dex (Fig. 3A). EId10 and EId23 cells grew normally until 48 h, but began to lose viability thereafter and 70–80% of the cells died at 120 h. A significant fraction of 15–20% of the cells died in the absence of dex, presumably because of the leakage of E1A and Id expression. g12 cells that express only E1A12S showed no significant morphological change after prolonged cultivation, although viability loss was observed in a minor fraction. The clones YId6 and YId24 that expressed both Id-1H and Id-2H mRNAs exhibited no viability loss. ΔEId23 and ΔEId2 cells that expressed Id-1H, Id-2H, and mutant E1A mRNAs showed no viability loss, indicating that the N-terminal region of E1A also is required for induction of apoptosis in 3Y1 cells carrying the p53 mutation. Electrophoretic analysis of EId10 and EId23 cell genomic DNA prepared after 96 h of dex treatment showed the ladder pattern characteristic of apoptosis (Fig. 3B). The fragmentation began to be observed after 72 h. No fragmentation was observed with g12 and YId24 cell genomic DNAs, suggesting that the expression of both E1A and Id proteins is required for induction of apoptosis in 3Y1 cells.
Figure 3.
Induction of apoptosis in EId10 and EId23 cells after coexpression of E1A12S, Id-1H, and Id-2H. (A) Cell viability. The cells were cultivated in the presence and absence of 1 μM dex. Cell viability determined by trypan blue exclusion is expressed as the percentage of the total cell number. (B) DNA fragmentation. DNA was isolated from both floating and adherent cells by the method of Hirt and subjected to electrophoresis. DNA was stained with ethidium bromide. (C) Annexin analysis of apoptotic cells. Sparse cultures of EId10 cells on coverslips were treated with dex for 72 h and 100 ng⋅ml−1 of hoechst 33342, a permeable DNA-specific fluorochrome was added to the culture for 15 min. The cells then were stained with annexin-V fluorescein (Boehringer Mannheim) and PI, an impermeable fluorochrome, in the absence (a and b) and presence (c and d) of 0.1% Triton X-100, which renders the cells permeable. Photographs were taken under a fluorescence microscope at a magnification of ×1,000 with the excitation (λex) between 455 nm and 500 nm (a and c) and with the λex between 330 nm and 450 nm (b and d). Annexin-V fluorescein shows the localization of phosphatidylserine at the outer layer of the plasma membrane (a) and at both inner and outer layers (c). Hoechst-stained nonpermeabilized cell DNA (b) and both Hoechst- and PI-stained permeabilized cell DNA (d).
Apoptosis also is characterized by a loss of membrane phospholipid asymmetry, resulting in the exposure of phosphatidylserine in the outer layer of plasma membrane (15). To confirm that the loss of cell viability is caused by apoptosis, EId10 cells treated with dex for 72 h were triple-stained with hoechst 33342, PI, and FITC-labeled annexin V that binds to phosphatidylserine. Both hoechst and PI intercalate to DNA, but only hoechst can permeate the active cell membrane. A significant proportion of the cells were annexin V positive (Fig. 3Ca), and only those positive cells showed chromatin condensation (Fig. 3Cb). When the EId10 cells were triple-stained in the presence of 0.1% Triton X-100, which makes cells permeable to PI and annexin V, all the cells were stained with hoechst, PI, and annexin V (Fig. 3C c and d). These results indicate that EId10 cells expressing E1A and Id proteins die through apoptosis.
Expression of Mutant Type p53 in EId10 and EId23 Cells.
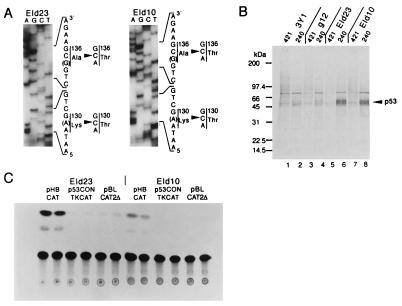
The 3Y1 cell line carries p53 mutations at codons 130 and 136 (8). To confirm that EId10 and EId23 cells still retain the same mutations, a portion of the p53 cDNAs carrying the mutation was amplified by RT-PCR and sequenced. The mutations originally detected were retained in EId10 and EId23 cells (Fig. 4A). Expression of mutant type p53 in EId10 and EId23 cells was examined by labeling with [35S]methionine, and p53 expressed was evaluated by immunoprecipitation of cell lysates with mAbs PAb240 and PAb421 that recognize the common mutant form of p53 and both wt and mutant forms of p53, respectively. The mutant form of p53 was efficiently immunoprecipitated from EId10 and EId23 cell lysates by PAb240 but inefficiently by PAb421 (Fig. 4B).
Figure 4.
Expression and functioning of mutated p53 in EId10 and EId23 cells. (A) Presence of p53 mutations in EId10 and EId23 cells. A portion of the p53 cDNA previously identified to carry mutations in 3Y1 cells (8) was amplified by PCR by using the upstream sense primer, from nucleotides 330 to 349 with the NaeI cohesive end and the downstream antisense primer from nucleotides 570 to 488 with the AflIII cohesive end. The amplified cDNAs were cloned into pBluescript II KS+ and sequenced. (B) Growing cells were labeled with [35S]methionine for 3 h, and aliquots of the cell lysates were immunoprecipitated with antibodies PAb240 and PAb421. The immunoprecipitates were subjected to electrophoresis followed by autoradiography. (C) Mutated p53-dependent transactivation of the heat shock protein 70 promoter. EId10 and EId23 cells were transfected with 20 μg of either pHBCAT, p53CONTKCAT, or pBLCAT2Δ in duplicate. CAT activities were assayed at 48 h posttransfection.
The functioning of mutant type p53 in EId10 and EId23 cells was analyzed by the expression of reporter plasmids p53CONTK-chloramphenicol acetyltransferase (CAT) and pHBCAT. The former contains the two p53 binding sequences upstream of the herpesvirus thymidine kinase promoter and is effectively transactivated by wt p53, whereas the latter containing the human heat shock protein 70 promoter was transactivated by mutated p53 (12). EId10 and EId23 cells were transfected with these reporters and pBLCAT2Δ that contain no promoter. As shown in Fig. 4C, the heat shock protein 70 promoter was expressed efficiently, whereas the promoter of p53CONTK-CAT was not, indicating that the mutant form of p53 is functioning in these cell lines.
Induction of Apoptosis by E1A and Ids Occurs After Accumulation of Cells in S Phase.
Cell cycle distribution of EId10 and EId23 cells was analyzed by flow cytometry after induction of E1A, Id-1H, and Id-2H (Fig. 5A). As controls, g12 and YId24 cells were similarly analyzed. Growing cultures of these cells were treated with 1 μM dex. At 0 h, g12 cells exhibited two distinct peaks of DNA contents representing G1 and G2/M cells. The S phase population observed between these peaks was minor and comprised of less than 10% of the total population. This profile of cell distribution was not altered until 48 h, when the cells became confluent. After 72 h, a proportion of the cells with DNA contents below 2n (sub-G1 population) increased slightly in consistency with a slight loss of viability after this time (Fig. 3A).
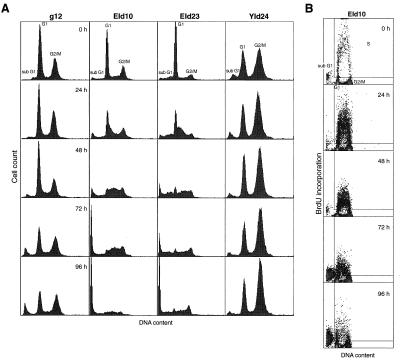
Figure 5.
Flow-cytometric analysis of cells expressing E1A12S, and/or Id-1H and Id-2H. (A) After treatment of cells with dex, DNA contents were examined by PI staining and flow cytometry. The cells with a DNA content less than that of G1 cells are shown by sub-G1. (B) Sparse cultures of EId10 cells were treated with dex and incubated with 100 μM BrdU for 2 h before fixation. The cells synthesizing DNA, estimated by incorporation of BrdU, are shown in the insets and the populations of cells in G1, G2/M and sub-G1 population are shown outside the insets.
In the case of EId10 cells, the S phase population began to accumulate within 24 h, causing the G1 peak to skew toward the G2/M peak. At 48 h, the population was distributed broadly between the G1 and G2/M peaks comprising more than 40% of the total population. A fraction of the S phase population began to be converted to the sub-G1 population after 48 h, while most of the population still remained as the broad peak at 72 h. At 96 h, the sub-G1 population increased markedly. Alteration in cell cycle distribution of EId23 cells was similar to that of EId10 cells, but proceeded slightly slowly. Quiescent YId24 cells at 0 h exhibited two peaks of DNA, but the profile did not alter after induction of Id-1H and Id-2H expression.
The ability of EId10 cells accumulated in S phase to synthesize DNA was analyzed by incorporation of BrdU (Fig. 5B). EId10 cells treated with dex were incubated with BrdU for 2 h before the cell harvest, and a fraction of cells that incorporated BrdU was assessed by flow cytometry using mouse anti-BrdU antibody and FITC-conjugated anti-mouse IgG. The distribution of the cells incorporating BrdU in the cell cycle is shown in the insets of Fig. 5B and that of the cells not incorporating BrdU is shown outside the insets. The incorporation of BrdU was greatly stimulated at 24 h and then decreased gradually. A fraction of the cells incorporated BrdU at 24 and 48 h showed DNA content of 4n representing the late S and G2 population, suggesting that the cells accumulated in S phase are capable of synthesizing DNA, but most of them undergo apoptosis before entering G2 phase.
Suppression of E1A-induced Apoptosis by Mutated p53 in p53 Null Mouse Cells Is Overcome by Ids.
To show the effects of wt and mutated p53 on the induction of apoptosis by E1A and Ids and to confirm that the induction of apoptosis in EId10 and EId23 cell lines is caused by coexpression of E1A and Ids, the mouse cerebellum cell line A40 established from a p53 null mouse was transfected with expression vectors for E1A and/or Id-1H, Id-2H together with the expression vector for CD20, a cell surface calcium binding protein. Aliquots of the cells were cotransfected with the expression vector encoding either wt p53 or mutated p53. The apoptotic cells were quantitatively assayed by using FACS by the accumulation of the sub-G1 population (Fig. 6). The cell damage caused by transfection procedure in control cells (−), to which only the CD20 expression vector was transfected (Fig. 6A), resulted in the increase in the sub-G1 population to 9.7% at 72 h. The sub-G1 population increased significantly to 13–14% by expression of E1A alone or E1A plus Ids at 72 h. No significant increase was observed when Ids alone were expressed.
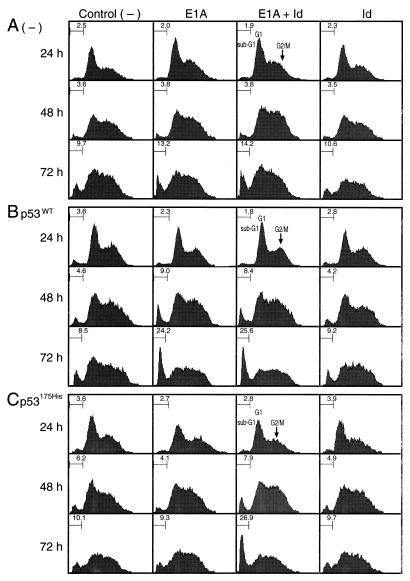
Figure 6.
Suppression of E1A-induced apoptosis by mutated p53 in A40 cells is overcome by Ids. Growing cultures of A40 cells (p53−/−) were transfected with either pHβAPr-E1A12S (E1A), pCMV-Id-1H plus pCMV-Id-2H (Id), or a combination of these three expression vectors (E1A + Id) together with pCMVCD20. Control (−), the cells were transfected with pCMVCD20 alone. (A) The expression vectors were transfected in the absence of p53. (B and C) The cells were simultaneously transfected with either pMoLV-wtp53 or pMoLV-p53His175. At the times indicated, the cells were stained with PI, and DNA contents of the cells were analyzed by flow cytometry. The cells were gated based on the expression of CD20.
When wt p53 was coexpressed (Fig. 6B), the sub-G1 population increased markedly both in the cells expressing E1A alone (24.2%) and in the cells expressing E1A and Ids (25.6%). The sub-G1 population therefore increased about 4-fold by coexpression with wt p53, when calculated after subtraction of the background value (≈10%). Coexpression of wt p53 had no effect in the cells expressing Ids alone. When mutated p53175His was coexpressed (Fig. 6C), the increase in the sub-G1 population induced by E1A alone was completely suppressed, whereas the increase induced by E1A and Ids was unaffected, indicating that the suppressive function of mutated p53 is overcome by Ids. Expression of p53175His alone (−) or p53175His plus Ids (Id) had no effect. Essentially the same results were obtained by cotransfection with another p53 mutant, p53205Cys (data not shown).
Association of Id-2H with E1A and E2A bHLH Proteins in Vivo.
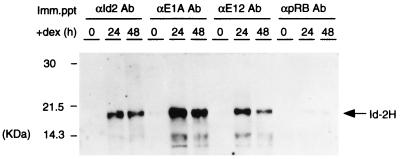
Among four mammalian Id proteins, Id-2 also binds to the retinoblastoma protein (pRB) (16). To characterize the partners to which Id-2H binds, the cell lysates were prepared from EId10 cells before (0 h) and after treatment with dex for 24 and 48 h. The lysates were immunoprecipitated with antibodies to Id-2, E1A, E2A (E12/E47) bHLH protein, and pRB and the precipitates were dissociated with 0.8% deoxycholate. The lysates then were analyzed by Western blotting with anti-Id-2 antibody, which crossreacts with human Id-2H. As shown in Fig. 7, Id-2H was nearly absent in the 0-h lysate, but induced after treatment of the cells with dex. Large amounts of Id-2H were detected in the immunoprecipitates prepared from the 24-h and 48-h lysates with antibodies to E1A and E2A (E12/E47), whereas Id-2H was scarcely detected in the immunoprecipitate prepared with anti-pRB antibody, suggesting that Id-2H predominantly associates with E1A and bHLH proteins in EId10 cells after induction of apoptosis.
Figure 7.
Id-2H associates with E1A and E12 bHLH protein in vivo. The cell lysates were prepared from EId10 cells treated with dex for 0, 24, and 48 h, and aliquots of 200 μg of protein were immunoprecipitated with antibodies to E1A, E2A (E12 and E47) and pRB. The precipitates were treated with 0.8% deoxycholate and analyzed by Western blotting with anti-Id2 antibody.
DISCUSSION
The present study showed that E1A is capable of inducing apoptosis in rat 3Y1 cells that express mutated p53 in collaboration with Id proteins. 3Y1 cells are very sensitive to coexpression of E1A and Ids, and the 3Y1 derivative cell lines, EId10 and EId23, undergo apoptosis when these proteins are expressed beyond thresholds in response to dex. Neither E1A nor Ids alone induced apoptosis. Induction of apoptosis in EId10 and EId23 cells is not caused by the selection of particular cells, because expression of E1A alone or E1A plus Ids in mouse A40 cells (p53−/−) by transient transfection resulted in the induction of apoptosis in a significant fraction of the cells. The induction of apoptosis in A40 cells by E1A alone or E1A plus Ids was stimulated by simultaneous expression of wt p53; however, apoptosis induced by E1A alone was completely suppressed by simultaneous expression of mutated p53, whereas apoptosis induced by E1A and Ids was stimulated the same as wt p53 (Fig. 6), suggesting that the suppressive function of mutated p53 is overcome by Ids. The result is consistent with the high sensitivity of EId10 and EId23 cells to coexpression of E1A and Ids in inducing apoptosis.
The induction of apoptosis by E1A in most cases requires the expression of wt p53 and its stabilization (17, 18). The N-terminal region of E1A together with CR1 binds to the p300/CREB binding protein (CBP) family, and this p300 binding site is essential for E1A to stabilize p53 (19–21) and to induce apoptosis (4, 5). p300/CBP functions as a transcriptional coactivator associating with a variety of transcription factors and may regulate the expression of genes required for cell proliferation, differentiation, and apoptosis (22). p300 also binds to p53, and the p300-p53 complex seems to modulate both p300- and p53-dependent transcription (19). E1A represses some enhancer-dependent transcription by counteracting the interaction between p300 and transcription factors (22).
Analysis of the immunoprecipitates by Western blotting with anti-Id-2 antibody (Fig. 7) showed that Id-2H bound preferentially to E1A and E2A and poorly to pRB, suggesting that Ids play a role in inducing apoptosis through interaction with either E1A, bHLH transcription factor, or both. Among various Id proteins of human, mouse, Xenopus, and Drosophila, amino acid sequences are well conserved in five regions (boxes 1–5) including the HLH structure in box 3 (3). The binding site of Ids to E1A must be located in these conserved regions. Enforced expression of E2A bHLH factors induces cell growth arrest (23, 24), but additional expression of Id overcomes this growth inhibition through formation of the heterodimer at the HLH region (25). Because both the N terminus of E1A (amino acids 1–36) and E1AΔ17–23 can bind to Ids, the binding sequence of E1A is likely to reside within the amino acid sequence from 1 to 16 or between 24 and 36.
Because both E1A and Ids are positive regulators of cell proliferation, the cells that expressed these proteins may enter S phase prematurely, and the S phase checkpoint may not operate normally. DNA synthesis induced by overexpression of E2F-1 in quiescent fibroblasts does not lead to a complete replication, but leads to massive cell death characteristics of apoptosis (26–28). This induction of apoptosis, however, depends on wt p53 and is subverted by mutated p53.
Among E2F family members, only E2F-1 has the cyclin A binding site and its heterodimeric partner DP1 is phosphorylated by cyclin A/cdk2 kinase during S phase. This phosphorylation inactivates the DNA binding activity of E2F-1/DP1 and is essential for normal progression of S phase. Prevention of phosphorylation by introduction of mutation into either cyclin A binding site of E2F-1 or phosphorylation site of DP1 resulted in accumulation of the cells in S phase, leading to apoptosis (29). The mammalian Id2 and Id3 also are phosphorylated by cyclin E/cdk 2 and cyclin A/cdk 2 at Ser 5 site during late G1 to early S (30). The phosphorylation dramatically alters the target HLH proteins to which Ids bind and seems to be required for promoting S phase progression.
Induction of apoptosis in cells by many anti-tumor agents correlates with the expression of both oncogenes and the wt p53 gene, and malignant cells expressing mutated p53 are usually resistant to the induction of apoptosis. Because p53 mutations are extremely common in human cancer, the present study offers a good system to analyze the function of mutated p53 and may provide a clue to how apoptosis can be induced in malignant cells expressing mutated p53.
Acknowledgments
We thank D. Baltimore for generously providing the E12 cDNA, N. Tsuchida for expression vectors encoding wild-type and mutated p53, E. Harlow for pCMVCD20, and Y. Taya for anti-pRB antibody. This work is supported by special coordination funds of the Science and Technology Agency of the Japanese government.
ABBREVIATIONS
- bHLH
basic helix–loop–helix
- dex
dexamethasone
- wt
wild type
- MMTV
mouse mammary tumor virus
- LTR
long terminal repeat
- RT
reverse transcriptase
- PI
propidium iodide
- BrdU
5-bromo-2′-deoxyuridine
- FITC
fluorescein isothiocyanate
- GST
glutathione S-transferase
- pRB
retinoblastoma protein
- CAT
chloramphenicol acetyltransferase
References
- 1.Nevins J R. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 2.Taylor D A, Kraus V B, Schwarz J J, Olson E N, Kraus W E. Mol Cell Biol. 1993;13:4714–4727. doi: 10.1128/mcb.13.8.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Norton J D, Deed R W, Craggs G, Sablitzky F. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 4.White E, Cipriani R, Sabbatini P, Denton A. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mymryk J S, Shire K, Bayley S T. Oncogene. 1994;9:1187–1193. [PubMed] [Google Scholar]
- 6.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 7.Kimura G, Itagaki A, Summers J. Int J Cancer. 1975;15:694–706. doi: 10.1002/ijc.2910150419. [DOI] [PubMed] [Google Scholar]
- 8.Ushijima T, Makino H, Nakayasu M, Aonuma S, Takeuchi M, Segawa K, Sugimura T, Nagao M. Jpn J Cancer Res. 1994;85:455–458. doi: 10.1111/j.1349-7006.1994.tb02379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 10.Lee F, Mulligan R, Berg P, Ringold G. Nature (London) 1981;294:228–232. doi: 10.1038/294228a0. [DOI] [PubMed] [Google Scholar]
- 11.Gunning P, Leavitt J, Muscat G, Ng S-Y, Kedes L. Proc Natl Acad Sci USA. 1987;84:4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsutsumi-Ishii Y, Tadokoro K, Hanaoka F, Tsuchida N. Cell Growth Differ. 1995;6:1–8. [PubMed] [Google Scholar]
- 13.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fadok V A, Voelker D R, Campbell P A, Cohen J J, Bratton D L, Henson P M. J Immunol. 1992;148:2207–2216. [PubMed] [Google Scholar]
- 16.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 17.Lowe S W, Ruley H E. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 18.Debbas M, White E. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 19.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 20.Querido E, Teodoro J G, Branton P E. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiou S K, White E. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shikama N, Lyon J, La Thangue N B. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01149-5. [DOI] [PubMed] [Google Scholar]
- 23.Crescenzi M, Fleming T P, Lassar A B, Weintraub H, Aaronson S A. Proc Natl Acad Sci USA. 1990;87:8442–8446. doi: 10.1073/pnas.87.21.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorrentino V, Pepperkok R, Davis R L, Ansorge W, Philipson L. Nature (London) 1990;345:813–815. doi: 10.1038/345813a0. [DOI] [PubMed] [Google Scholar]
- 25.Peverali F A, Ramqvist T, Saffrich R, Pepperkok R, Barone M V, Philipson L. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Levine A J. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan B, Lee W-H. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin X-Q, Livingston D M, Kaelin W G, Adams P D. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krek W, Xu G, Livingston D M. Cell. 1995;83:1149–1158. doi: 10.1016/0092-8674(95)90141-8. [DOI] [PubMed] [Google Scholar]
- 30.Hara E, Hall M, Peters G. EMBO J. 1997;16:332–342. doi: 10.1093/emboj/16.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]