Abstract
The evolutionarily conserved protein EB1 originally was identified by its physical association with the carboxyl-terminal portion of the adenomatous polyposis coli (APC) tumor suppressor protein, an APC domain commonly mutated in familial and sporadic forms of colorectal neoplasia. The subcellular localization of EB1 in epithelial cells was studied by using immunofluorescence and biochemical techniques. EB1 colocalized both to cytoplasmic microtubules in interphase cells and to spindle microtubules during mitosis, with pronounced centrosome staining. The cytoskeletal array detected by anti-EB1 antibody was abolished by incubation of the cells with nocodazole, an agent that disrupts microtubules; upon drug removal, EB1 localized to the microtubule-organizing center. Immunofluorescence analysis of SW480, a colon cancer cell line that expresses only carboxyl-terminal-deleted APC unable to interact with EB1, demonstrated that EB1 remained localized to the microtubule cytoskeleton, suggesting that this pattern of subcellular distribution is not mediated by its interaction with APC. In vitro cosedimentation with taxol-stabilized microtubules demonstrated that a significant fraction of EB1 associated with microtubules. Recent studies of the yeast EB1 homologues Mal3 and Bim1p have demonstrated that both proteins localize to microtubules and are important in vivo for microtubule function. Our results demonstrate that EB1 is a novel component of the microtubule cytoskeleton in mammalian cells. Associating with the mitotic apparatus, EB1 may play a physiologic role connecting APC to cellular division, coordinating the control of normal growth and differentiation processes in the colonic epithelium.
During mitotic cell division, the interphase network of cytoplasmic microtubules is transformed into a bipolar spindle consisting of a complex and dynamic array of microtubules nucleating from two centrosomes and extending to the chromosomes. Some microtubules attach to chromosomes through the kinetochores, whereas others overlap with each other in the midzone, generating the typical fusiform shape. This complex structure assembles and disassembles rapidly and efficiently as cells proceed through mitosis (1–3). Considerable progress has been made in understanding the role of the microtubule-based motor proteins dynein and kinesin in morphogenesis (4, 5). Less is known about the role of nonmotor microtubule-associated proteins, but these also are postulated to be important, probably through the regulation of microtubule dynamics.
EB1 is a 30- to 35-kDa protein of unknown function that was isolated in a yeast two-hybrid screen by its binding to the carboxyl-terminal domain of the adenomatous polyposis coli (APC) tumor suppressor protein (6), a domain that is deleted in the majority of familial and sporadic forms of colon carcinoma (7). The role of the carboxyl-terminal domain of APC has not been defined and, whereas carboxyl-terminal mutations of APC predispose to the development of colonic cancer, other genetic changes appear to be necessary for the manifestation of the transformed phenotype. It has been hypothesized that a defect in chromosome segregation may be an early event in colorectal tumorigenesis, leading to genetic instability, critical for the development of all colorectal cancers (8).
We undertook a systematic study of EB1 subcellular localization during the cell cycle by using both biochemical and immunofluorescence techniques. Using mAbs specific for EB1, we demonstrated that EB1 decorated part of the microtubule cytoskeleton during interphase with pronounced staining of the centrosome. During cell division, EB1 localized to the mitotic apparatus. This microtubule localization was abolished in the presence of the microtubule-destabilizing drug nocodazole; upon drug removal, the microtubule distribution of EB1 was recovered and EB1 fluorescence concentrated at the microtubule-organizing center. These results suggest that EB1 is associated with the microtubule network and may be involved in microtubule polymerization and spindle function.
MATERIALS AND METHODS
Cell Culture and Antibodies.
The African green monkey kidney cell line CV-1 was obtained from the American Type Culture Collection and grown in DMEM (GIBCO/BRL) supplemented with 10% heat-inactivated fetal bovine serum (Sigma), 100 units/ml of penicillin (GIBCO/BRL), and 0.1 mg/ml of streptomycin (GIBCO/BRL), termed DMEM-10%. The colon cancer cell line SW480 (American Type Culture Collection) was grown in DMEM-10% media at 37°C in a 5% C02 incubator. The Jurkat T cell line J77 was grown in RPMI medium 1640 (GIBCO/BRL) supplemented with 10% heat-inactivated fetal bovine serum, 10 mM Hepes (pH 7.5), 2 mM glutamine (GIBCO/BRL), and 50 μM 2-mercaptoethanol (Sigma), termed RPMI-10%.
The murine anti-human EB1 mAbs GD10 and EA3 (Oncogene Research Products, Cambridge, MA) and the rat monoclonal anti-α-tubulin termed YL1/2 (Serotec) were used for immunoblotting, immunoprecipitation, and immunofluorescence experiments. The secondary antibodies donkey anti-rat- fluorescein isothiocyanate and donkey anti-mouse-rhodamine were purchased from Jackson ImmunoResearch.
Immunoprecipitation and Immunoblotting.
CV-1 and J77 cells (1 × 107) were lysed in 1 ml of buffer A (30 mM Hepes, pH 7.5/150 mM NaCl/1% Triton X-100/1 mM phenylmethylsulfonyl fluoride/1 mM Na3VO4/10 μg/ml of leupeptin/10 μg/ml of aprotinin) and incubated on ice for 15 min; lysates were clarified by centrifugation at 14,000 × g for 10 min at 4°C. Protein G-Sepharose beads (Pierce) precoated with anti-EB1 mAb were added to the CV-1 lysate. After a 2-hr incubation at 4°C, the beads were washed four times with buffer A and solubilized in SDS sample buffer. Immunoprecipitated proteins and cell lysates were separated by electrophoresis on 10% SDS polyacrylamide gels, transferred to poly(vinylidene difluoride) membranes (Millipore), and probed with anti-EB1 mAb. The antibody-labeled protein bands were detected by autoradiography after enhanced chemiluminescence (Amersham).
Immunofluorescence.
For immunofluorescence experiments, CV-1 and SW480 cells were incubated in trypsin/EDTA (0.25% trypsin/1 mM EDTA, GIBCO/BRL) for 5 min, harvested from tissue culture dishes, and seeded onto sterile 22-mm2 glass coverslips in six-well plates (Falcon) to reach 70–80% confluence after 48 hr. Several fixation procedures (3% formaldehyde in PBS, 4% paraformaldehyde in PBS, 100% acetone, and 100% methanol) were tested to investigate the EB1 subcellular distribution; because the most robust and reproducible pattern was obtained by methanol fixation, this method was used in all subsequent experiments. Cells were fixed in methanol at −20°C for 5 min and washed three times in PBS to allow cell rehydration. In some experiments living cells grown on glass coverslips were treated with 10 μM nocodazole (Sigma) in DMEM-10% media for 1 hr and either fixed immediately or washed twice in DMEM-10%, media replaced and fixed in methanol 5, 10, or 20 min later. After fixation, the cells were incubated in PBS containing 2% BSA and 0.02% sodium azide (NaN3) for 10 min to reduce nonspecific antibody binding and washed twice in PBS. Primary antibodies diluted in PBS-2% BSA-0.02% NaN3 (1:10 optimized dilutions for EB1 antibodies and 1:100 dilution for anti-α-tubulin antibody) or diluent alone were added to each coverslip and incubated for 30 min at room temperature in a humidified chamber. Cells were washed extensively in PBS containing 0.2% gelatin, followed by incubation in appropriate secondary antibodies previously diluted in PBS-2% BSA for 30 min at room temperature. For double-labeling experiments, the primary and secondary antibodies were incubated sequentially; equivalent results were obtained regardless of the order in which primary antibodies were used. DNA was counterstained with 0.5 μg/ml of 4,6-diamidino-2-phenylindole (Sigma) in PBS, and the coverslips were mounted in glycerol containing 1 μg/ml of p-aminophenylenediamine (Sigma) to reduce fluorescence bleaching. Controls to demonstrate specificity and lack of crossreactivity of secondary antibodies also were performed; incubating the coverslips with secondary antibodies alone showed no fluorescence, and in double-label experiments exclusion of either primary antibody did not affect the distribution of fluorescence of the second detecting antibody (data not shown). Cells were examined by using a confocal laser scanning microscope LSM 410 (Zeiss). Images were printed with a Fujix Pictography 3000 color printer (Fujifilm) using Adobe PhotoShop software (Adobe Systems, Mountain View, CA).
Microtubule Binding Assay.
In vitro microtubule binding assays were performed (9, 10) by using aliquots of J77 cytosolic lysate containing endogenous EB1. Taxol-stabilized microtubules were prepared from purified bovine brain tubulin that was precleared of aggregates by ultracentrifugation (100,000 × g, 10 min at 4°C) immediately before use. Taxol (Sigma) was added sequentially to 0.1, 1.0, and 10 μM at 5-min intervals to a solution of 14 μM tubulin, incubated at 37°C in reassembly buffer BRB-80 (80 mM K Pipes, pH 6.8/1 mM MgCl2/1 mM EGTA) supplemented with 1 mM GTP, 100 μg/ml of BSA, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and 10 μg/ml each of leupeptin and aprotinin. Cytosolic J77 lysate was precleared as above and aliquots of 50 μg were incubated with or without taxol-polymerized microtubules for 15 min at room temperature. The mixture was divided into 50-μl aliquots that were each ultracentrifuged at 100,000 × g, during 10 min, through 80 μl of a 50% sucrose cushion made up in BRB-80 supplemented with 1 mM GTP, 10 μM taxol, and 100 μg/ml of BSA. Supernatants and pellets were collected and separated by SDS/PAGE, transferred to poly(vinylidene difluoride), and probed with antisera against EB1.
RESULTS
Colocalization of EB1 with Cellular Microtubules and the Centrosome.
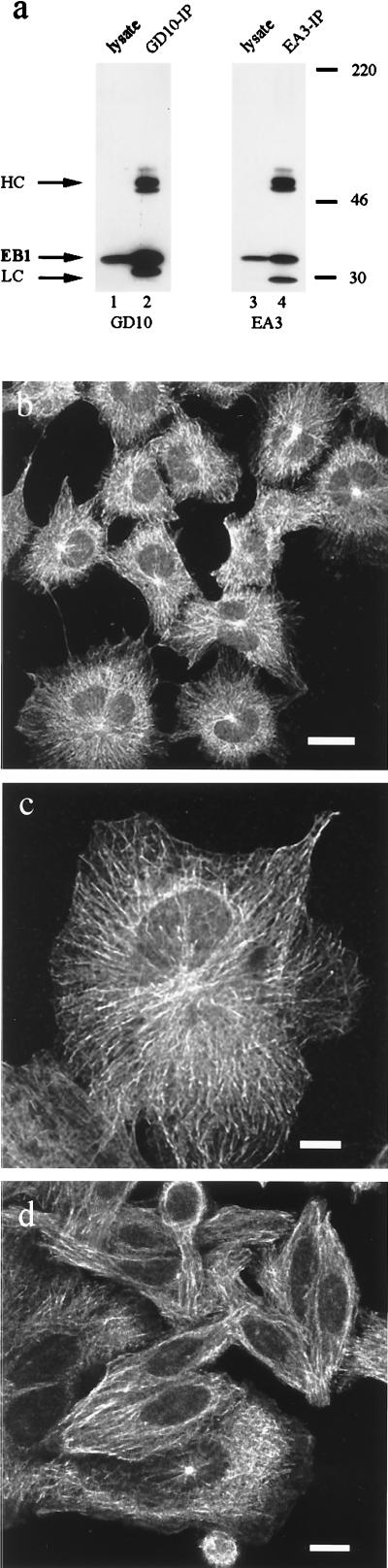
Indirect immunofluorescence was used to investigate the intracellular distribution of EB1 by using two different mAbs directed against the human EB1 protein (6). The specificity of these antibodies was demonstrated by immunoprecipitation and immunoblotting analysis: a single band was detected both in cytosolic lysates (Fig. 1a, lanes 1 and 3) and upon immunoprecipitation (Fig. 1a, lanes 2 and 4). The epithelial cell lines CV-1 (chosen because of its large size and flat morphology) and SW480 were grown to near confluence and prepared for indirect immunofluorescence. Different fixation procedures were compared to optimize immunofluorescence analysis. Methanol fixation at −20°C for 5 min gave robust and reproducible detection of EB1. For both the GD10 and EA3 antibodies, fluorescence was largely localized in the cytoplasm, although minimal fluorescence also was detected in the nucleus (Fig. 1b); no nuclear or cytoplasmic staining was observed with secondary antibody alone (data not shown). In interphase cells, EB1 decorated fibers throughout the cytoplasm with brighter staining surrounding the nucleus (Fig. 1b). The protein was concentrated at the microtubule-organizing center (Fig. 1c), but diffuse fluorescence in the cytoplasm also was detected. These results demonstrated that EB1 is associated with the microtubule cytoskeleton.
Figure 1.
Subcellular localization of EB1 in mammalian cells. (a) Specificity of the anti-EB1 mAbs GD10 and EA3 for EB1 in cell lysates (lanes 1 and 3) and immunoprecipitation (lanes 2 and 4). Cell lysates and immunoprecipitated proteins from CV-1 cells, prepared as described, were separated in 10% SDS-polyacrylamide gels, transferred to poly(vinylidene difluoride) membranes, probed with anti-EB1 antibodies (GD10 and EA3), and detected by ECL; heavy chain (HC) and light chain (LC) are indicated. Interphase CV-1 (b and c) and SW480 cells (d) were stained with GD10 antibody after methanol fixation at −20°C. A secondary donkey anti-mouse rhodamine-conjugated antibody was used, and the samples were analyzed by confocal microscopy. [Bars: 25 μm (b) and 10 μm (c and d).]
The low protein expression of APC has rendered immunofluorescence and localization experiments difficult. This limitation has been overcome by transient overexpression of APC (11), with the caveat that overexpression itself may modify the subcellular localization of proteins. Transiently overexpressed APC has been shown to localize with the microtubule cytoskeleton. We investigated whether the association of EB1 with microtubules was mediated by APC. Using SW480 cells, a colon cancer cell line that expresses only a truncated form of APC that is unable to interact with EB1 (ref. 6, and data not shown), the cellular distribution of EB1 was unchanged (Fig. 1d). Therefore the carboxyl terminus of APC is not required for EB1 association to microtubules.
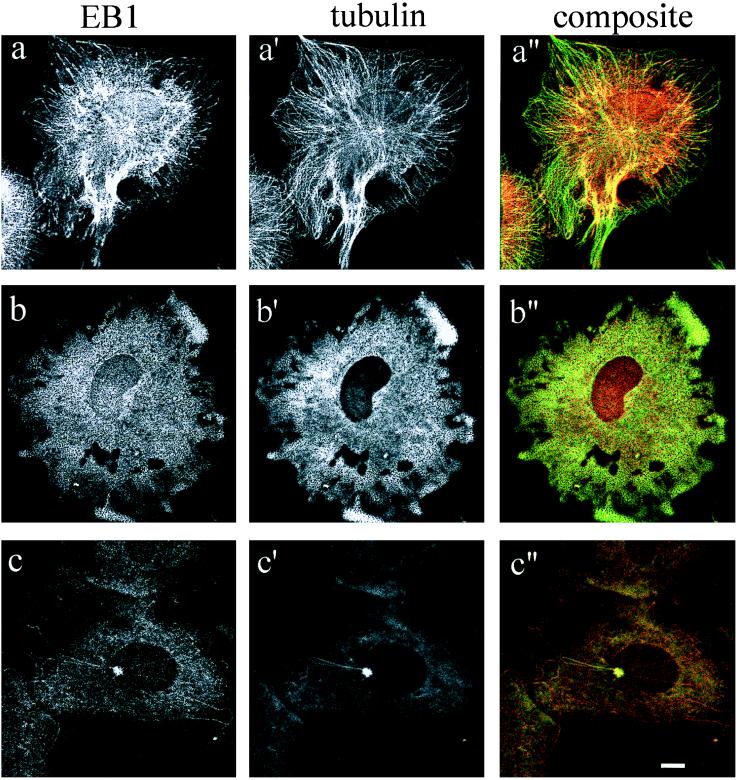
To analyze the EB1-microtubule association, CV-1 cells were subjected to dual immunofluorescence using a rat monoclonal anti-α-tubulin antibody and the mouse monoclonal anti-EB1 antibody GD10. CV-1 cells were treated with the microtubule-destabilizing drug nocodazole to detect changes in the EB1 immunofluorescence pattern upon microtubule disruption. Confocal microscopy images of cells before nocodazole treatment (Fig. 2a) resolved areas of overlap (yellow) between fibers stained by anti-EB1 antibody (red) and fibers stained by anti-α-tubulin antibody (green), demonstrating that EB1 colocalizes with a subpopulation of microtubules. The colocalization signal was particularly prominent at the centrosome; the dramatic intensity of overlap signal at the centrosome represents either a high density of microtubule fibers in this area or a direct association of EB1 with the centrosome. Disruption of the microtubule cytoskeleton by nocodazole treatment completely abolished the EB1 staining pattern, confirming that EB1 is associated with the microtubule network (Fig. 2b).
Figure 2.
Colocalization of EB1 with cytoplasmic microtubules and the centrosome. Unsynchronized CV-1 cells were fixed in methanol and processed for dual immunofluorescence with antibodies directed against EB1 (GD10) and α-tubulin (YL1/2), in the presence or absence of the microtubule destabilizing drug nocodazole (10 μM). (a, a′, and a′′) Control untreated cells. (b, b′, and b′′) Cells treated with nocodazole for 1 hr and fixed in methanol at −20°C. (c, c′, and c′′) Cells fixed in methanol 5 min after nocodazole removal. Staining was analyzed by confocal microscopy. (Bar, 10 μm.)
EB1 Associates With Microtubules Upon Nocodazole Removal.
To further study the interaction between EB1 and the microtubule cytoskeleton, EB1 localization in CV-1 cells was followed during a nocodazole washout period as microtubules repolymerized. After 1-hr incubation in 10 μM nocodazole at 37°C, the drug was removed and replaced by prewarmed DMEM-10% media, and the cells were incubated for 5, 10, or 20 min at 37°C. The cells were fixed in methanol and prepared for indirect immunofluorescence with anti-α-tubulin and anti-EB1 antibodies. We could not resolve the centrosome clearly until approximately 1 min after nocodazole removal, at which time EB1 was associated with the centrosome. Five minutes after drug removal, the microtubule arrays began to repolymerize, nucleating from the microtubule-organizing center. Strikingly, EB1 fluorescence was concentrated on 90% of these newly formed microtubules (Fig. 2c), suggesting a predominant association with new microtubules and consistent with involvement of EB1 in microtubule polymerization. Ten minutes after drug removal, the entire assembly of microtubule fibers and colocalization with EB1 was restored (data not shown).
EB1 Associates with Microtubules in Vitro.
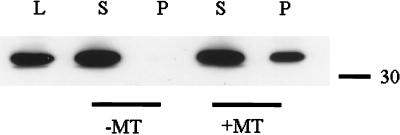
Based on the association of EB1 with the microtubule cytoskeleton, we investigated the binding of EB1 to microtubules in vitro by using sedimentation analysis. Purified brain tubulin was taxol-polymerized in BRB80 buffer in the presence of GTP and incubated with aliquots of J77 cytosolic cell lysate containing endogenous EB1. After incubation, the reaction mixture was sedimented; supernatants and pellets were collected and analyzed by SDS-polyacrylamide gels. Consistent with our immunofluorescence results, approximately 40% of the EB1 present in the extract bound to microtubules (Fig. 3) with an affinity binding within the micromolar concentration (data not shown). This finding indicates low affinity binding of EB1 to microtubules under the conditions tested. Binding also was observed by using 35[S]-EB1 translated in rabbit reticulocyte lysate, but at lower levels than observed with the J77 cell lysate (data not shown), perhaps, in part, because of the presence of endogenous EB1 in the reticulocyte lysate.
Figure 3.
In vitro EB1 microtubule binding. Cell extracts from J77 prepared in buffer A (see Materials and Methods) were incubated with taxol-stabilized microtubules. Supernatants (S) and pellets (P) were collected and separated by SDS-polyacrylamide gel, transferred to poly(vinylidene difluoride), and probed with the anti-EB1 antibody GD10. Microtubule binding was demonstrated by cosedimentation of EB1 with the microtubule pellet. Whole-cell lysate (L) demonstrated endogenous EB1 mobility.
Tubulin undergoes several posttranslational modifications that generate different microtubule populations during the cell cycle. One of these modifications is the reversible removal of the carboxyl-terminal tyrosine residue in the α-tubulin subunit, a process that has been related to changes in the microtubule stability (12, 13). Different populations of microtubules may interact preferentially with motor proteins or other microtubule-associated proteins. Because the majority of brain tubulin is derived from a stable population of microtubules containing predominantly detyrosinated α-tubulin, we investigated whether EB1 would bind preferentially to a population of microtubules containing tyrosinated tubulin. We used tubulin purified from HeLa cells, known to provide a source of dynamic, predominantly tyrosinated α-tubulin (14), and performed the microtubule binding assay as above. Binding of EB1 to microtubules from HeLa cells did not show enhanced EB1 protein present in the microtubule pellet compared with binding to brain tubulin (data not shown).
EB1 Localizes to the Mitotic Spindle During Cell Division.
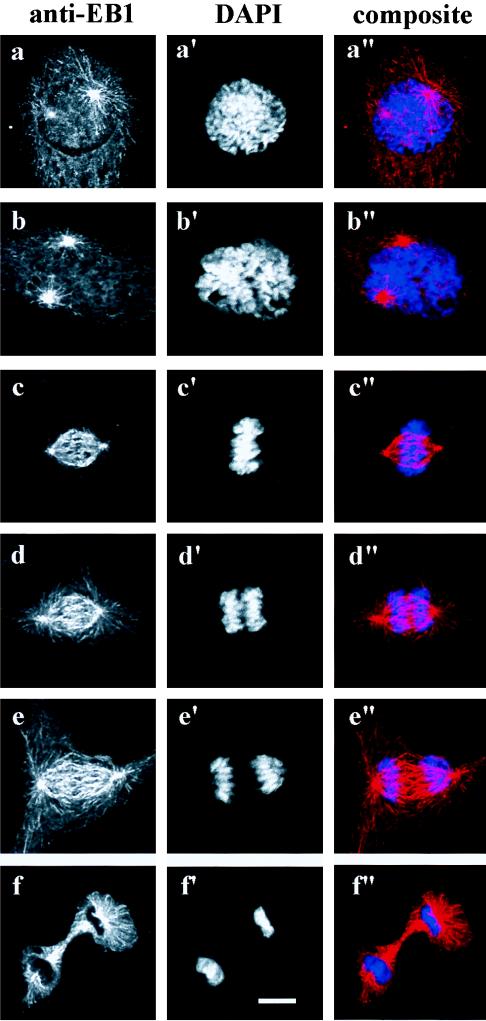
The cellular localization of EB1 was examined during mitosis. CV-1 cells were fixed in methanol, stained with anti-EB1 antibody, and counterstained with 4,6-diamidino-2-phenylindole to detect DNA. EB1 localization during the stages of mitosis were visualized (Fig. 4). During prophase and prometaphase EB1 fluorescence was associated with the centrosomes and attached microtubules (Fig. 4 a and b). As cells progressed into metaphase, EB1 immunofluorescence along cytoplasmic microtubules disappeared, leaving only the prominent staining of the spindle fibers (Fig. 4c). The cytoplasmic microtubular array reappeared again in anaphase and telophase (Fig. 4 d and e). EB1 fluorescence was distributed uniformly along the elongating spindle during anaphase and telophase. During cytokinesis, EB1 was located at the midbody and to astral microtubules (Fig. 4f).
Figure 4.
EB1 localizes to the mitotic apparatus during cell division. Unsynchronized CV-1 cells were fixed in methanol and stained with the anti-EB1 antibody GD10 and a secondary donkey anti-mouse rhodamine-conjugated antibody. DNA was counterstained with 4,6-diamidino-2-phenylindole (DAPI). (a, a′, and a′′) Prophase. (b, b′, and b′′) Prometaphase. (c, c′, and c′′) Metaphase. (d, d′, and d′′) Anaphase. (e, e′, and e′′) Telophase. (f, f′, and f′′) Cytokinesis.
DISCUSSION
EB1 was identified by its ability to interact with APC, a tumor suppressor protein that has been shown to associate with microtubules and promote microtubule assembly in vitro (15). We have examined the subcellular distribution of EB1 by indirect immunofluorescence and confocal microscopy, using mAbs specific for EB1. Our results indicate that EB1 decorates the centrosome and the microtubule cytoskeleton throughout the cell cycle.
Previous studies performed on RKO, a human colorectal cancer cell line, and the mouse fibroblast cell line NIH 3T3 have shown that overexpressed full-length, but not truncated, APC associated to microtubules (11). We analyzed the subcellular distribution of EB1 in SW480, a colon cancer cell line that expresses a carboxyl-terminal deleted form of APC that is unable to interact with either EB1 or microtubules; in these cells EB1 localization to microtubules and the centrosome was preserved (Fig. 1d), demonstrating that the cellular distribution of EB1 does not depend on APC. Although EB1 does not require the carboxyl-terminal domain of APC to associate with microtubules, the converse is not necessarily true: it is possible that APC is linked to microtubules by EB1, or that a third component mediates both EB1 and APC binding to microtubules.
Immunofluorescence and cell fractionation studies have demonstrated that full-length, but not truncated, APC is located in the nucleus (16), suggesting a physiologic connection between APC and cell division. We also detected a prominent EB1 signal in the nucleus (Fig. 2); perhaps EB1 and APC are cotransported to the nucleus. Interestingly, EB1 was concentrated at the midbody during telophase (Fig. 3f), where full-length APC also has been shown to localize after transient overexpression (15).
The localization of EB1 at the microtubule-organizing center suggests a functional or structural role of the protein in microtubule nucleation and polymerization. EB1 localized to growing microtubules after nocodazole removal, suggesting that EB1 may participate in the generation of new polymeric tubulin (Fig. 2c). Whether the involvement of EB1 in microtubule dynamics depends on motor proteins or other microtubule-associated proteins is under investigation. The idea that EB1 may stabilize polymerizing microtubules is supported by the fact that fungal cells lacking EB1-related proteins are sensitive to microtubule-depolymerizing drugs (17, 18).
Using a microtubule copelleting assay, we found that a fraction of the total EB1 pool interacts with microtubules, consistent with our immunofluorescence experiments (Fig. 3). The predicted amino acid sequence of EB1 does not contain motifs common to known microtubule-associated proteins. Either the interaction uses a new binding motif or the EB1-microtubule interaction is indirect; the interaction may require posttranslational modifications of either EB1 or tubulin. The EB1-related proteins therefore may be a novel class of microtubule-associated proteins.
By using separate screening procedures, we showed that homologues of EB1 found in fission yeast, Mal3 (17), and budding yeast, Bim1p, (18) were necessary for normal microtubule function. Cells lacking Mal3 protein were viable but demonstrated an increased rate of loss of a nonessential minichromosome as well as hypersensitivity to the microtubule-destabilizing drug thiabendazole (TBZ). Expression of human EB1 rescued the TBZ-hypersensitivity phenotype of the Mal3 deletion strain. Bim1p was isolated by its interaction with α-tubulin in a yeast two-hybrid screen. Deletion of Bim1p produced a defect in karyogamy, the microtubule-driven process whereby haploid nuclei are fused in zygotes. Additionally, bim1Δ cells displayed hypersensitivity to the microtubule-depolymerizing drug benomyl and aberrant spindle morphology. Overexpression of Mal3 protein compromised spindle assembly and function, and overexpression of Bim1p in wild-type cells was lethal. Mal3 and Bim1p localized in vivo to cytoplasmic and nuclear microtubules, consistent with the cellular distribution of the mammalian protein described here. More recently, a new checkpoint mechanism requiring Bim1p has been shown that allows correction of the nuclear orientation defect of act5 mutants before cytokinesis (19).
Although the interaction of EB1 to microtubules does not depend on expression of wild-type APC, our data do not rule out regulation of EB1 binding by APC or regulation of APC binding by EB1. Together with the yeast data, our findings suggest that EB1 may play a physiological role in microtubule function and may mediate the interaction of APC with the mitotic apparatus, together coordinating the control of the normal growth and differentiation processes in the colonic epithelium.
Acknowledgments
We thank Jen-Yeu Wang and Aime Levesque for technical assistance.
ABBREVIATION
- APC
adenomatous polyposis coli
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hyman A A, Karsenti E. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- 2.Merdes A, Cleveland D. J Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stearns T. J Cell Biol. 1997;138:957–960. doi: 10.1083/jcb.138.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton N R, Goldstein L S. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sawin K E, Endow S A. BioEssays. 1993;15:399–407. doi: 10.1002/bies.950150606. [DOI] [PubMed] [Google Scholar]
- 6.Su L-K, Burrell M, Hill D E, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler K W. Cancer Res. 1995;55:2972–2977. [PubMed] [Google Scholar]
- 7.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–626. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 9.Butner K A, Kirschner M. J Cell Biol. 1991;115:717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goode B L, Feinstein S C. J Cell Biol. 1994;124:769–782. doi: 10.1083/jcb.124.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith K J, Levy D, Maupin P, Pollard T D, Vogelstein B, Kinzler K. Cancer Res. 1994;54:3672–3675. [PubMed] [Google Scholar]
- 12.Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. J Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreis T E. EMBO J. 1987;6:2597–2606. doi: 10.1002/j.1460-2075.1987.tb02550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapin S J, Bulinski J C. Methods Enzymol. 1991;196:254–264. doi: 10.1016/0076-6879(91)96024-l. [DOI] [PubMed] [Google Scholar]
- 15.Munemitsu S, Souza B, Muller O, Albert I, Rubinfel B, Polakis P. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- 16.Neufeld K L, White R L. Proc Natl Acad Sci USA. 1997;94:3034–3039. doi: 10.1073/pnas.94.7.3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beinhauer J D, Hagan I M, Hegemann J H, Fleig U. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz K, Richards K, Botstein D. J Mol Biol. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muhua L, Adames N R, Murphy M D, Shields C R, Cooper J A. Nature (London) 1998;393:487–491. doi: 10.1038/31014. [DOI] [PMC free article] [PubMed] [Google Scholar]