Abstract
The filamentous fungus Neurospora crassa is a model laboratory organism, but in nature is commonly found growing on dead plant material, particularly grasses. Using functional genomics resources available for N. crassa, which include a near-full genome deletion strain set and whole genome microarrays, we undertook a system-wide analysis of plant cell wall and cellulose degradation. We identified approximately 770 genes that showed expression differences when N. crassa was cultured on ground Miscanthus stems as a sole carbon source. An overlap set of 114 genes was identified from expression analysis of N. crassa grown on pure cellulose. Functional annotation of up-regulated genes showed enrichment for proteins predicted to be involved in plant cell wall degradation, but also many genes encoding proteins of unknown function. As a complement to expression data, the secretome associated with N. crassa growth on Miscanthus and cellulose was determined using a shotgun proteomics approach. Over 50 proteins were identified, including 10 of the 23 predicted N. crassa cellulases. Strains containing deletions in genes encoding 16 proteins detected in both the microarray and mass spectrometry experiments were analyzed for phenotypic changes during growth on crystalline cellulose and for cellulase activity. While growth of some of the deletion strains on cellulose was severely diminished, other deletion strains produced higher levels of extracellular proteins that showed increased cellulase activity. These results show that the powerful tools available in N. crassa allow for a comprehensive system level understanding of plant cell wall degradation mechanisms used by a ubiquitous filamentous fungus.
Keywords: cellulase, secretome, transcriptome
Plant biomass, primarily composed of lignocellulose, is widely viewed as a potential feedstock for the production of liquid fuels and other value-added materials (1). However, the principal barriers to production of liquid fuels from lignocellulose are the high costs of pretreatment and conversion of insoluble polysaccharides to fermentable sugars (2). One approach to depolymerization of plant cell wall polysaccharides involves using hydrolytic enzymes produced by bacteria and fungi (3). Toward achieving this goal, the filamentous fungus, Hypocrea jecorina (Trichoderma reesei) has been engineered, largely through random mutagenesis and screening, to produce elevated amounts of cellulases (4).
Neurospora crassa is a well-known model organism that has been used for >90 years to study genetics, biochemistry, and fungal biology (5). Many N. crassa isolates have been recovered from sugar cane, which is closely related to Miscanthus, an attractive crop for biofuel production (6–8). Although it was shown to degrade cellulose >30 years ago (9, 10), relatively little has been reported on plant biomass utilization by N. crassa. The N. crassa genome is predicted to contain twice as many cellulases as H. jecorina (11), as well as many hemicellulases and other enzymes involved in plant biomass degradation. Genetic and molecular tools to manipulate N. crassa are extensive (5) as are genomic resources, including whole genome microarrays and a near-full genome deletion strain set (12). N. crassa is the only example of a model organism that also happens to be a proficient degrader of plant cell walls.
In this study, we exploit functional genomic resources to perform a systems analysis of the N. crassa transcriptome associated with complex plant biomass and pure cellulose utilization. In addition, the secretome of N. crassa grown under identical conditions was analyzed using a shotgun proteomics approach. We evaluated strains containing deletions in genes encoding proteins identified from overlapping transcriptome and secretome datasets for their ability to use cellulose and for cellulase activity. From this analysis, we identified known proteins involved in plant cell wall degradation, but also proteins of unknown function that affect cellulose degradation and cellulase activity. Taken together, these data begin to unravel the functionally distinct strategies used by N. crassa to degrade plant cell walls and highlight how a systems biology approach using genomic resources is a powerful tool to identify industrially important components associated with plant cell wall degradation.
Results
Transcriptome Analysis of N. crassa Grown on Miscanthus and Avicel.
Growth and cellulase activity of N. crassa (FGSC 2489) cultured on minimal medium with crystalline cellulose (Avicel) as the sole carbon source was similar to that of H. jecorina (QM9414) (Fig. S1); N. crassa completely degraded Avicel in approximately 4 days. N. crassa also grew rapidly on ground Miscanthus stems, suggesting functional cellulase and hemicellulase degradative capacity. To determine the transcriptome associated with plant cell wall deconstruction, we used full genome microarrays (13–15) to monitor gene expression profiles during growth of N. crassa on ground Miscanthus stems. RNA sampled from N. crassa grown for 16 h on sucrose was compared to RNA from N. crassa grown on Miscanthus medium at 16 h, 40 h, 5 days, and 10 days (Fig. 1 and Dataset S1, p 1).
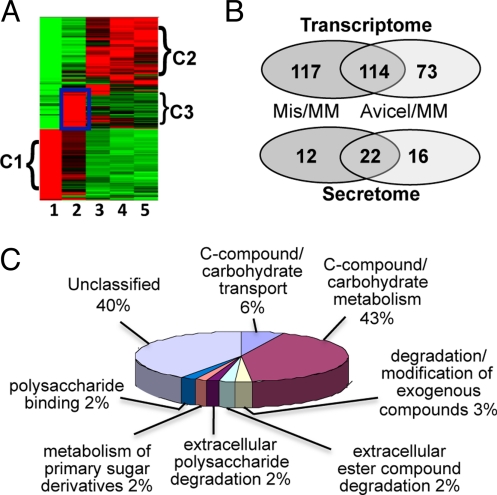
Fig. 1.
Transcriptional profiling of N. crassa grown on Miscanthus and Avicel. (A) Hierarchical clustering analysis of 769 genes showing expression differences in Miscanthus culture. Red indicates higher relative expression and green indicates lower relative expression. Lane 1: A 16 h N. crassa culture grown in sucrose minimal medium. Lane 2: A 16 h culture with Miscanthus as a sole carbon source. Lanes 3–5: Expression profiles from cultures grown on Miscanthus for 40 h, 5 days, and 10 days. The C3 cluster showed increased expression levels of most of the cellulase and hemicellulase genes (boxed). (B) Overlap in expression profiles between the N. crassa Miscanthus versus Avicel grown cultures (Top). Overlap of proteins in culture filtrates detected by tandem mass spectrometry (Bottom). (C) Functional category analysis (16) of the 231 genes that showed a significant enrichment (P < 0.001) in relative expression levels in Miscanthus cultures.
A total of 769 N. crassa genes showed a statistically significant difference in relative expression level among the four Miscanthus samples as compared to the sucrose sample (Dataset S1, p 3). Hierarchical clustering showed that these genes fell into three main clusters (Fig. 1A). The first cluster of genes (C1; 300 genes) showed the highest expression levels in minimal medium with sucrose. Functional category (FunCat) analysis (16) of these genes showed an enrichment for ribosomal proteins and other functional categories associated with primary metabolism (Dataset S1, p 4). The second cluster (C2) included 327 genes that showed the highest expression levels in Miscanthus cultures at later time points (40 h to 10 days; Fig. 1A). Within this group were 89 genes that showed a high relative expression level in Miscanthus cultures at all time points. FunCat analysis (16) of the remaining 238 genes showed one functional category (C-compound and carbohydrate metabolism) was slightly enriched (Dataset S1, p 5).
A third cluster of 142 genes showed the highest relative expression level after 16 h of growth of N. crassa on Miscanthus (C3, Fig. 1A; Dataset S1, p 3). FunCat analysis (16) of these 142 genes plus the 89 genes that showed high expression levels in Miscanthus cultures at all time points (C3+ cluster; total 231 genes) showed an enrichment for proteins involved with carbon metabolism, including predicted cellulases and hemicellulases (Fig. 1C; Dataset S1, p 6). Of the 23 predicted cellulase genes in the N. crassa genome, 18 showed significant increases in expression levels during growth on Miscanthus (Table 1), particularly at the 16 h time point (Fig. S2). Five genes showed an increase in expression level >200-fold {cbh-1 [CBH(I)], NCU07340; gh6-2 [CBH(II)-like gene], NCU09680; gh6–3 (NCU07190), and two GH61 genes (gh61-4; NCU01050 and NCU07898)}.
Table 1.
Predicted cellulase genes in Neurospora crassa
| Gene | GH family | CBM1 | SP | MS | EL Miscanthus | EL Avicel |
|---|---|---|---|---|---|---|
| NCU00762 | 5 | Yes | Yes | Both | 29.6 | 31.5 |
| NCU03996 | 6 | No | No | ND | ND | ND |
| NCU07190 | 6 | No | Yes | Both | 526.0 | 119 |
| NCU09680 | 6 | Yes | Yes | Both | 230.9 | 251.3 |
| NCU04854 | 7 | No | Yes | ND | 32.9 | 10.8 |
| NCU05057 | 7 | No | Yes | Both | 8.7 | 7.4 |
| NCU05104 | 7 | No | Yes | ND | 11.6 | NC7 |
| NCU07340 | 7 | Yes | Yes | Both | 426.4 | 382.2 |
| NCU05121 | 45 | Yes | Yes | avi | 8.6 | 17.2 |
| NCU00836 | 61 | Yes | Yes | ND | 91.2 | 31 |
| NCU01050 | 61 | No | Yes | Both | 206.7 | 382.1 |
| NCU01867 | 61 | Yes | Yes | ND | 2.2 | NC |
| NCU02240 | 61 | Yes | Yes | avi | 193.5 | 84 |
| NCU02344 | 61 | No | Yes | ND | 8.1 | 4.1 |
| NCU02916 | 61 | Yes | Yes | ND | 85.2 | 17.7 |
| NCU03000 | 61 | No | Yes | ND | NC | ND |
| NCU03328 | 61 | No | Yes | ND | 26.4 | 23.8 |
| NCU05969 | 61 | No | Yes | ND | ND | 12.7 |
| NCU07520 | 61 | No | Yes | ND | ND | ND |
| NCU07760 | 61 | Yes | Yes | ND | 3.7 | NC |
| NCU07898 | 61 | No | Yes | Both | 376.3 | 230 |
| NCU07974 | 61 | No | Yes | ND | NC | NC |
| NCU08760 | 61 | Yes | Yes | Both | 107.5 | 44.7 |
GH, glycoside hydrolase; CBM1, carbohydrate binding module; SP, signal peptide prediction; MS, mass spectrometry analysis; EL, relative expression level; ND, not detected; NC, no change.
Plant cell walls are complex structures composed of cellulose microfibrils, hemicellulose, lignin, pectin, cutin, and protein. Thus, we compared expression profiles of N. crassa grown on Miscanthus to expression profiles of N. crassa grown on Avicel, a pure form of crystalline cellulose (Dataset S1, p 2). Over 187 genes showed a significant increase in relative expression level during growth of N. crassa on Avicel. Of these genes, 114 overlapped with the 231 genes in the C3+ cluster (Fig. 1B). FunCat analysis of the 114-overlap gene set showed a clear enrichment for genes predicted to be involved in carbon metabolism (Dataset S1, p 6). Within this gene set, there was a further enrichment for secreted proteins (53 of the 114 gene products). Of the 53 genes, 32 encode predicted proteins with annotation suggesting a role in plant cell wall degradation, while 16 encode putative or hypothetical proteins. The remaining 61 genes encode predicted intracellular proteins, including 10 predicted major facilitator superfamily transporters (NCU00801, NCU00988, NCU01231, NCU04963, NCU05519, NCU05853, NCU05897, NCU06138, NCU08114, and NCU10021) and 23 putative or hypothetical proteins.
Of the 117 genes within the Miscanthus-specific cluster (Fig. 1B), 37 encoded proteins predicted to be secreted. Nine predicted hemicellulases or enzymes related to the degradation of hemicellulose were identified (NCU00710, NCU04265, NCU04870, NCU05751, NCU05965, NCU09170, NCU09775, NCU09923, and NCU09976) (Table 2 and Dataset S1, p 7). The remaining 80 Miscanthus-specific genes encode predicted intracellular proteins, including genes involved in the metabolism of pentose sugars (for example, NCU00891, xylitol dehydrogenase and NCU00643, a predicted arabinitol dehydrogenase), a predicted sugar transporter (NCU01132), and 48 proteins of unknown function.
Table 2.
Predicted hemicellulase genes in N. crassa
| Gene | GH family | CBM | SP | MS | EL Miscanthus | EL Avicel |
|---|---|---|---|---|---|---|
| NCU05924 | 10 | No | Yes | Both | 149.3 | 55.9 |
| NCU08189 | 10 | No | Yes | Both | 94.4 | 39.8 |
| NCU04997 | 10 | Yes | Yes | ND6 | ND | ND |
| NCU07130 | 10 | No | Yes | ND | ND | ND |
| NCU02855 | 11 | No | Yes | Avi | 364.1 | 10.2 |
| NCU07225 | 11 | Yes | Yes | Both | 33.5 | 11.4 |
| NCU08087 | 26 | No | Yes | ND | ND | ND |
| NCU07326 | 43 | No | Yes | Both | 426.6 | 104.5 |
| NCU01900 | 43 | No | No | ND | 26 | 10.03 |
| NCU05965 | 43 | No | Yes | ND | 5.4 | ND |
| NCU09170 | 43 | No | Yes | ND | 16.7 | ND |
| NCU09652 | 43 | No | No | ND | 95.4 | 12.24 |
| NCU00852 | 43 | No | Yes | ND | ND | ND |
| NCU06861 | 43 | No | No | ND | NC7 | NC |
| NCU02343 | 51 | No | Yes | Mis | 174.6 | 6.6 |
| NCU00972 | 53 | No | No | ND | 15.6 | 9.02 |
| NCU09775 | 54 | No | Yes | Mis | 48.3 | ND |
| NCU07351 | 67 | No | Yes | ND | ND | ND |
| NCU05955 | 74 | Yes | Yes | Both | 50.5 | 19.9 |
GH, glycoside hydrolase; CBM, carbohydrate-binding module; SP, signal peptide; MS, mass spectrometry; EL, relative expression level; ND, not detected; NC, no change.
Secretome Analysis of N. crassa Grown on Miscanthus and Avicel.
Lignocellulose degradation by fungi requires the secretion of proteins associated with depolymerization of cell wall constituents (3). To compare with transcriptional profiling data, we analyzed the secretome of N. crassa using a shotgun proteomics approach (Fig. 1B). Supernatants from 3- and 7-day-old Miscanthus and Avicel cultures were digested with trypsin and analyzed by liquid chromatography nano-electrospray ionization tandem mass spectrometry (MS) (SI Materials and Methods); datasets from the 3- and 7-day samples showed no significant difference. Secreted proteins that bound to phosphoric acid swollen cellulose (PASC) were enriched and also analyzed by MS.
A total of 50 proteins were identified with confidence by tandem MS (Dataset S2). There were 34 proteins detected in the Miscanthus grown N. crassa cultures, while 38 proteins were identified from Avicel grown cultures; 22 proteins were detected in both samples. Of these 22 proteins, 21 were predicted to be secreted based on computational analyses and 19 showed increased expression levels in both the Miscanthus and Avicel grown cultures (Dataset S1). The overlap dataset included eight of the 23 predicted cellulases in N. crassa (Table 1). There were also five predicted hemicellulases, a predicted β-glucosidase (gh3-4; NCU04952), five proteins with predicted activity on carbohydrates, and two proteins with unknown function (NCU07143 and NCU05137) (Dataset S2).
There were 16 proteins only identified with confidence in the Avicel culture, and 14 of these were predicted to be secreted (Dataset S2), including two predicted cellulases (gh61-1; NCU02240 and gh45-1; NCU05121), one xylanase (gh11-1; NCU02855), one predicted protease (NCU04205), three other proteins with predicted activity on carbohydrates [NCU08909, NCU05974, and gh30-1 (NCU04395)], three Neurospora-specific proteins of unknown function, and four conserved hypothetical proteins, including one protein with a cellulose-binding domain (NCU09764). Twelve proteins were specific for culture filtrates of Miscanthus cultures and seven of these had predicted secretion signals (Dataset S2). Three of the five predicted intracellular proteins were conserved hypothetical proteins. The remaining two included a predicted glyoxal oxidase (NCU09267, identified from the N. crassa Miscanthus transcriptome) and a nucleoside diphosphate kinase (ndk-1; NCU04202, not identified in the N. crassa transcriptome). The seven proteins predicted to be secreted included three predicted esterases (NCU04870, NCU05159, and NCU08785), two predicted xylanases (GH51; NCU02343 and GH54; NCU09775), a predicted β-xylosidase (gh3-7; NCU09923), and a conserved hypothetical protein (NCU05751).
Many plant cell wall degrading enzymes contain a cellulose-binding module (CBM), which aids in attachment of the enzyme to the substrate (17). Within the N. crassa genome, 19 genes are predicted to encode proteins with a CBM1 domain (18). Sixteen of these genes showed an increase in relative gene expression in Miscanthus-grown cultures (Dataset S1). From the 50 proteins identified by MS, 11 contain a CBM1 domain (Dataset S2). We used PASC to enrich for proteins that bind to cellulose (see Materials and Methods). Nine proteins that bound to PASC from the supernatant of Miscanthus-grown N. crassa cultures and eight proteins from Avicel supernatant were identified by MS; seven proteins were identified in both (Dataset S2). These included NCU00206, a predicted cellobiose dehydrogenase; gh5-1 (NCU00762), a predicted endoglucanase; NCU05955, a predicted GH74 xyloglucanase; gh11-2 (NCU07225), a predicted endoxylanase; cbh-1 (NCU07340); gh61-5 (NCU08760), a predicted endoglucanase, and gh6-2 (NCU09680), a predicted cellobiohydrolase 2.
Characterization of Extracellular Proteins and Cellulase Activity in Strains Containing Deletions in Genes Identified in the Overlap of the Transcriptome/Secretome Datasets.
Of the 22 extracellular proteins detected in both the Miscanthus and Avicel grown cultures, homokaryotic strains containing deletions in 16 genes are available (12). None of these 16 deletion strains have been previously characterized with respect to plant cell wall or cellulose degradation by N. crassa. The 16 deletion strains were grown in media containing sucrose or Avicel as a carbon source. All strains showed a wild-type phenotype on sucrose medium. The total secreted protein, endoglucanase activity, β-glucosidase activity, and aggregate Avicelase activity of Avicel-grown culture filtrates were measured after 7 days and compared to the wild-type strain from which all mutants were derived (Fig. 2 and Table S1). SDS/PAGE of unconcentrated culture supernatants assessed the relative abundance of secreted proteins.
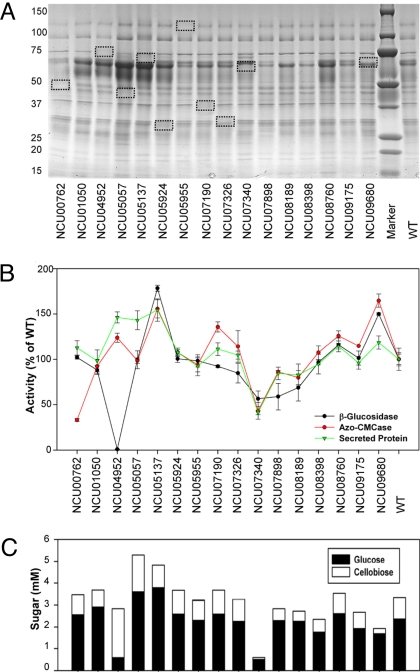
Fig. 2.
Protein profile and enzymatic activity of culture supernatants from strains containing deletions of genes encoding secreted proteins identified by MS. (A) SDS/PAGE of proteins present in the culture filtrates of 16 deletion strains as compared to WT when grown on Avicel for 7 days. Missing protein bands that correspond to the deleted genes are marked with boxes. (B) Total secreted protein, azo-CMCase, and β-glucosidase activity assays performed on 16 deletion strains and the WT parental strain (FGSC 2489) using the same sample from A. (C) Cellulase activity of the culture filtrates from the 16 deletion strains using the same samples as in A. Bars, standard deviation. Glucose (black) and cellobiose (white) were measured after 8 h of incubation at 40 °C. Bars, standard deviation.
There were growth deficiencies on Avicel for strains containing deletions of two predicted exoglucanases, Δcbh-1 (NCU07340) and Δgh6-2 (NCU09680), and a predicted β-glucosidase, Δgh3-4 (NCU04952). The phenotype of the cbh-1 mutant was the most severe; after 7 days in culture much of the Avicel remained, while in the wild-type strain all of the Avicel was degraded. SDS/PAGE of extracellular proteins from 10 of the 16 deletion strains showed an altered protein profile with loss of a single band, allowing assignment of a particular protein band to a predicted gene (Fig. 2A, boxes, and Fig. S3). These included NCU00762 (gh5-1), NCU04952 (gh3-4), NCU05057 (gh7-1), NCU05137, NCU05924 (gh10-1), NCU05955, NCU07190 (gh6-3), NCU07326, NCU07340 (cbh-1), and NCU09680 (gh6-2).
For the majority of the deletion strains, the total secreted protein, endoglucanase, β-glucosidase, and Avicelase activities of the culture supernatants were similar to wild-type (WT) (Fig. 2 B and C and Table S1). Deviations from this trend were seen with Δgh5-1 (NCU00762), Δgh3-4 (NCU04952), ΔNCU05137, Δcbh-1 (NCU07340), and Δgh6-2 (NCU09680). In Δgh5-1 (NCU00762), Δgh3-4 (NCU04952), and Δcbh-1 (NCU07340), Avicelase, endoglucanase, or β-glucosidase activities were lower than the corresponding WT activities. In particular, the deletion of NCU04952 eliminated all β-glucosidase activity from the culture supernatant, as evidenced by PNPGase activity and by higher levels of cellobiose and lower levels of glucose in the Avicelase enzyme assays (Fig. 2 B and C). Unlike a WT strain, culture filtrates from ΔNCU04952 were completely unable to hydrolyze cellobiose to glucose, consistent with loss of β-glucosidase activity. Despite lowering endoglucanase activity, the culture filtrate from Δgh5-1 (NCU00762) showed no significant deficiency in Avicelase activity relative to the WT strain (Fig. 2C). As expected, mutations in cbh-1 (NCU07340) resulted in lower Avicelase activity. A strain containing a deletion of NCU09680, encoding a CBH(II)-like protein (gh6-2), also showed reduced cellobiose accumulation (Fig. 2C).
Mutations in three strains resulted in an increased level of secreted proteins, especially CBH(I) (Fig. 2A); Δgh3-4 (NCU04952), Δgh7-1 (NCU05057), and a hypothetical protein gene, ΔNCU05137. The ΔNCU05137 mutant also showed increased endoglucanase, β-glucosidase, and Avicelase activity (Fig. 2 B and C). NCU05137 is highly conserved in the genomes of a number of filamentous ascomycete fungi, including other cellulolytic fungi, but notably does not have an ortholog in H. jecorina (Fig. S4). It is possible that the increase in CBH(I) levels observed in Δgh3-4, Δgh7-1, and ΔNCU05137 could be due to increased secretion, protein stability or feedback that results in increased expression of cbh-1. To differentiate these possibilities, we compared the profile of extracellular proteins produced by ΔNCU05137 and Δgh3-4 (NCU04952) with gene expression levels of cbh-1 (NCU07340) and gh6-2 (CBH(II); NCU09680) as assayed by quantitative RT-PCR (Fig. S5). The ΔNCU05137 and Δgh3-4 strains showed a higher level of CBH(I) protein as early as 3 days in an Avicel-grown culture and higher expression levels of both cbh-1 and gh6-2 at 3 days of growth, while expression of both of these genes decreased significantly in the WT strain (Fig. S5). Sustained expression of cbh-1 and gh6-2 genes in the ΔNCU05137 and Δgh3-4 mutants could be responsible for the observed increase in CBH(I) and CBH(II) protein levels.
Discussion
Degradation of plant biomass requires production of many different enzymes, which are regulated by the type and complexity of the available plant material (Fig. 3) (19). Here, we report on the systematic analyses of plant cell wall degradation by a cellulolytic fungus, which includes transcriptome, secretome, and mutant analyses. Our profiling data shows that N. crassa coordinately expresses a host of extracellular and intracellular proteins when challenged by growth on Miscanthus or Avicel (Fig. 3). The most highly expressed genes encode proteins predicted to be involved in the metabolism of plant cell wall polysaccharides, many of which were identified by MS analyses. The genomes of filamentous fungi have a large number of predicted glycosyl hydrolases (≈200) with varying numbers of predicted cellulases, from 10 in H. jecorina (11) to 60 in Podospora anserina (20). A comparison between our results and a cDNA expression/Northern analysis of 8 endoglucanases and 7 GH3/β-glucosidases in H. jecorina (21) showed complete overlap with our profiling data, with the exception of one ortholog of a β-glucosidase (cel3e = NCU05577). However, a recent transcriptome/secretome study on the white rot basidiomycete fungus, Phanerochaete chrysosporium (22) showed little overlap. These data suggest that different fungi may use different gene sets for plant cell wall degradation. One aspect all of these studies have in common is the high number of uncharacterized genes/proteins associated with cellulose degradation. Using the functional genomics tools available with N. crassa, we can address both function and redundancy of plant cell wall degrading enzyme systems to create optimal enzyme mixtures for industrial production of liquid fuels from lignocellulose biomass.
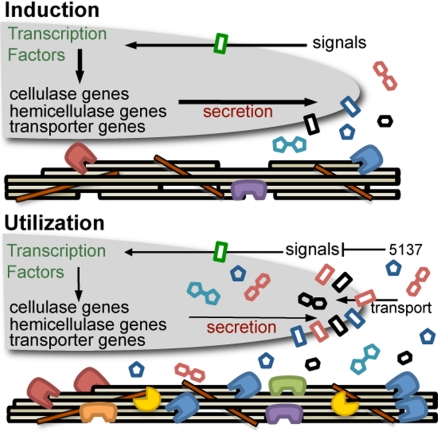
Fig. 3.
Model of plant cell wall deconstruction in N. crassa. Induction: Extracellular enzymes expressed at low levels generate metabolites that signal N. crassa to dramatically increase the expression level of genes encoding plant cell wall degrading enzymes. Utilization: Extracellular enzymes and transporters specific for translocation of cell wall degradation products enable N. crassa to use plant cell material for growth. Some extracellular proteins may generate metabolites that modulate gene expression of cellulases and hemicellulases during the utilization phase. Double red hexagon (cellobiose), double teal pentagon (xylobiose), black hexagon (glucose), and blue pentagon (xylose). Blue, CBH(I); red, CBH(II); purple, EG2; green, EG1; orange, EG6; and yellow, xylanase. Additional cellulolytic enzymes not shown. Thickness of arrows indicates relative strength of response.
N. crassa cellobiohydrolase(I) (CBHI) is the most highly produced extracellular protein during growth on Avicel or Miscanthus, and deletion of this gene causes the most severe growth deficiencies on cellulosic substrates. By contrast, in H. jecorina, deletion of cbhII caused the most severe phenotype (23–25). In N. crassa, deletion of cellobiohydrolase(II) also causes growth deficiencies on cellulosic substrates, but to a much lesser extent than CBH(I), suggesting that exoglucanase activity in N. crassa is predominantly from CBH(I) and that endoglucanases and other CBHs do not compensate for the loss of CBH(I). The three most highly produced endoglucanases are proteins encoded by NCU05057, NCU00762, and NCU07190. These proteins have homology to endoglucanases EG1, EG2, and EG6, respectively. Deletion of these genes did not affect growth on Avicel, although differences in secreted protein levels and endoglucanase activity were observed. Unexpectedly, in the ΔNCU05057 strain, extracellular protein levels were much higher, especially CBH(I). The loss of catalytic activity from NCU05057 may result in compensation by higher expression of the other cellulases, particularly CBH(I), or the products of NCU05057 may, at some level, repress cellulase production. We conclude that no one endoglucanase in N. crassa is required for growth on crystalline cellulose and that different endoglucanases have functional redundancies.
The glycoside hydrolase family 61 enzymes are greatly expanded in N. crassa compared to H. jecorina (11). These enzymes have poorly defined biological function, but their general conservation and abundance in cellulolytic fungi suggests an important role in plant cell wall metabolism. Genes for 10 of the 14 GH61 enzymes were identified in the N. crassa transcriptome, suggesting that these enzymes are used during growth on cellulosic biomass. The four GH61 deletion strains tested showed only small differences compared to wild-type in the secreted protein levels, endoglucanase, and total cellulase activities. However, analyses of additional GH61 mutants and the capacity to create strains containing multiple mutations in N. crassa via sexual crosses will address redundancy and expedite functional analysis of this family.
In addition to predicted cellulase genes, genes encoding hemicellulases, carbohydrate esterases, β-glucosidases, β-xylosidases, and other proteins predicted to have activity on carbohydrates were identified in the N. crassa transcriptome from both Miscanthus and Avicel. Expression of hemicellulase genes even when N. crassa was grown on bacterial cellulose (Fig. S6) indicates that cellulose is the primary inducer of genes encoding plant cell wall degrading enzymes. However, genes encoding some hemicellulases and carbohydrate esterases were only expressed during growth on Miscanthus. Similarly, in other cellulolytic fungi such as H. jecorina and Aspergillus niger, genes encoding some cellulases and hemicellulases are coordinately regulated, while others are differentially regulated (26). As expected, deletions of noncellulase genes had little effect on growth on Avicel or cellulase activity, with the exception of NCU05137. The ΔNCU05137 strain secreted more protein, had higher cellulase activity and showed higher expression of cbh-1 [CBH(I)] and gh6-2 [CBH(II)] than WT. NCU05137 encodes a secreted protein that lacks homology to proteins of known function, but is highly conserved in other cellulolytic fungi (Fig. S4). NCU05137 also has distant homologs of unknown function in a number of bacterial species. We hypothesize that the NCU05137 protein may affect signaling processes associated with the regulation of cellulase gene expression in N. crassa (Fig. 3). Similarly, mutations in gh3-4 (NCU04952) also increased CBH(I) activity. Deletion of Δgh3-4 completely removed PNPGase activity and resulted in cellobiose accumulation in in vitro cellulase assays. These data suggest that NCU04952 encodes the primary extracellular β-glucosidase in N. crassa.
Extracellular degradation of plant cell walls results in the formation of soluble carbohydrates that are subsequently transported into the cell (Fig. 3). In this study, we identified 10 genes encoding permeases/transporters whose expression increased significantly when N. crassa was grown on Miscanthus or Avicel. The major degradation products by cellulases and hemicellulases in vitro are cellobiose, glucose, xylobiose, and xylose. Some of these transporters may be functionally redundant or capable of transporting oligosaccharides. Construction of downstream processing strains capable of transporting oligosaccharides by heterologous expression of N. crassa transporters may improve industrial fermentation of biomass hydrolysis products. None of these transporters or what they may transport has been characterized at the molecular or functional level in any filamentous fungus.
Many genes that increased in expression level during growth on Miscanthus and Avicel encode proteins of unknown function and are conserved in other cellulolytic fungi. By assessing the phenotype of only 16 strains, we identified a mutation in a protein of unknown function that significantly affected cellulase activity. The well understood genetics and availability of functional genomic resources make N. crassa an ideal model organism to assess biological function of these proteins, examine regulatory aspects of cellulase and hemicellulase production, and dissect redundancies and synergies between extracellular enzymes involved in the degradation of plant cell walls.
Materials and Methods
Strains.
The sequenced N. crassa strain (FGSC 2489) was used for transcriptional profiling. Information on strains and growth conditions are in SI Materials and Methods. Stem and leaf tissues from Miscanthus gigantaeus were harvested from field-grown plants at the University of Illinois, Urbana–Champaign (2007) at the end of the growing season, air dried, and milled to a particle size of 100 μm.
Enzyme Activity Measurements.
Total extracellular protein content was determined by a Bio-Rad DC Protein Assay kit (Bio-Rad). Endoglucanase activity in culture supernatants was measured with an azo-CMC kit (Megazyme SCMCL). β-glucosidase activity was measured by mixing 10-fold diluted culture supernatant with 500 μM 4-nitrophenyl β-d-glucopyranoside in 50 mM sodium acetate buffer, pH 5.0, for 10 min at 40 °C (SI Materials and Methods). Avicelase activity was measured by mixing 10-fold diluted culture supernatant with 50 mM sodium acetate, pH 5.0, and 5 mg/mL Avicel at 40 °C. Supernatants were analyzed for glucose content using a coupled enzyme assay with glucose oxidase/peroxidase (SI Materials and Methods). Cellobiose concentrations were determined using a coupled enzyme assay with cellobiose dehydrogenase (CDH) from Sporotrichum thermophile (SI Materials and Methods).
RNA Isolation, Microarray Analysis, and Signal Peptide predictions.
Mycelia were harvested by filtration and flash frozen in liquid nitrogen. Total RNA was isolated using TRIzol (14, 15). Microarray hybridization and data analysis were as described in ref. 14 (SI Materials and Methods). Normalized expression values were analyzed using BAGEL (Bayesian analysis of gene expression levels) (27, 28). Profiling data are available at (www.yale.edu/townsend/Links/ffdatabase/).
Protein Gel Electrophoresis.
Except where otherwise noted, unconcentrated culture supernatants were treated with 5× SDS loading dye and boiled for 5 min before loading onto Criterion 4–15% Tris·HCl polyacrylamide gels. Coomassie dye was used for staining.
Mass Spectrometry and Secretome Analysis.
Trypsin-digested proteins (SI Materials and Methods) were analyzed using a tandem MS connected in-line with ultraperformance liquid chromatography (UPLC). Peptides were separated using a nanoAcquity UPLC (Waters) equipped with C18 trapping (180 μm × 20 mm) and analytical (100 μm × 100 mm) columns and a 10-μL sample loop. The column was connected to a NanoEase nanoelectrospray ionization (nanoESI) emitter mounted in the nanoflow ion source of a quadrupole time-of-flight mass spectrometer (Q-Tof Premier, Waters). Data resulting from LC-MS/MS analysis of trypsin-digested proteins were processed using ProteinLynx Global Server software (version 2.3, Waters). The processed data were searched against the N. crassa database (Broad Institute; http://www.broad.mit.edu/annotation/genome/neurospora/Home.html).
Supplementary Material
Acknowledgments.
We thank Spencer Diamond for technical assistance and Dr. Monica Schmoll for gift of Hypocrea jecorina. Miscanthus gigantaeus was a gift from Drs. Frank Dohleman and Steve Long (University of Illinois at Urbana–Champaign, Urbana, IL). We thank Dr. Raphael Lamed and Chris Phillips for their comments on the manuscript. W.T.B. is a recipient of a National Science Foundation predoctoral fellowship. This work was supported by National Institutes of Health program project Grant GM068087 (to N.L.G.) and a grant from Energy Biosciences Institute to N.L.G. and Drs. John W. Taylor and Tom Bruns (University of California, Berkeley). LC-MS instrumentation was acquired with National Institutes of Health support (1S10RR022393-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906810106/DCSupplemental.
References
- 1.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- 2.Himmel ME, et al. Biomass recalcitrance: Engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 3.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seidl V, et al. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327. doi: 10.1186/1471-2164-9-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis RH, Perkins DD. Neurospora: A model of model microbes. Nat Rev Genet. 2002;3:397–403. doi: 10.1038/nrg797. [DOI] [PubMed] [Google Scholar]
- 6.Perkins DD, Turner BC, Barry EG. Strains of Neurospora collected from nature. Evol. 1976;30:281–313. doi: 10.1111/j.1558-5646.1976.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 7.Pandit A, Maheshwari R. Life-history of Neurospora intermedia in a sugar cane field. J Biosci. 1996;21:57–79. [Google Scholar]
- 8.Smith ML, et al. Vegetative incompatibility in the het-6 region of Neurospora crassa is mediated by two linked genes. Genetics. 2000;155:1095–1104. doi: 10.1093/genetics/155.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberhart BM, Beck RS, Goolsby KM. Cellulase of Neurospora crassa. J Bacteriol. 1977;130:181–186. doi: 10.1128/jb.130.1.181-186.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero MD, Aguado JLG, Ladero M. Cellulase production by Neurospora crassa on wheat straw. Enzyme Microb Technol. 1999;25:244–250. [Google Scholar]
- 11.Martinez D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 12.Dunlap JC, et al. Enabling a community to dissect an organism: Overview of the Neurospora functional genomics project. Adv Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasuga T, Glass NL. Dissecting colony development of Neurospora crassa using mRNA profiling and comparative genomics approaches. Eukaryot Cell. 2008;7:1549–1564. doi: 10.1128/EC.00195-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian C, Kasuga T, Sachs MS, Glass NL. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot Cell. 2007;6:1018–1029. doi: 10.1128/EC.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasuga T, et al. Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res. 2005;33:6469–6485. doi: 10.1093/nar/gki953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruepp A, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linder M, Teeri TT. The cellulose-binding domain of the major cellobiohydrolase of Trichoderma reesei exhibits true reversibility and a high exchange rate on crystalline cellulose. Proc Natl Acad Sci USA. 1996;93:12251–12255. doi: 10.1073/pnas.93.22.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantarel BL, et al. The carbohydrate-active Enzymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouws H, Wattenberg A, Zorn H. Fungal secretomes—Nature's toolbox for white biotechnology. Appl Microbiol Biotechnol. 2008;80:381–388. doi: 10.1007/s00253-008-1572-5. [DOI] [PubMed] [Google Scholar]
- 20.Espagne E, et al. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 2008;9:R77. doi: 10.1186/gb-2008-9-5-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foreman PK, et al. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem. 2003;278:31988–31997. doi: 10.1074/jbc.M304750200. [DOI] [PubMed] [Google Scholar]
- 22.Vanden Wymelenberg A, et al. Transcriptome and secretome analyses of Phanerochaete chrysosporium reveal complex patterns of gene expression. Appl Environ Microbiol. 2009;75:4058–4068. doi: 10.1128/AEM.00314-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seiboth B, Messner R, Gruber F, Kubicek CP. Disruption of the Trichoderma reesei cbh2 gene coding for cellobiohydrolase II leads to a delay in the triggering of cellulase formation by cellulose. J Gen Microbiol. 1992;138:1259–1264. [Google Scholar]
- 24.Suominen PL, Mantyla AL, Karhunen T, Hakola S, Nevalainen H. High frequency one-step gene replacement in Trichoderma reesei. II. Effects of deletions of individual cellulase genes. Mol Gen Genet. 1993;241:523–530. doi: 10.1007/BF00279894. [DOI] [PubMed] [Google Scholar]
- 25.Seiboth B, Hakola S, Mach RL, Suominen PL, Kubicek CP. Role of four major cellulases in triggering of cellulase gene expression by cellulose in Trichoderma reesei. J Bacteriol. 1997;179:5318–5320. doi: 10.1128/jb.179.17.5318-5320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stricker AR, Mach RL, de Graaff LH. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei) Appl Microbiol Biotechnol. 2008;78:211–220. doi: 10.1007/s00253-007-1322-0. [DOI] [PubMed] [Google Scholar]
- 27.Townsend JP. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinformatics. 2004;5:54. doi: 10.1186/1471-2105-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Townsend JP, Hartl DL. Bayesian analysis of gene expression levels: Statistical quantification of relative mRNA level across multiple strains or treatments. Genome Biol. 2002;3:71. doi: 10.1186/gb-2002-3-12-research0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.