Abstract
Stable, single alpha-helix (SAH) domains are widely distributed in the proteome, including in myosins, but their functions are unknown. To test whether SAH domains can act as levers, we replaced four of the six calmodulin-binding IQ motifs in the levers of mouse myosin 5a (Myo5) with the putative SAH domain of Dictyostelium myosin MyoM of similar length. The SAH domain was inserted between the IQ motifs and the coiled coil in a Myo5 HMM construct in which the levers were truncated from six to two IQ motifs (Myo5–2IQ). Electron microscopy of this chimera (Myo5–2IQ-SAH) showed the SAH domain was straight and 17 nm long as predicted, restoring the truncated lever to the length of wild-type (Myo5–6IQ). The powerstroke (of 21.5 nm) measured in the optical trap was slightly less than that for Myo5–6IQ but much greater than for Myo5–2IQ. Myo5–2IQ-SAH moved processively along actin at physiological ATP concentrations with similar stride and run lengths to Myo5–6IQ in in-vitro single molecule assays. In comparison, Myo5–2IQ is not processive under these conditions. Solution biochemical experiments indicated that the rear head did not mechanically gate the rate of ADP release from the lead head, unlike Myo5–6IQ. These data show that the SAH domain can form part of a functional lever in myosins, although its mechanical stiffness might be lower. More generally, we conclude that SAH domains can act as stiff structural extensions in aqueous solution and this structural role may be important in other proteins.
Keywords: ATPase, electron microscopy, optical trap, single alpha helix
Myosins form a superfamily of motor proteins, which are ubiquitous and responsible for a wide range of movement in cells. They all contain three basic components: (i) a motor domain that binds actin and hydrolyzes ATP to generate force, (ii) a lever that contains “IQ motifs” to which light chains (generally calmodulin) bind, and (iii) a tail, which directs the cellular function of each myosin. There are 12 classes of myosins in the human genome, of which 11 are the so-called “unconventional myosins” (1, 2). One of the best characterized of the unconventional myosins is myosin 5a (Myo5). This dimeric myosin has a long lever (six IQ motifs), which allows it to bind to sites on actin 36 nm apart (the spacing of the actin filament helical pseudo-repeat) and thus “walk” straight along actin, taking 36-nm steps (3, 4). Its high duty ratio (3–5) enhances its ability to take several steps along actin before falling off (i.e., to behave processively).
We recently challenged the convention that the myosin lever always consists solely of IQ motifs plus their light chains. We showed that the predicted coiled-coil domain of myosin 10 actually forms a stable single α-helical (SAH) domain, using a synthetic peptide (6). SAH domains lack the hydrophobic seam found in coiled-coil α-helices, and are highly charged (rich in E, K, and R residues), with many (i, i ± 4) and (i, i ± 3) stabilizing intrahelical ionic bonds between either K and E, or R and E (2). We further showed that the lever lengths of expressed myosin 10 monomers, or much rarer dimers, were consistent with that of the three IQ domains present in this myosin plus 15 nm contributed by the adjacent SAH domain (6). By sequence homology, we also proposed that myosins in classes 6 and 7 and a myosin from Dictyostelium discoideum, MyoM, contain a SAH domain rather than coiled coil downstream of their IQ motifs that would increase the size of their powerstrokes, and enable them to take 36 nm steps along actin if dimerized. A SAXS analysis of myosin 6 has recently suggested that its predicted coiled-coil domain is instead consistent with a SAH domain long enough to allow dimeric molecules to take 36-nm steps if artificially dimerized via the inclusion of a leucine zipper (7). The putative SAH domains of myosin 7 and MyoM have not yet been studied, although myosin 7a from Drosophila was recently shown to be monomeric as expressed in Sf9 cells (8, 9).
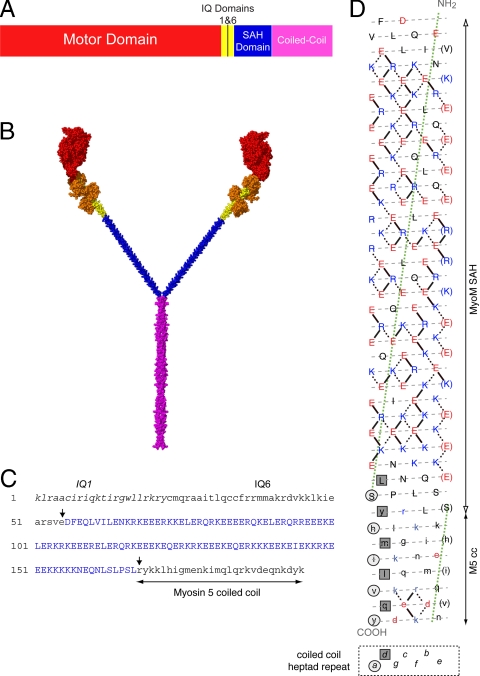
To directly test whether the SAH domain can function as a mechanical lever, we have characterized a chimeric Myo5, in which the lever consists of two IQ motifs from Myo5 and the putative SAH domain from MyoM (Fig. 1). We show this is similar in length to that of the parental, wild-type Myo5–6IQ molecule. A previously characterized Myo5 with only two IQ motifs (Myo5–2IQ) has a powerstroke of only 8.9 nm (cf. 25.2 nm in Myo5–6IQ), and unlike the wild-type Myo5–6IQ, it is processive only at low ATP concentrations (10). The putative SAH domain from MyoM, which has the characteristic pattern of charged residues found in this type of domain (Fig. 1D and refs. 2 and 6), has a predicted length of 16.8 nm (calculated from the number of residues (112), and the rise per residue of an α-helix (0.15 nm). Thus, it is slightly longer than the four deleted IQ motifs (14.4 nm) that it replaces, and the total length of the full-length lever in the Myo5–2IQ-SAH chimera (24 nm) is expected to be slightly longer than in the Myo5–6IQ (21.6 nm). If the SAH domain can function as part of the lever, then the properties of this chimera should be similar to those of Myo5–6IQ, and if not, its properties should be similar to those of Myo5–2IQ.
Fig. 1.
Diagram of the Myo5–2IQ-SAH chimera and the SAH domain of MyoM. (A) The domain arrangement of the chimeric Myo5–2IQ-SAH HMM construct. (B) An atomic model of the chimera molecule shown in spacefill using the same color scheme as in (A) with the addition of bound CaM molecules shown in orange. The construction of this model is described in SI Text. (C) The sequence in the chimera from the start of IQ1 to the start of the coiled-coil domain in Myo5a, to show the boundaries (arrowed) between Myo5a sequence (lowercase) and the inserted MyoM SAH domain (blue, uppercase). The original Myo5–2IQ construct used to generate the chimera has been described in ref. 10. (D) A heptad net representation of the MyoM SAH domain (uppercase) and the first 27 residues of the coiled coil of Myo5 HMM (lowercase). The helix has been cut beside one of the seven 18/1 helices (that would be designated a-g in an α-helical coiled coil) and opened flat to give a 2-D representation of the amino acid residues and their potential interactions in the helix, the N terminus at the top; acidic residues are shown in red, basic in blue, all others in black. The preferred ionic interactions (see ref. 2) are shown as solid lines. In addition alternative/other interactions are shown as dotted lines. Every seventh residue is repeated on the right of the plot (in brackets) so that all interactions can be shown. The gray dashed lines indicate the path of the polypeptide backbone; the green dotted line defines the orientation of the α-helix axis. The light gray circles indicate “a” positions and the dark gray squares indicate “d” positions that form the hydrophobic seam in the coiled coil of myosin 5. The last few residues of the SAH domain (uppercase) could, in principle, form a coiled coil in register with the coiled coil of Myo5 HMM, but there is no hydrophobic seam in the majority of the SAH domain, suggesting that it would not form coiled coil.
Results
Electron Microscopy Reveals the SAH Domain of MyoM.
Electron microscopy, by using rotary or unidirectional shadowing, confirmed that the putative SAH domain of MyoM did form a SAH domain in the GFP-Myo5–2IQ-SAH chimera and not a coiled coil as originally predicted (11, 12). Attempts to visualize the chimera by negative staining were unsuccessful, probably because the SAH domain is too thin to be delineated by stain. The heads of the chimera are strikingly longer than the parent Myo5–2IQ HMM (Fig. 2A) and the SAH domain was revealed as a thin structure between the coiled-coil tail and the head domain of the parent Myo5–2IQ HMM in contrast to the lever of Myo5–6IQ (Fig. 2A). The SAH domain did not become incorporated into the coiled coil, because the measured length of the observed coiled coil matches both that predicted for the number of residues (175) of Myo5 coiled coil included in the chimera and the length previously measured in Myo5–2IQ HMM by this method (Table 1) (6).
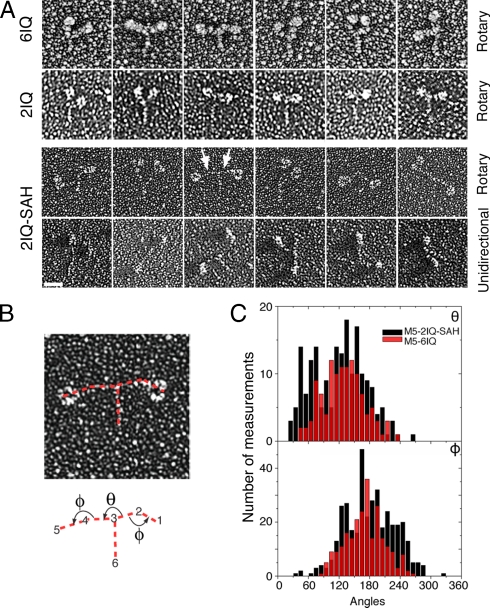
Fig. 2.
Electron Micrographs and analysis of rotary shadowed molecules of the Myo5–2IQ-SAH chimera. (A) A gallery of single molecules of rotary shadowed Myo5–6IQ and Myo5–2IQ, together with those for GFP-Myo5–2IQ-SAH HMM chimera as seen in either rotary or unidirectional shadowing. The arrows indicate the positions of the SAH domains in a molecule of the chimera. (Scale bar, 25 nm.) (B) Diagram of the chimera showing how measurement of the angles between domains was carried out. Red dotted lines indicate the lines between which the angles were measured between the Myo5–2IQ head and the SAH domain (Φ), and between the SAH domains (θ). To measure the angles for each molecule, SPIDER was used to select the tip of the head, head-SAH junction, SAH-SAH junction and the end of the coiled coil in the order shown. The angles were always measured in the anticlockwise direction as indicated by the arrows. (C) Distribution of the angles for the chimera, as indicated in part (B) (black) and for Myo5–6IQ (red).
Table 1.
Average length measurements of chimera molecules from rotary shadowed preparations compared to expected lengths
| Length (nm) | Motor + 2IQ domains | SAH domain | Motor + IQ + SAH domain | Coiled-coil domain |
|---|---|---|---|---|
| Expected | 15.2 | 16.8 | 32.0 | 26.0 |
| Measured | 16.6 ± 2.9 | 17.5 ± 4.6 | 34.1 ± 5.1 | 25.9 ± 6.7 |
n = 376 molecules except for coiled-coil domain measurements, for which n = 188. The motor is expected to be 8 nm long, and 2 IQ domains, 7.2 nm long, making the total expected length of motor + 2IQ to be 15.2 nm. Values shown are mean ± SD.
Measurements of the Myo5–2IQ-SAH chimera from the rotary shadowed images indicate that the SAH domain extends the lever close to that of Myo5–6IQ. The length of the motor domain + 2IQs was 16.6 ± 2.9 nm, while that of the SAH domain was 17.5 ± 4.6 nm (n = 376), consistent with the predicted values of 15.2 and 16.8 nm, respectively (Table 1). Inspection of the individual molecules indicated that the SAH domain itself is fairly rigid with little bending apparent along its length. The mean angle (Φ) between the SAH domain and the head domain was 180° ± 48° (mean ± SD, n = 376) as expected (i.e., an unbroken line between SAH domain and the head). The broad distribution of angles (Fig. 2C) could arise both from some flexibility at the junction between the SAH domain and IQ6 and from different views of the curved Myo5–2IQ head structure (Fig. 1B). Evidence for the latter is the similar breadth of motor-lever angles measured in Myo5–6IQ HMM [170.8° ± 37.1° (n = 274)]. The angle (θ) between the two SAH domains has a broad non-Gaussian distribution centered at ≈120°. This suggests that there is considerable flexibility at the SAH domain-coiled coil junction. The range is broader than that measured for Myo5–6IQ HMM [128.6° ± 40.6° (n = 117)] for the angle between the levers.
Powerstroke Size of Myo5–2IQ-SAH Chimera Is Similar to Wild-Type Myo5.
Optical tweezers measurements showed that the size of the powerstroke for the chimera was close to that of Myo5–6IQ (Fig. 3F) and much larger than that for Myo5–2IQ. The powerstroke for Myo5–2IQ-SAH was 21.5 ± 2.0 nm (mean ± SEM, Fig. 3F) during single (nonprocessive) interactions of Myo5–2IQ-SAH with actin. Myo5–6IQ and Myo5–2IQ have powerstrokes of 25.2 ± 0.6 nm and 8.9 ± 0.3 nm, respectively (10). Thus, the SAH domain has increased the mechanically effective length of the lever. The powerstroke of the chimera also exceeds that of a construct containing four IQ motifs (15.4 ± 0.5 nm) and is larger than that measured for Myo5–6IQ-2Ala, a construct in which two alanine residues were inserted between IQs 3 and 4 of Myo5–6IQ (10.2 ± 0.5 nm) (10). This indicates that the lever in the Myo5–2IQ-SAH chimera is stiffer than that of the Myo5–6IQ-2Ala construct. We conclude that the SAH domain contributes substantially to the mechanical lever of the chimera.
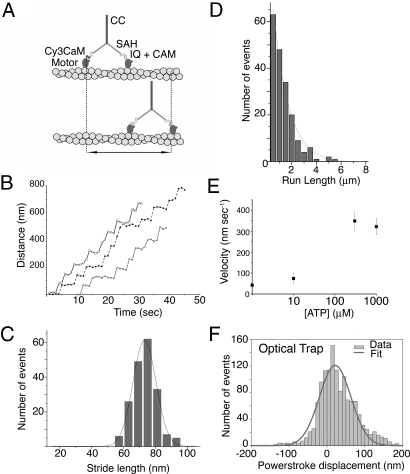
Fig. 3.
Stride length data for Myo5–2IQ-SAH chimera from TIRF microscopy and FIONA, and powerstroke measurements from optical trap assays. (A) Diagram showing the movement that is measured for singly labeled heads, in this assay. (B) Example traces of stride length data for molecules that are labeled by Cy3 calmodulin bound to a single head show that the Myo5–2IQ-SAH chimera is able to move processively along actin. Because movement is stochastic, velocities can vary significantly even within a single run. (C) Histogram of stride lengths fitted with a Gaussian distribution for Cy3 labeled chimera with a label on one head. The mean value is 70.3 ± 7.5 nm (n = 188). Experiments were performed using 300 nM ATP. (D) Run lengths at 1 mM ATP (40 mM KCl) for the Myo5–2IQ-SAH chimera. n = 188, and the run length constant is 1,214 ± 52 nm. The line is an exponential fit to the data. (E) The ATP dependence of velocity of the Myo5–2IQ-SAH chimera on actin in TIRFM assays. n = 40–80 for each measurement. (F) The distribution of powerstroke displacements measured for Myo5–2IQ-SAH chimera in the optical trap, fitted by a Gaussian distribution with a mean value of 21.5 nm. The stiffness of the trap (0.015–0.030 pN·nm−1) implies the stroke is developed against a force of 0.3–0.7 pN. The breadth (S.E.M., 2.0 nm) largely derives from the imposed sine-wave oscillation imposed on the actin filament (see Methods).
Stride Length of Myo5–2IQ-SAH Chimera Is Similar to Wild-Type Myo5.
We next measured the stride lengths of the chimera in in-vitro single molecule motility assays by using TIRF microscopy (TIRFM). Movement of single molecules of the Myo5–2IQ-SAH chimera on actin was visualized by exchanging a Cy3-labeled calmodulin in place of one of the native calmodulins per molecule of the chimera. This shows the stride distance of the labeled head (double arrow, Fig. 3A) rather than the walking step for each head; movement is observed for every second step taken.
We found that the stride length of the Cy3-calmodulin-labeled Myo5–2IQ-SAH chimera (Fig. 3C) was 70.3 ± 7.5 nm (mean ± SD, n = 188). This value is close to that reported in the same type of assay for the Myo5–6IQ (75.3 ± 9.3 nm), and much larger than that for Myo5–2IQ (22.9 ± 6.4 nm) (13). In addition, the distribution of stride lengths (as indicated by the standard deviation) is similar for Myo5–2IQ-SAH and Myo5–6IQ. In contrast, although the stride length in Myo5–6IQ-2Ala was also similar, the distribution of stride lengths was much broader (67.5 ± 20.3 nm) suggestive of increased flexibility (10). Thus, the behavior of the chimera in processive runs is consistent with the powerstroke measurements in showing that the lever in this molecule is stiffer than that in the Myo5–6IQ-2Ala construct.
The results from TIRFM assays also showed that inserting the SAH domain into Myo5–2IQ restored robust processivity as expected if the SAH domain forms part of the lever. At high (1 mM) ATP concentrations, the Myo5–2IQ-SAH chimera moved processively in the TIRFM assay (Fig. 3B), whereas Myo5–2IQ does not and is only weakly processive at low ATP concentrations (10). The run length constant for the Myo5–2IQ-SAH chimera obtained from the exponential fit (Fig. 3D) was 1214 ± 52 nm (n = 818) at 1 mM ATP. This is only slightly lower than that previously measured for Myo5–6IQ (1380 nm) (13). Using the run length constant and average step lengths to calculate the number of steps taken, the Myo5–2IQ-SAH chimera takes 34 steps of 35.2 nm each compared to Myo5–6IQ, which takes 36 steps of 37.7 nm. These values can be compared to those for Myo5–6IQ-2Ala and 4IQ constructs (10), which take 22 or 30 steps, respectively, calculated from their run length constants (738 and 763 nm) and step lengths (33.8 and 25.1 nm). Therefore, the insertion of the SAH domain has substantially restored the processivity of Myo5–2IQ. The velocity of the Myo5–2IQ-SAH chimera increased with ATP concentration (Fig. 3E) as expected, and it saturated at a value of 350 ± 44 nm·s−1 at 300 μM ATP. This equates to ≈10 forward steps per second.
ADP Release from Myo5–2IQ-SAH Chimera on Actin Is Not Gated.
The results from EM, optical trap and TIRFM all indicate that the SAH domain is able to act as a lever in the Myo5–2IQ-SAH chimera. To fully restore function, we might expect that the kinetics of ADP release from the Myo5–2IQ-SAH chimera would be “gated” as found for wild-type Myo5–6IQ HMM. In wild-type Myo5–6IQ HMM, intramolecular mechanochemical gating inhibits the rate of ADP dissociation from the lead head by a factor of >30 when the trail head is bound to actin. This helps the molecule to remain attached to actin under stall forces, and inhibits futile cycles or backwards steps by the lead head (4, 14, 15).
Measurements of the actin-activated ATPase of the Myo5–2IQ-SAH chimera showed that Vmax was 9.3 ± 0.8 s−1·head−1 and the KATPase was 2.2 ± 0.7 μM actin. The value for Vmax compares well to that for Myo5–6IQ HMM (10.5 ± 1.5 s−1·head−1). However, the value for the KATPase is 6- to 7-fold higher than the value measured for Myo5–6IQ HMM (0.32 ± 0.09 μM) and is more similar to the value measured for Myo5 S-1 (3.6 ± 1.0 μM). This suggests that the two heads in the chimera might not be gated by intramolecular strain.
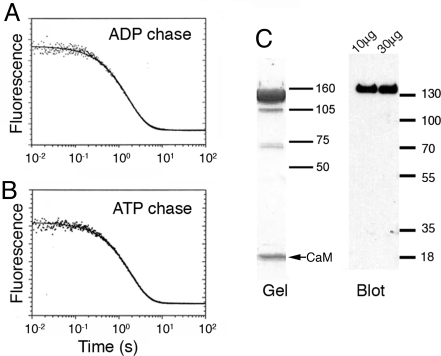
To investigate whether there is gating present during the ATP hydrolysis cycle of the Myo5–2IQ-SAH, we performed double-mixing stopped-flow measurements to determine the rate of ADP release following the powerstroke using deac-aminoATP as the substrate. Regardless of whether ADP or ATP is used as the chase (see Methods), we observed a single exponential transient of similar magnitude with rate constants of 0.58 s−1 using an ADP chase and 0.52 s−1 using an ATP chase for Myo5–2IQ-SAH (Fig. 4 A and B). For comparison, the previously reported transient for Myo5–6IQ HMM was a double exponential yielding rate constants of 0.50 and 0.015 s−1 using an ADP chase, and a single exponential yielding 0.36 s−1 using an ATP chase (15, 16). The results for Myo5–2IQ-SAH indicate that ADP dissociation from the two heads is not gated. The lack of gating did not originate from a large proportion of single-headed molecules due to cleavage during expression or purification, as SDS gels showed a major band for the HMM heavy chain and little evidence of S1 or S2. In addition, Western blots using anti-FLAG (detecting the C terminus of the heavy chain) labeled the HMM heavy chain, with no evidence of the “S2” heavy chain (MW ≈20 kDa) or “SAH-S2” heavy chain (≈32 kDa) expected if the sample contained single-headed HMM (Fig. 4C). The lack of gating is unlikely to arise from binding of the two heads of Myo5–2IQ-SAH to different actin filaments, as there was no evidence of bundling of actin filaments in negative stained specimens of Myo5–2IQ-SAH bound to actin during ATP hydrolysis (Fig. S1).
Fig. 4.
Product dissociation from the acto-Myo5–2IQ-SAH-ADP-Pi complexes. (A) Myo5–2IQ-SAH chimera (0.4 μM) was mixed with 0.5 μM deac-aminoATP, held for 20 s in a delay line, then mixed with hexokinase-glucose treated phalloidin-actin and 2 mM ADP. Experimental conditions: 40 mM KCl, 10 mM Mops, 3 mM MgCl2, 1 mM EGTA, and 1 mM DTT, pH 7.5, 20 °C. Final concentrations in the cell: 0.11 μM Myo5–2IQ-SAH active sites, 0.14 μM deac-aminoATP, 11.1 μM actin, and 1.1 mM ADP. The solid line through the data are the best fit to a single exponential equation: I(t) = 0.079e−0.58t + C. (B) Experimental conditions were similar to those in (A) except that a 2 mM ATP chase replaced ADP: I(t) = 0.080e−0.52t + C. (C) SDS gel and Western blot (using an anti-FLAG antibody) of the Myo5–2IQ-SAH chimera used in these experiments. The major band on the gel and the blot shows that the purified protein mainly consists of full-length molecules. Two different loadings (10 μg and 30 μg) of protein were used in the blot. (Size markers in kDa.)
Discussion
Properties of the Myo5–2IQ-SAH Chimera.
The addition of the MyoM SAH domain into Myo5–2IQ HMM restores its function close to that of the wild-type Myo5–6IQ HMM. Myo5 with a lever that consists of only two IQs is not processive at high ATP concentrations, but inserting the 17 nm SAH domain restores the processive behavior, restores the powerstroke close to wild-type and allows steps along actin with similar lengths to wild-type. We did not observe any marked deformation in the SAH domains of rotary shadowed molecules, suggesting that it is little deformed by thermal forces. We found that the powerstroke size for the chimera is much bigger than that measured for the Myo5–6IQ-2Ala construct when measured in the optical trap under a mild load. Therefore, the lever in the chimera must be stiffer than that in the Myo5–6IQ-2Ala construct, in which two alanine residues inserted into the middle of the lever (between IQs 3 and 4) are known to increase the flexibility of the lever. However, ADP release from the lead head is not gated in the chimera suggesting that the SAH domain is less stiff than a canonical “IQ+calmodulin” lever.
The powerstroke of the chimera (21.5 nm) was somewhat smaller than that of Myo5–6IQ (25.2 nm), however, the bending stiffness of the SAH domain is probably less than that of the IQ-calmodulin lever (≈1,500 pN·nm2) (16). This suggests that the powerstroke size of the chimera will be more sensitive to load than Myo5–6IQ. The bending stiffness of SAH domains has not yet been directly measured, but the SAH domain (≈10 nm) in myosin 6 was predicted to have a persistence length of ≈12 nm (7), and the bending stiffness (EI) of model SAH domains was calculated as 154 pN·nm2, in part from MD simulations (17), thus about an order of magnitude less than the IQ-calmodulin lever. The powerstroke of the chimera was measured at a trap stiffness of 0.015–0.030 pN·nm−1, during which the peak load on the chimeric lever will have been 0.32–0.65 pN. If the lever region approximates to a cantilever bending mode then its stiffness, κ, measured at a distance, L, along its length is given by κ = 3EI/L3. Using EI = 154 pN·nm2 for the 17 nm long SAH domain, its bending stiffness would be 0.1 pN·nm−1, and the peak load would bend it by 3.4–6.9 nm. This is reasonably consistent with the 3.7-nm reduction in powerstroke observed compared to that measured for Myo5–6IQ. Therefore, the evidence indicates that the SAH domain can extend the lever of myosins in a functional way as we proposed earlier (6).
Implications of Ungated ADP Release in the Chimera.
Unlike Myo5–6IQ, ADP release from the lead head in the Myo5–2IQ-SAH chimera was not gated, and yet the chimera can move processively. This suggests that gating is not essential for processivity. It has been suggested that the processivity of motors increases as the duty ratio (i.e., the fraction of the mechanical cycle that the motors spend bound to actin) increases (4). Therefore, the key point is how long the detached head takes to carry out its diffusive search for the next binding site on actin. If this search time is short, then it is less likely that the remaining attached head will detach before the free head rebinds. This search time has been estimated to be ≈10 ms for Myo5–6IQ (4, 18), and the rate constants for ATP hydrolysis on the detached head and for actin activation of phosphate release from the lead head (84 s−1 and 200 s−1, respectively) (19) suggest a similar searching period. In the Myo5–2IQ-SAH chimera, this search time must be short enough for this motor to be processive even at high ATP concentrations. As we found that the powerstroke size of the chimera is similar to that of Myo5–6IQ, then the area over which the free head has to carry out its diffusive search, and therefore the search time should be similar. The median number of steps (34) we estimate per processive run can be used to calculate a duty ratio of 0.855 for the chimera (4). With an ATPase of ≈10 s−1·head−1, this implies an average attachment duration of 85.5 ms and search duration of 14.5 ms.
The observed lack of ADP release gating in the chimera might be expected to reduce its ability to perform processive runs, but run lengths are actually only slightly reduced compared to Myo5–6IQ. In Myo5–6IQ, ADP release between states 2–6 and 3–7 (Fig. 5) is inhibited by strain (short dotted arrows) and the motor is more likely to proceed to state 4, resulting in forward movement. However, in the chimera, in which there is no gating, ADP dissociates at the same rate from trail and lead heads (solid arrows in Fig. 5). As a result moving from state 2, motors will be partitioned equally between states 3 and 6 (solid arrows, Fig. 5). At physiological concentrations of ATP (≈1 mM) the lead head of state 6 will be expected to bind ATP to produce a futile cycle (6→1→2), which does not produce forward movement but hydrolyzes ATP. Such futile cycles can account for the higher ATPase observed (≈10 ATP s−1·head−1) compared to the rate of forward movement in TIRFM assays (≈5 steps s−1 by each head). In a small percentage of the cycles, dissociation of ADP from either the trail head of state 6 or the lead of state 3 would produce state 7 which could be detached by binding ATP to both heads (state 8). The net effect is a small decrease in the run lengths, which is what we observed for the chimera. Thus, the net effect of the lack of gating is not a large reduction in processivity, but in an increase in futile cycles.
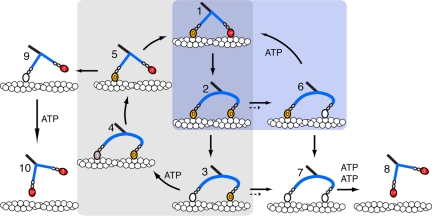
Fig. 5.
Diagram of the principal states of the ATP hydrolysis pathway of actin-Myo5–2IQ-SAH. T = ATP, D = ADP, P = phosphate. In the levers, the 2 IQ motifs with bound calmodulins are shown as ellipses, and the SAH domain by a blue rod, which is shown curved where it is distorted by intramolecular stresses. In wild-type Myo5–6IQ, ADP dissociation from the lead head (converting state 2 to 6, rate constant k2,6) is much slower than from the trail head and the principal pathway is the processive mechanism (1→2→3→4→5→1; gray shaded area) where one step is made for each ATP molecule hydrolyzed. ADP dissociation from the trail head (2→3) is rate limiting and the other steps on the processive path are at least 10-fold faster. Side paths that might lead to dissociation of wild-type Myo5–6IQ occur only rarely because ADP dissociates mainly from the trail head; that is, k2,3 ≫ k2,6 and k3,4 ≫ k3,7. Runs terminate mainly by release of the attached head (5→9→10) during the search phase of the cycle (5→1). For the Myo5–2IQ-SAH chimera, ADP dissociation from the trail and lead heads occurs at the same rate (k2,3 ≈ k2,6) and state 2 thus proceeds equally to states 3 and 6. Dissociation of ADP from the lead head would mostly lead to a “futile cycle” (6→1→2; blue shaded area) in which the lead head would consume a molecule of ATP without producing movement along actin. Processivity would be decreased slightly by the dissociation (7→8) that would occur upon state 7 binding two molecules of ATP, but usually binding of ATP to state 7 would allow a return to state 3 of the cycle. Short dotted arrows between 2 and 6 and between 3 and 7 indicate the slower rate of release of ADP from the lead head of Myo5–6IQ, compared to the chimera (solid arrows). Note that the canonical two IQ lever of the lead head is shown as post-powerstroke in slates 2, 3, 4, 6, and 7, unlike in wild type Myo5-6IQ.
The structural basis of the ungated ADP release in the chimera is not clear. It may be a consequence of the lower bending stiffness of the SAH domain compared to the canonical lever structure. It is estimated that there is 3.6 pN intramolecular stress exerted on the two heads (in opposite directions) when both are attached to actin with ADP in the active site (20). As discussed above this would be sufficient to induce considerable curvature into the two SAH domains, and this could allow the 2IQ lever of the lead head to execute its powerstroke and open the gate on ADP release (Fig. 5). We have so far been unsuccessful in our attempts to test this conjecture by electron microscopy of the chimera-actin complex. For nonmuscle myosin 2, there is strong gating by actin of ADP release. However, our initial EM has shown the two motors attached to adjacent actin subunits with both levers apparently in post-powerstroke orientation (21). Thus, it is not necessary to hold the lead head in a pre-power- stroke conformation to inhibit ADP release in myosin 2, and the structural basis of gating remains obscure.
In conclusion, the replacement of four IQ motifs in the canonical lever of Myo5 by a similar length of SAH domain results in a processive molecule that moves along actin in TIRFM assays and takes similar sized stride length to wild-type Myo5. The lack of ADP release gating in this chimera supports the idea that such gating is not absolutely required for processivity. The optical trap data indicate that the SAH domain augments the working stroke, indicating that it contributes to the movements driven by myosins that contain it. Therefore, myosin levers need to be redefined as the converter plus the IQ motifs/light chains plus the SAH domain, if present. Different myosins take 36-nm steps using different strategies. Myo5 does this by having a long stiff lever composed of six IQ motifs. Myosin 10 dimers may possibly take 36-nm steps because its lever, consisting of three IQ motifs plus a SAH domain, was shown to be approximately the same length as that of Myo5 (6). It remains to be discovered what the relative merits of these two stratagems are, and how they may be important for their cellular roles. More widely, our data indicate that SAH domains in other proteins may act not only as spacers, but also as levers.
Methods
Myosin 5 Chimera.
A chimeric ‘HMM’ like construct was made (Myo5–2IQ-SAH) in which residues D931 to L1042 (112) residues of the predicted SAH domain of Dictyostelium MyoM were inserted downstream of IQ motif 6 in a Myo5–2IQ construct (10) which contains IQ1 and IQ6 (Fig. 1) and upstream to 175 residues of coiled-coil sequence from Myo5 (i.e., between E916 and R917 of the mouse myosin 5a sequence). The construct was C-terminally FLAG tagged, and expressed and purified as described in ref. 10. A second construct was also made in which the coding sequence for eGFP was fused to the N terminus of the chimera (eGFP-Myo5–2IQ-SAH) with the linker sequence TCC GGA CTC AGA TCT CGA GCT CAA GCT TCG AAT TTT GCG GCC GCA AGG CCT encoding 17 aa (SGLRSRAQASNFAAARP) between eGFP and Myo5. The cDNA encoding MyoM was a kind gift from Thierry Soldati (Department of Biochemistry, University of Geneva).
Electron Microscopy.
For metal shadowing EM, the Myo5–2IQ-SAH chimera was typically diluted to between 0.1 and 0.25 mg·mL−1 in 0.5 M ammonium acetate, pH 7.4, 0.5 mM MgCl2, and 50% (vol/vol) glycerol, applied to freshly cleaved mica squares and allowed to deposit for approximately 20 s. Excess sample was wicked off and the mica centrifuged for 1 min at 8,500 × g. The samples were then dried under high vacuum before shadowing. Unidirectional and rotary shadowing used an angle of 1 in 10. After platinum shadowing, samples were coated with carbon. Replicas were floated off onto water and mounted on copper mesh EM grids. Electron micrographs were recorded on a JEOL 1200EX at a nominal magnification of ×30,000 and digitized at a resolution of 0.648 nm/px, calibrated using paramyosin filaments (22).
Optical Trapping.
A dual-bead optical tweezers-based force transducer, as described in refs. 23 and 24, was used to measure the mechanical transitions made by the Myo5–2IQ-SAH chimera using the three-bead assay (25, 26). Details are provided in the SI Text.
TIRF Microscopy.
Stride lengths, velocities and run lengths were obtained from TIRF microscopy data as described in refs. 10 and 13. Solutions used in these experiments were 20 mM Mops, pH 7.4, 5 mM MgCl2, 0.1 mM EGTA, 40 mM KCl, 1 mM ATP (varied for ATP dependence measurements), 1 μM CaM, 50 mM DTT, and an oxygen scavenger system as described in ref. 27.
Transient Kinetic Experiments.
Stopped-flow measurements were performed as described in refs. 19, 28, and 29. Details are provided in SI Text. Solutions for steady state measurements were the same as the in vitro TIRFM assays, except that oxygen scavenger was omitted.
Supplementary Material
Acknowledgments.
We thank Dr. Thierry Soldati (University of Geneva) for the MyoM cDNA used to generate the chimera, Dr. Martin Webb (National Institute for Medical Research) for supplying deac-aminoATP, and Dr. Stan Burgess (University of Leeds) for help with image analysis. This work was supported by Biotechnology and Biological Sciences Research Council BB/C004906/1 (to M.P. and P.J.K.), an Underwood fund grant (to M.P., P.J.K., and J.R.S.), a Wellcome Trust vacation studentship VS/07/LEE/A4 (to P.J.K. and S.M.J.), National Institutes of Health Grant EB00209 (to H.D.W.) and National Institutes of Health/National Institute on Deafness and Other Communication Disorders Grant 5R03DC009335 (to E.F.). J.R.S. was supported by intramural funds from National Heart, Lung, and Blood Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909851106/DCSupplemental.
References
- 1.Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peckham M, Knight PJ. When a predicted coiled coil is really a single α-helix, in myosins and other proteins. Soft Matter. 2009;5:2493–2503. [Google Scholar]
- 3.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 4.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4:59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 5.De La Cruz EM, Wells AL, Rosenfeld SS, Ostap EM, Sweeney HL. The kinetic mechanism of myosin V. Proc Natl Acad Sci USA. 1999;96:13726–13731. doi: 10.1073/pnas.96.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280:34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 7.Spink BJ, Sivaramakrishnan S, Lipfert J, Doniach S, Spudich JA. Long single alpha-helical tail domains bridge the gap between structure and function of myosin VI. Nat Struct Mol Biol. 2008;15:591–597. doi: 10.1038/nsmb.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umeki N, et al. The tail binds to the head-neck domain, inhibiting ATPase activity of myosin VIIA. Proc Natl Acad Sci USA. 2009;106:8483–8488. doi: 10.1073/pnas.0812930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, et al. A FERM domain autoregulates Drosophila myosin 7a activity. Proc Natl Acad Sci USA. 2009;106:4189–4194. doi: 10.1073/pnas.0808682106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto T, et al. Neck length and processivity of myosin V. J Biol Chem. 2003;278:29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 11.Oishi N, Adachi H, Sutoh K. Novel Dictyostelium unconventional myosin, MyoM, has a putative RhoGEF domain. FEBS Lett. 2000;474:16–22. doi: 10.1016/s0014-5793(00)01564-7. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz EC, Geissler H, Soldati T. A potentially exhaustive screening strategy reveals two novel divergent myosins in Dictyostelium. Cell Biochem Biophys. 1999;30:413–435. doi: 10.1007/BF02738122. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto T, Yildiz A, Selvin PR, Sellers JR. Step-size is determined by neck length in myosin V. Biochemistry. 2005;44:16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 14.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279:40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto T, Webb MR, Forgacs E, White HD, Sellers JR. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455:128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilfan A. Elastic lever-arm model for myosin V. Biophys J. 2005;88:3792–3805. doi: 10.1529/biophysj.104.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivaramakrishnan S, Spink BJ, Sim AY, Doniach S, Spudich JA. Dynamic charge interactions create surprising rigidity in the ER/K alpha-helical protein motif. Proc Natl Acad Sci USA. 2008;105:13356–13361. doi: 10.1073/pnas.0806256105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn AR, Spudich JA. Dynamics of the unbound head during myosin V processive translocation. Nat Struct Mol Biol. 2007;14:246–248. doi: 10.1038/nsmb1206. [DOI] [PubMed] [Google Scholar]
- 19.Forgacs E, et al. Switch 1 mutation S217A converts myosin V into a low duty ratio motor. J Biol Chem. 2009;284:2138–2149. doi: 10.1074/jbc.M805530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veigel C, Schmitz S, Wang F, Sellers JR. Load-dependent kinetics of myosin-V can explain its high processivity. Nat Cell Biol. 2005;7:861–869. doi: 10.1038/ncb1287. [DOI] [PubMed] [Google Scholar]
- 21.Kovacs M, Thirumurugan K, Knight PJ, Sellers JR. Load-dependent mechanism of nonmuscle myosin 2. Proc Natl Acad Sci USA. 2007;104:9994–9999. doi: 10.1073/pnas.0701181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott A, Offer G, Burridge K. Electron microscopy of myosin molecules from muscle and non-muscle sources. Proc R Soc Lond B Biol Sci. 1976;193:45–53. doi: 10.1098/rspb.1976.0030. [DOI] [PubMed] [Google Scholar]
- 23.Molloy JE, et al. Single-molecule mechanics of heavy meromyosin and S1 interacting with rabbit or Drosophila actins using optical tweezers. Biophys J. 1995;68:298S–303S. 303S–305S. [PMC free article] [PubMed] [Google Scholar]
- 24.Veigel C, Bartoo ML, White DC, Sparrow JC, Molloy JE. The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys J. 1998;75:1424–1438. doi: 10.1016/S0006-3495(98)74061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 26.Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Movement and force produced by a single myosin head. Nature. 1995;378:209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 27.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 28.Forgacs E, et al. Kinetic mechanism of myosinV-S1 using a new fluorescent ATP analogue. Biochemistry. 2006;45:13035–13045. doi: 10.1021/bi060712n. [DOI] [PubMed] [Google Scholar]
- 29.Forgacs E, et al. Kinetics of ADP dissociation from the trail and lead heads of actomyosin V following the power stroke. J Biol Chem. 2008;283:766–773. doi: 10.1074/jbc.M704313200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.