Abstract
Leukotrienes are now established contributors to the inflammatory process in asthma and leukotriene modifiers are mainstays in the therapy of asthma. This review focuses on published association studies implicating the role of leukotriene pathway genes in asthma pathogenesis and treatment response, specifically focusing on those genetic variants associated with asthma affection status, the development of aspirin exacerbated respiratory disease, and pharmacogenetic response. While published studies have been limited by small sample sizes and lack of independent replication, multiple loci within multiple leukotriene pathway genes have now been associated in more than one study related to asthma or asthma treatment response. Those specific variants include two variants in ALOX5 that are both associated with response to 5-LO inhibition and to leukotriene receptor antagonists, variants in the two established cysteinyl leukotriene receptor antagonists, CYSLTR1 and CYSLTR2 that are both associated with asthma susceptibility in at least two independent populations, and a LTC4S promoter polymorphism that has been associated with asthma affection status and with asthma exacerbated respiratory disease. Desite these successes, genetic investigations into this pathway remain in their formative stages. Future studies aimed at providing a broader scope of investigation through increased sample sizes and through genome-wide approaches are needed.
Keywords: Leukotrienes; SNP; asthma, 5-lipoxygenase; zileuton; montelukast; pharmacogenetics
Introduction
Leukotrienes now have an established or evolving role in a wide variety of inflammatory diseases, including asthma, allergic rhinitis, atherosclerotic cardiovascular disease, inflammatory bowel disease, multiple sclerosis, and cancer (recently reviewed by Werz, et al)1. A genetic basis has been postulated for susceptibility to each of these diseases. Each of these medical conditions is syndromic, i.e., caused by more than one molecular defect. As outlined below there is reason to believe that there is substantial heritability of the leukotriene driven component of asthma and allergy and their response to treatment.
Medications that interrupt the leukotriene pathway have an established role in the therapy of asthma and allergic rhinitis, and have postulated benefits for each of the aforementioned diseases. Leukotriene modifiers are the only orally-administered class of the three most commonly prescribed classes of asthma medications and, thus, are easier for patients to administer than inhaled medications. Currently, two classes of leukotriene modifier drugs have been approved for asthma treatment, the leukotriene Cys-LT1 receptor antagonists (e.g., montelukast, zafirlukast, and pranlukast) and the 5-lipoxygenase (5-LO) inhibitor (zileuton). Among patients with asthma treated with these agents there is substantial inter-individual variability in the therapeutic response; a significant proportion of patients fail to demonstrate a salutary treatment response2. For instance, in one study evaluating the use of inhaled fluticasone vs. oral montelukast in a cross-over clinical trial design, 55% of enrolled asthmatics failed to improve their FEV1 by 7.5% on either medication2. While no heritability studies exist for response to leukotriene modifiers, since the intra-individual response to treatment in patients with asthma is highly repeatable3, a genetic basis for the heterogeneity of this therapeutic response is plausible. The study of the influence of heritable factors on drug treatment response represents one branch of the general field of pharmacogenetics.
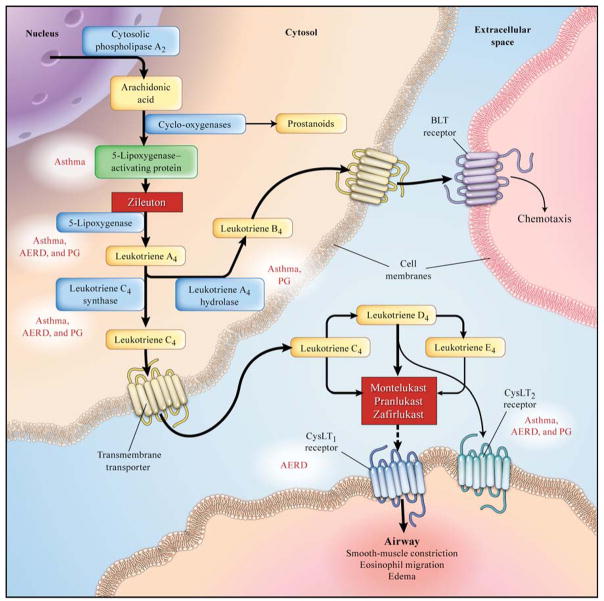
In this review, we shall evaluate the current state of human genetic studies as they pertain to both disease susceptibility and drug treatment response of the leukotriene pathway of asthma. To date, nearly all of the human genetic and pharmacogenetic association studies in these areas have focused on variants in genes that are integral to the leukotriene pathway itself (Figure 1). This includes genes encoding for 5-lipoxygenase activating protein (ALOX5AP), 5-lipoxygenase (ALOX5), leukotriene C4 synthase (LTC4S), leukotriene A4 hydrolase (LTA4H), and the two established receptors for cys-leukotrienes, CYSLTR1 and CYSLTR2 that are known to be involved in asthma and allergy. Following the review of the recent asthma genetic and pharmacogenetic studies and their limitations, we will provide some thoughts on the future of genetics as it relates to leukotriene biology.
Figure 1.
Overview of the leukotriene pathway, demonstrating the key enzymes encoded by genes with variants associated with asthma, aspirin intolerant asthma, or the pharmacologic response to leukotriene modifier therapy. Highlighted boxes in red demonstrate known genetic associations. AERD = association with aspirin exacerbated respiratory disease, PG = pharmacogenetic association. (Modified from Drazen, et al, N Engl J Med, 1999)
Leukotriene Genetics of Asthma Susceptibility
Asthma affects an estimated 300 million individuals worldwide4 and remains the leading disease resulting in childhood school absences and hospitalizations. As noted, asthma has a clear genetic component and has been known to cluster in families for over 3 centuries. However, asthma is a complex genetic disease, in that no single gene is causal by itself. Instead, it likely results from the influence of multiple genetic, environmental, and developmental factors.
Several recent genetic studies have sought to identify variants associated with a global asthma diagnosis. For instance, Kedda, et al5, evaluated an LTC4S promoter polymorphism (-444 A/C) previously associated with aspirin exacerbated respiratory disease (AERD) and asthma severity in an Australian population, comprised of 604 asthmatics and 458 controls. While this variant was not associated with AERD or with asthma severity in their cohort, the variant C allele was nominally associated with asthma affection (minor allele frequency of 0.30 vs. 0.26 for asthmatics vs. controls, respectively) (p = 0.04)5. No formal report of genetic modeling was given, although the raw data do support a possible dominant model of association. In a study focusing on the ALOX5AP and LTA4H genes, Holloway, et al6 noted nominal associations of single nucleotide polymorphisms (SNPs) in both genes (rs10507391 and rs9315050 in ALOX5AP and rs1978331 in LTA4H) in 341 families containing asthmatic probands. However there were additional associations with DNA variants noted and the authors identified a specific risk haplotype for the LTA4H gene with much stronger associations with asthma (p = 0.0006). Although this study did not have a replication population, the effects of the strongest ALOX5AP and LTA4H SNPs appeared to be additive, with an odds ratio of 2.17 (95% CI: 1.41–3.32) of developing asthma.
While the above studies focused primarily on association, Kalayci, et al7 initially interrogated the SP-1 binding motif in the promoter region of the ALOX5 gene for its functional effect, noting that eosinophilic expression of ALOX5 was significantly lower in the individuals homozygous for the variant genotype (in the case of the SP-1 binding motif, five “GGGCGG” repeats are the wild-type). Effects of the promoter genotype were further assessed via measurement of leukotriene C4 directly from the supernatants of the eosinophils, with the authors noting significantly lower leukotriene production related to mutant genotype status (6 ± 1 pg/ml for the homozygous variant allele vs. 21 ± 5 pg/ml for the homozygous wild type). The authors followed their expression work with an association study conducted in 624 asthmatics, relating the microsatellite to asthma severity. Those homozygous for variant SP-1 binding motif alleles were much more likely to be classified as severe asthmatics (univariate p = 0.008). Overall, using a dominant model, any variation in the ALOX5 SP-1 binding motif from the most commonly occurring 5 repeat form was associated with asthma severity (OR 3.65, 95% CI 1.15–11.61).
A second study focusing initially on functional variation was performed by Thompson, et al, who interrogated the coding region of CYSLTR1 for coding variants in a population of Tristan da Cunha subjects8. Two missense mutations, G300S and I206S were identified. Given a relatively high minor allele frequency (~15%) and functional studies demonstrating increased potency of LTD4 on the response of CYSLT1 receptors resulting from the 300S variant (EC50 of 46 nmol/l ± 44.7% vs. 5.6 nmol/l ± 45.8% for wild type vs. 300S variant allele for inositol phosphate accumulation) association studies were performed. Allelic associations revealed a strong association of the variant 300S allele with atopy (p = 0.0001) and with asthma (p = 0.0001), although the latter was confined to women.
Although most genetic studies of asthma susceptibility related to the leukotriene pathway have been limited by the failure to report association in a second, independent, cohort, Pillai, et al9 evaluated asthma susceptibility related to variation in the CYSLTR2 gene in two independent populations. Using family-based data and transmission disequilibrium testing (TDT) testing, a significant undertransmission of the G allele (encoding for Valine) of the coding polymorphism, Met201Val with asthma affection status was noted in a population of 359 families (p = 0.003). This association was replicated by the authors in a second cohort of 384 families participating in the Genetics of Asthma International Network study (p = 0.04). While intriguing due to its non-synonymous nature, the frequency of this SNP was only 0.03 in each population, indicating that very few subjects homozygous for the mutant allele could have been included in their study.
In contrast, in a study of common (allele frequency ≥0.05) coding variants, 129 SNPs located in 105 candidate asthma genes were genotyped by Hong, et al in 170 asthma cases and 347 controls from the city of Anqing, China10. One SNP, rs320995, met criteria for significance following correction for multiple comparisons (nominal p = 0.00004). This synonymous CYSLTR1 SNP was also associated in a second sample of 202 asthma cases and 332 asthma controls, as well as a pooled analysis using three additional studies that had genotyped this CYSLTR1 variant (allelic OR 1.3, 95% CI 1.1–1.5 for the five included studies)10, although none of the other studies met strict criteria for association.
Overall, the weight of the evidence suggests that genetic variants in the leukotriene pathway likely play a relatively minor role, if any, in overall asthma susceptibility. The studies to date have found associations of multiple pathway genes with asthma, yet few report either a strong functional basis for their reported associations or statistical replication of the association in an independent population. The recent studies on CYSLTR1 and CYSLTR2, while compelling due to their replication, are still limited by their low effect size (CYSLTR1) or low risk allele frequency (CYSLTR2). Nonetheless, each of the reported associations is biologically plausible and may yet be confirmed.
The Genetics of Aspirin Exacerbated Respiratory Disease
In addition to the general asthma susceptibility studies, many genetic association studies have focused on the specific relationship of the leukotriene pathway to aspirin exacerbated respiratory disease (AERD). The latter association is due to the known relationship between cyclo-oxygenase inhibition (which aspirin does effectively), the enhaced basal and aspirin induced leukotriene biosynthesis which occurs in this condition and the effectiveness of anti-leukotriene treatment in ameliorating the disorder in experimental settings. (Figure 1). In cases of modest severity, AERD asthmatics demonstrate falls in FEV1 shortly aspirin ingestion. Symptoms begin 1–2 hours after aspirin ingestion, persist for 3–4 hours, and are often also accompanied by tearing, sneezing, itchy eyes, runny nose, and, often, severe nausea and vomiting. Histologically, there is an intense eosinophilic inflammation of the upper and lower airways. Baseline urinary LTE4 excretion is higher in patients with aspirin sensitivity than in control patients with asthma, increases approximately fivefold following aspirin ingestion before returning to baseline as the aspirin response resolves11. While no formal heritability studies have been conducted, a familial clustering of aspirin intolerance has been observed, with one report noting that five of nine siblings developed either urticaria or bronchospasm upon exposure to aspirin12. In addition, since the increase in leukotriene production is a consistent finding in AERD, it is tenable that genetic variation in the leukotriene pathway may lead to an increased susceptiblity to this form of asthma.
Most of the recent studies on the genetics of AERD have been published by a group of investigators in Korea, where the prevalence of this disease in particularly high. These investigators have compiled phenotypic data from over 100 aspirin intolerant asthmatics, comparing them with comparable numbers of aspirin tolerant asthmatics and with normal controls. In their initial study, while they were unable to replicate an association of the aforementioned LTC4S promoter polymorphism (-444 A/C) with AERD, they did note the presence of five common haplotypes in ALOX5 (encoded for by positional variants at −1708[G/A], +21[C/T], +270[G/A], and +1728[A/G], respectively). There was a significant over representation of ALOX5 haplotype 1 [G-C-G-A] in AERD asthmatics (OR 5.0, 95% CI. 1.54–17.9 vs aspirin tolerant asthma and OR 4.5, 95% CI 1.1–18.4 vs. normal controls)13. A follow-up study focused on variants in CYSLTR1. There, a weak association was noted with three promoter SNPs and AERD, but only in males14. An additional analysis focused on the role of CYSLTR215. As compared to the Pillai study8, which focused on a rare coding variant, these investigators focused on common SNPs discovered via resequencing. The DNA sequence analysis of 24 Korean, 50 Caucasian, and 50 African American subjects identified four common CYSLTR2 SNPs that were associated with AERD in the Korean cases vs, controls. Haplotypic analysis revealed an association of a CYSLTR2 haplotype with greater decline in FEV1 following asthma provocation. While none of these studies has been replicated, they imply a mechanistic role for variation in the leukotriene pathway with susceptibility to AERD.
Pharmacogenetics of Leukotriene Modifiers
In the United States in 2007 the societal cost of asthma was approximately $20 billion dollars; the single largest direct expenditure for asthma care remains medication costs. As noted, there are large inter-individual variations in the phenotypic responses to leukotriene modifiers2, with a substantial proportion of patients failing to manifest a salutary therapeutic response when treated. It stands to reason that if it were possible to determine in advance which patients with asthma would have a positive therapeutic response to treatment with leukotriene pathway modifiers, that this information could be of clinical and economic value.
Pharmacogenetics has traditionally been divided into four categories based upon the effects of genetic variability on the pharmacologic properties of a drug. Those categories include variation related to pharmacokinetics, pharmacodynamics, idiosyncratic reactions, and disease pathogenesis. Pharmacokinetics studies the effect of the body upon an administered drug, including consideration of the absorption, distribution, tissue localization, biotransformation, and excretion of drugs. Compared to the other major classes of asthma medications, leukotriene modifiers are uniquely suited to pharmacokinetically based pharmacogenetic studies, since they are administered orally. Pharmacodynamics is the study of the biochemical and physiological consequences of the administration of a drug and its mechanism of action, i.e., the effect of a drug at its therapeutic target. In this category, the genetic variation is typically present at the site of the target or one of the downstream participants in the target’s mechanistic pathway, thereby modulating the effects of the drug. The majority of the asthma pharmacogenetics studies reported to date fall, including most of the leukotriene pathway pharmacogenetic reports, into this category. The idiosyncratic category of pharmacogenetic response to drugs includes the individuals that experience an adverse drug reaction to a therapeutic agent that could not be anticipated based upon its known actions. The final pharmacogenetic category is that of genetic factors influencing disease pathogenesis. Heritable factors that influence the natural history of asthma progression may also affect the ability of an individual to respond to therapy.
As noted, the oral administration of the leukotriene modifiers makes them the only class of commonly prescribed asthma medications with the potential to undergo significant metabolism in the liver, so called “first pass effects”. Thus genetic variants that impact drug metabolism and/or drug transport could impact a drug’s therapeutic effects. Metabolism for both the 5-LO inhibitors and for leukotriene receptor antagonists is largely mediated through the cytochrome p450 system, with CYP2C9 and CYP3A4 responsible for metabolism of both classes (CYP1A2 also plays a role in the metabolism of 5-LO inhibitors). Studies have yet to examine the role of variation in drug metabolizing enzymes to leukotriene modifier response. Nonetheless, it has now been shown that montelukast demonstrates significant inter-individual variability in plasma levels. This variability appears to be mediated at least in part via the organic anion transporter, OATP2B1, which is encoded by the gene SLCO2B1 (solute carrier organic anion transporter family, member 2B1). The non-synonymous SLCO2B1 SNP, rs12422149, which encodes for a change from arginine to glutamine at position 312, has recently been associated with variation in plasma montelukast levels, with heterozygous individuals demonstrating an ~30% reduction in levels vs. those harboring the wild type genotype16. Clinically, there was concordance with the drug level data in that those harboring the variant associated with higher drug levels also had a significant improvement in asthma symptoms one and six months following the initiation of montelukast therapy; these data were obtained retrospectively from a large clinical trial comparing the efficacy of low-dose theophylline, montelukast and placebo16. There were no symptom differences between genotypes pre-randomization. This is the first study indicating that variability in drug levels resulting from genetic differences in drug transport can affect the clinical response to montelukast. This study also supports the prominence that the OATP genes in specific17, and drug transporter genes in general, have gained with regard to clinically meaningful variation in drug pharmacokinetics.
The ALOX5 SP1-binding motif has also been studied closely in asthma pharmacogenetics. In a one of the earliest asthma pharmacogenetic studies, a clinical drug trial of the zileuton-like 5-lipoxygenase inhibitor, ABT-761, asthmatics with at least one wild-type allele had an average improvement in their FEV1 of 19%, while patients with any two of the mutant alleles had an average decrement in their FEV1 of 1% 18.
Lima, et al19, genotyped 28 SNPs in 5 leukotriene pathway genes in subjects participating in a clinical trail comparing montelukast vs. low dose theophylline vs. placebo. The primary analysis was performed on 61 Caucasians taking montelukast. Outcomes included FEV1 change and presence of exacerbations over 1, 3, and 6 months. At 6 months, two SNPs were associated with change in FEV1, one in the MRP1 gene (rs119774) and the second in ALOX5 (rs2115819). Variation in one LTA4H SNP (rs2660845) was associated with a 4-fold increase in the risk of at least one exacerbation over the 6 month follow-up, whereas variation in the aforementioned ALOX5 SP1-binding motif and LTC4S rs730012 were associated with a 73% and a 76% reduction in exacerbations, respectively. The association of non-wild type ALOX5 SP-1 binding motif repeat alleles with increased exacerbations was reported in one additional study20. However, that study, conducted by Telleria, et al20, noted that the wild-type (those homozygous or heterozygouse for at least one copy the 5-repeat promoter polymorphism) was associated with fewer exacerbations, and improved FEV1 outcomes (consistent with the findings of Drazen and colleagues21). While this may appear to be in conflict with the findings of Lima, et al, the latter authors compared homozygous wild types vs. heterozygous wild types in their primary analyses, making generalization difficult. Klotsman, et al22, analyzed 25 SNPs in 10 candidate genes in a population of 174 asthmatics taking montelukast over 12 weeks. Associations with differential change in peak flow rates were noted with two ALOX5 and two CYSLTR2 SNPs.
While most of the recent asthma pharmacogenetics literature has focused on leukotriene receptor antagonists, 5-lipoxygenase inhibitors have gained increasing attention due to the evolving role of the leukotriene pathway in other diseases. Given that these two agents act at different parts of the same pathway, overlap between loci determining response to each of these therapies is plausible. Tantisira, et al23, recently genotyped 26 of the variants in the Lima, et al study19, relating them to longitudinal FEV1 in a population of 577 asthmatics participating in a trial of intermittent zileuton vs. continuous release zileuton vs. placebo. Of the six SNPs associated with zileuton response, two, ALOX5 rs2115819 and ABCC1 rs119774 had been demonstrated by Lima and colleagues to be associated with FEV1 response to montelukast. This suggests that genetic variation within the leukotriene pathway can affect the therapeutic response to multiple agents.
The primary limitations to the leukotriene pharmacogenetic studies to date have paralleled those of asthma affection status - sample size and failure to replicate in an independent population. Each of the leukotriene receptor antagonist studies has reported association results of a single ethnic population contained fewer than 100 individuals (Klotsman, et al, combined two separate clinical trials in their results with only 138 total Caucasians analyzed). And while findings from a montelukast study have been “replicated” in a trial using zileuton, aside from the ALOX5 SP-1 promoter polymorphism, these replications have not been independently confirmed in independent clinical trial populations of either medication. All in all, the data to date suggest that there are likely multiple leukotriene pathway pharmacogenetic loci modulating the therapeutic response to these agents and that ALOX5 likely modulates at least part of the response. Nonetheless, future studies will require both larger sample sizes and replication across multiple additional cohorts before predictive modeling can begin to be considered.
Summary
Genetic studies have indicated a role for variability of genes in the leukotriene pathway in disease susceptibility and medication response. Although the studies to date have been generally limited by small sample sizes and lack of independent replication, multiple loci within multiple genes have now been associated with more than one association related to asthma or asthma treatment response (Table I). This strongly supports a heritable basis involving the leukotriene pathway for these asthma-related phenotypes. While most of the studies have focused on the historical relationship with asthma, many of the association findings from the asthma literature are now being tested in the context of other diseases for which inflammation due to enhanced leukotriene production has been implicated. This has broad implications for future studies focused on understanding the mechanistic basis of genetic variation within the leukotriene pathway. In addition, while most studies to date have focused on a relatively small group of pathway specific genes, future studies in this area will likely include a broader range of genes whose expression is modulated via leukotrienes. These genes may be identified via studies using microarray, integrative genomic, comparative genomic, and genome-wide association approaches. With the addition of reported relationships that require initial replication, it is clear that genetic and pharmacogenetic investigations involving the leukotriene pathway are in their formative stages.
Table I.
Leukotriene Pathway Loci Implicated in Multiple Asthma or Asthma Pharmacogenetic Studies
| Gene | Variant | Associations | Reference |
|---|---|---|---|
| ALOX5 | SP-1 Binding Motif | Asthma Severity | Kalayci, et al7 |
| SP-1 Binding Motif | Response to 5-LO Inhibition | Drazen, et al21 | |
| SP-1 Binding Motif | Response to Leukotriene Receptor Antagonists | Lima, et al19 | |
| SP-1 Binding Motif | Response to Leukotriene Receptor Antagonists | Telleria, et al20 | |
| ALOX5 | rs2115819 | Response to Leukotriene Receptor Antagonists | Lima, et al19 |
| rs2115819 | Response to 5-LO Inhibition | Tantisira, et al23 | |
| CYSLTR2 | Met201Val | Asthma Affection in Two Populations | Pillai, et al9 |
| CYSLTR1 | rs320995 | Asthma Affection in Two Populations, with Meta- analysis in Five Studies | Hong, et al10 |
| LTC4S | -444 A/C promoter | Asthma Affection | Kedda, et al5 |
| -444 A/C promoter | AERD | Kawagishi, et al24 |
Acknowledgments
This work was supported by NIH: U01:HL65899, K23:HG3983, and R01:HL92197.
Abbreviations
- SNP
single nucleotide polymorphism
- 5-LO
5-lipoxygenase
- ALOX5AP
the 5-lipoxygenase activating protein gene
- ALOX5
the 5-lipoxygenase gene
- LTC4S
leukotriene C4 synthase
- LTA4H
leukotriene A4 hydrolase
- CYSLTR1
cysteinyl leukotriene receptor 1
- CYSLTR2
cysteinyl leukotriene receptor 2
- TDT
transmission disequilibrium testing
- AERD
aspirin exacerbated respiratory disease
- SLCO2B1
solute carrier organic anion transporter family, member 2B1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Werz O, Steinhilber D. Therapeutic options for 5-lipoxygenase inhibitors. Pharmacol Ther. 2006;112:701–18. doi: 10.1016/j.pharmthera.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol. 2005;115:233–42. doi: 10.1016/j.jaci.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Drazen JM, Silverman EK, Lee TH. Heterogeneity of therapeutic responses in asthma. Br Med Bull. 2000;56:1054–70. doi: 10.1258/0007142001903535. [DOI] [PubMed] [Google Scholar]
- 4.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–78. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 5.Kedda MA, Shi J, Duffy D, Phelps S, Yang I, O’Hara K, et al. Characterization of two polymorphisms in the leukotriene C4 synthase gene in an Australian population of subjects with mild, moderate, and severe asthma. J Allergy Clin Immunol. 2004;113:889–95. doi: 10.1016/j.jaci.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Holloway JW, Barton SJ, Holgate ST, Rose-Zerilli MJ, Sayers I. The role of LTA4H and ALOX5AP polymorphism in asthma and allergy susceptibility. Allergy. 2008;63:1046–53. doi: 10.1111/j.1398-9995.2008.01667.x. [DOI] [PubMed] [Google Scholar]
- 7.Kalayci O, Birben E, Sackesen C, Keskin O, Tahan F, Wechsler ME, et al. ALOX5 promoter genotype, asthma severity and LTC production by eosinophils. Allergy. 2006;61:97–103. doi: 10.1111/j.1398-9995.2006.00979.x. [DOI] [PubMed] [Google Scholar]
- 8.Thompson MD, Capra V, Takasaki J, Maresca G, Rovati GE, Slutsky AS, et al. A functional G300S variant of the cysteinyl leukotriene 1 receptor is associated with atopy in a Tristan da Cunha isolate. Pharmacogenet Genomics. 2007;17:539–49. doi: 10.1097/FPC.0b013e328012d0bf. [DOI] [PubMed] [Google Scholar]
- 9.Pillai SG, Cousens DJ, Barnes AA, Buckley PT, Chiano MN, Hosking LK, et al. A coding polymorphism in the CYSLT2 receptor with reduced affinity to LTD4 is associated with asthma. Pharmacogenetics. 2004;14:627–33. doi: 10.1097/00008571-200409000-00007. [DOI] [PubMed] [Google Scholar]
- 10.Hong X, Zhou H, Tsai HJ, Wang X, Liu X, Wang B, et al. Cysteinyl leukotriene receptor 1 gene variation and risk of asthma. Eur Respir J. 2009;33:42–8. doi: 10.1183/09031936.00057708. [DOI] [PubMed] [Google Scholar]
- 11.Israel E, Fischer AR, Rosenberg MA, Lilly CM, Callery JC, Shapiro J, et al. The pivotal role of 5-lipoxygenase products in the reaction of aspirin-sensitive asthmatics to aspirin. Am Rev Respir Dis. 1993;148:1447–51. doi: 10.1164/ajrccm/148.6_Pt_1.1447. [DOI] [PubMed] [Google Scholar]
- 12.Settipane GA, Pudupakkam RK. Aspirin intolerance. III. Subtypes, familial occurence, and cross-reactivity with tartarazine. J Allergy Clin Immunol. 1975;56:215–21. doi: 10.1016/0091-6749(75)90092-5. [DOI] [PubMed] [Google Scholar]
- 13.Choi JH, Park HS, Oh HB, Lee JH, Suh YJ, Park CS, et al. Leukotriene-related gene polymorphisms in ASA-intolerant asthma: an association with a haplotype of 5-lipoxygenase. Hum Genet. 2004;114:337–44. doi: 10.1007/s00439-004-1082-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Oh JM, Kim YS, Palmer LJ, Suh CH, Nahm DH, et al. Cysteinyl leukotriene receptor 1 promoter polymorphism is associated with aspirin-intolerant asthma in males. Clin Exp Allergy. 2006;36:433–9. doi: 10.1111/j.1365-2222.2006.02457.x. [DOI] [PubMed] [Google Scholar]
- 15.Park JS, Chang HS, Park CS, Lee JH, Lee YM, Choi JH, et al. Association analysis of cysteinyl-leukotriene receptor 2 (CYSLTR2) polymorphisms with aspirin intolerance in asthmatics. Pharmacogenet Genomics. 2005;15:483–92. doi: 10.1097/01.fpc.0000166456.84905.a0. [DOI] [PubMed] [Google Scholar]
- 16.Mougey EB, Feng H, Castro M, Irvin CG, Lima JJ. Absorption of montelukast is transporter mediated: a common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenet Genomics. 2009;19:129–38. doi: 10.1097/FPC.0b013e32831bd98c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niemi M. Role of OATP transporters in the disposition of drugs. Pharmacogenomics. 2007;8:787–802. doi: 10.2217/14622416.8.7.787. [DOI] [PubMed] [Google Scholar]
- 18.Drazen JM, Yandava CN, Dube L, Szczerback N, Hippensteel R, Pillari A, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22:168–70. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 19.Lima JJ, Zhang S, Grant A, Shao L, Tantisira KG, Allayee H, et al. Influence of Leukotriene Pathway Polymorphisms on Response to Montelukast in Asthma. Am J Respir Crit Care Med. 2005 doi: 10.1164/rccm.200509-1412OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telleria JJ, Blanco-Quiros A, Varillas D, Armentia A, Fernandez-Carvajal I, Jesus Alonso M, et al. ALOX5 promoter genotype and response to montelukast in moderate persistent asthma. Respir Med. 2008;102:857–61. doi: 10.1016/j.rmed.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Drazen JM, Yandava CN, Dube L, Szczerback N, Hippensteel R, Pillari A, et al. Pharmacogenetic association between ALOX5 promoter genotype and the response to anti-asthma treatment. Nat Genet. 1999;22:168–70. doi: 10.1038/9680. [DOI] [PubMed] [Google Scholar]
- 22.Klotsman M, York TP, Pillai SG, Vargas-Irwin C, Sharma SS, van den Oord EJ, et al. Pharmacogenetics of the 5-lipoxygenase biosynthetic pathway and variable clinical response to montelukast. Pharmacogenet Genomics. 2007;17:189–96. doi: 10.1097/FPC.0b013e3280120043. [DOI] [PubMed] [Google Scholar]
- 23.Tantisira KG, Lima J, Sylvia J, Klanderman B, Weiss ST. 5-Lipoxygenase pharmacogenetics in asthma: overlap with Cys-leukotriene receptor antagonist loci. Pharmacogenet Genomics. 2009 doi: 10.1097/FPC.0b013e328326e0b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawagishi Y, Mita H, Taniguchi M, Maruyama M, Oosaki R, Higashi N, et al. Leukotriene C4 synthase promoter polymorphism in Japanese patients with aspirin-induced asthma. J Allergy Clin Immunol. 2002;109:936–42. doi: 10.1067/mai.2002.124466. [DOI] [PubMed] [Google Scholar]