Abstract
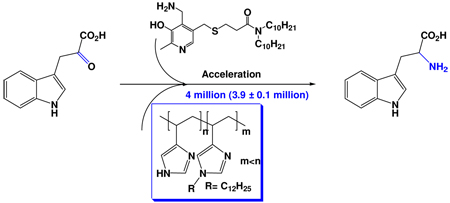
Free radical polymers of 4-vinylimidazole and copolymers with 1-dodecyl-4-vinylimidazole were used as enzyme mimics to transaminate pyruvic acid to alanine, phenylpyruvic acid to phenylalanine, and indole-3-pyruvic acid to tryptophan in water at pH 7.5 and 20 °C, using pyridoxamine carrying hydrophobic sidechains as coenzyme mimics. The best enzyme mimic accelerated the transamination of indole-3-pyruvic acid by 4-million fold, relative to the rate without the polymer, a higher rate ratio than we had previously achieved with a polyaziridine-based enzyme mimic. The properties of various polyvinylimidazoles were compared, including those prepared with the RAFT modification of the polymerization process.
We have studied enzyme mimics for many years,1 including those that performed transamination of ketoacids to amino acids. Our systems normally focused on the first step, the reaction of a pyridoxamine with the ketoacid to form a pyridoxal and the amino acid, although we have also shown how this can be part of a complete catalytic cycle by using decarboxylative transamination of 2,2-disubstituted glycines to convert the pyridoxal back to the pyridoxamine form.2 In our recent work we used polyaziridines to mimic the transaminase enzyme itself.3 Klotz,4 Suh5 and Kirby6 had used those polymers to mimic hydrolytic enzymes.
We used commercially available polyaziridines, as did Klotz, Suh and Kirby, and compared the results with different sizes of the polymers and with different N-alkylations of the nitrogens using alkyl groups from methyl to dodecyl, and with different amounts of such alkylations. We used either covalently attached pyridoxamine derivatives or, even better, reversibly bound pyridoxamines with hydrophobic sidechains as coenzyme mimics.7 We saw that the most effective polymers had a limited amount of dodecylation; with enough we achieved a hydrophobic core in the polymer, while with too much the materials were insoluble in water.
We used water as the solvent to take advantage of hydrophobic binding of substrates and cofactors into the polymer, and to take advantage of the higher rates achieved when the reaction was performed in an interior non-aqueous core of the polymer, as with natural enzymes. We achieved a rate increase as much as 350,000 when we compared the rate of transamination of hydrophobic phenylpyrucvic to form phenylalanine compared with the same process and same pyridoxamine in water without the polymer. An even higher acceleration was observed with indolepyruvic acid to form tryptophan.
We also used polyaziridines with thiazolium and with imidazolium coenzyme mimics carrying hydrophobic sidechains to catalyze the benzoin condensation of two benzaldehyde molecules in water.8 In the transaminase mimics the polymer furnished general acid and general base catalysis of the reaction using the partially protonated main chain amino groups. However, the benzoin condensation was not subject to general acid or base catalysis, but simply to electrostatic catalysis by positive charges on the amino groups of polyaziridines. In this report we will describe the contrasting results and even better catalyses of the transamination reactions using a polymer series different from the polyaziridines.
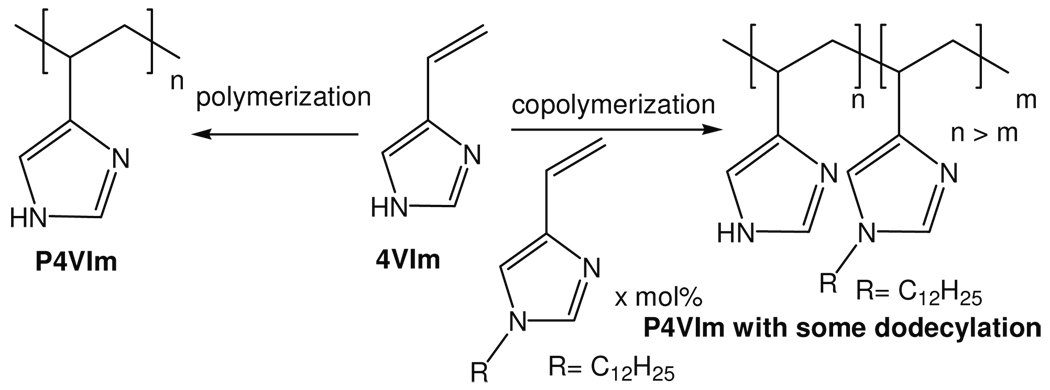
Overberger9 has studied polyvinylimidazoles (PVIm), from radical polymerization of either 4-vinylimidazole or 1-vinylimidazole. We were attracted to these compounds since they also incorporate base (imidazole) and acid (imidazolium) catalytic species, and have sidechains large enough to have some hydrophobic binding character of their own. We synthesized the polymer of 4-vinylimidazole (Scheme 1) according to the Overberger procedure (see Supporting Information) and characterized the methanol soluble fraction by MALDI-TOF mass spectroscopy. Using 1 mol% AIBN for the polymers listed in Table 1 (except for 5 mol% AIBN for the polymers in entries 9 and 10 of the table, as noted) we obtained a polymer mixture ranging from 20 to 40 mers, along with some methanol insoluble larger polymers (for the mass spectroscopy plot, see Supporting Information) and used the entire polymer mixture to catalyze the reaction of pyridoxamine (A) (Scheme 2) with pyruvic acid to form alanine and pyridoxal in water. The rate of the reaction at pH 7.5 and 20 °C was followed by the appearance of the u.v. spectrum of the product pyridoxal (see table 1 for the data and the reaction conditions), as we have described previously.2 We saw that the reaction was 100-fold faster with a 37.5 mg/L PVIm mixture (entry 2) than without the polymer (entry 1).
Scheme 1.
Synthesis of P4VIm and partially dodecylated P4VIm
Table 1.
Kinetic study of transamination reaction under various conditionsa
| entry | pyridoxamine | polymer | ktransamination (min−1) | krelative |
|---|---|---|---|---|
| 1 | A | no polymer | (1.4 ± 0.3) × 10−6 | 1.00b |
| 2 | A | P4VIm | (1.4 ± 0.2) × 10−4 | 100 |
| 3 | B | P4VIm | (7.0 ± 0.2) × 10−4 | 500 |
| 4 | C | P4VIm | (1.3 ± 0.1) × 10−2 | 9,300 |
| 5 | D | P4VIm | (3.3 ± 0.2) × 10−2 | 23,600 |
| 6 | E | P4VIm 0% dodecylated | (1.7 ± 0.1) × 10−1 | 121,400 |
| 7 | E | P1VIm 0% dodecylated | (31.2 ± 0.1) × 10−3 | 22,300 |
| 8 | E | PEI 0% dodecylated | (1.2 ± 0.1) × 10−2 | 8,600b |
| 9 | E | P4VIm 0% dodecylatedc | (1.4 ± 0.1) × 10−1 | 100,000 |
| 10 | E | P4VIm 0% dodecylatedd | (1.5 ± 0.1) × 10−1 | 107,200 |
| 11 | E | P4VIm 2.3% dodecylated | (1.2 ± 0.1) × 10−1 | 85,700 |
| 12 | E | P4VIm 4.5% dodecylated | (1.3 ± 0.1) × 10−1 | 92,900 |
| 13 | E | P4VIm 10.1% dodecylated | (1.1 ± 0.1) × 10−1 | 78,600 |
| 14 | E | P4VIm 14.8 % dodecylated | (6.9 ± 0.1) × 10−2 | 49,300 |
| 15 | E | P4VIm 30 % dodecylated | - | - |
Reaction conditions: 1.5 × 10−4 mol/L pyridoxamine, 37.5 mg/L polymer, 5.0 × 10−3 mol/L pyruvic acid, 2.0 × 10−3 mol/L EDTA, T = 20 °C, pH = 7.5.
Data from ref # 2.
polymerization was done with 5 mol% of AIBN instead of the normal 1 mol%.
polymerization was done with 5 mol% of AIBN instead of the normal 1 mol% and 8 mol% chain transfer agent (CTA) reagent.
Scheme 2.
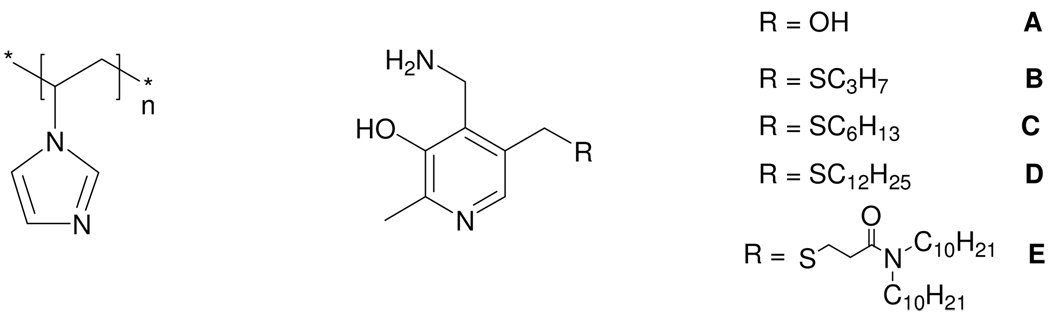
Structures of P1VIm (entry 7) and pyridoxamine derivatives
We then turned to the use of hydrophobic derivatives of pyridoxamine, and to copolymers of 4-vinylimidazole with 1-dodecylated-4-vinylimidazole (obtained from 4-vinylimidazole and dodecyl iodide. The NMR spectrum indicates that is 83% 4-vinyl-1-dodecylimidazole and 17% 5-vinylimidazole.). We prepared the derivatives of pyridoxamine (B–E) (Scheme 2) as we have described previously,2 and examined them with simple PVIm (entries 3–6) and also used cofactor E with copolymers of 4-vinylimidazole and its dodecylated monomer (entries 11–15) in Table 1. We also examined the use of the RAFT modified radical polymerization of 4-vinylimidazole, which normally affords a more monodisperse polymer, and saw by MALDI-TOF that this changed somewhat the distribution of polymer but did not lead to significantly less polydispersity. The modified (entry 10) and the unmodified polymer (entry 9) had similar rates with cofactor E. We found that simple PVIm with cofactor E was 14-fold more effective as a catalyst (entry 6) than was our polyaziridine without dodecyl groups under the same conditions (entry 8). The poly-1-vinylimidazole was also prepared (see SI) and the transamination rate was determined to be 22,300-fold (entry 7) which is only 2.6-fold more effective than polyaziridine (entry8).
Our copolymers with dodecyl groups are more effective with phenylpyruvic acid than with simple pyruvic acid. This reflects the better binding of a hydrophobic substrate into the more hydrophobic polymer. The reaction of phenylpyruvic acid was too rapid to follow by the u.v. method under our conditions, so we determined the selectivity of a competition reaction with 30:30:1 of (phenylpyruvic acid) to (pyruvic acid) to the pyridoxamine derivative forming a phenylalanine/alanine ratio that was determined by HPLC, as previously.2 We observed 3/1 for the reaction using simple PVIm and (19 ± 1)/1 for reaction with the copolymer having 4.5% of dodecylated monomer. The latter is slightly higher than the 14/1 ratio we had seen with the dodecylated polyaziridine previously.2 Since amination (entry 12) by this imidazole copolymer was 92,900-fold more rapid than was reaction without polymer, and since we had shown previously that simple pyridoxamines aminate pyruvic acid and phenylpyruvic acid at the same rate,10 this means that the acceleration of amination of phenylpyruvic acid by the polymer with cofactor E is (1.77 ± 0.1) × 106, almost two million-fold. The amination of indolepyruvic acid, forming tryptophan, was accelerated under the same conditions by four million fold (3.9 ± 0.1 × 106 fold).
The use of these polyvinylimidazoles in the benzoin condensation with thiazolium and imidazolium coenzyme mimics is under investigation. We are also pressing forward the attempt to produce polymers with smaller polydispersity, including the use of oxazoline ionic polymerization as we have used previously with phenylalanine derivatives.11 Since those ionic polymers are not only almost monodisperse but also isotactic, retaining the chirality of the parent amino acids, analogs derived from histidine could well be polyimidazoles with useful chiral selectivities for products and starting materials.
Supplementary Material
Acknowledgment
We thank the NIH and NSF for financial support of this work. Dr. R.S. has an FQRNT (Fonds Québécois de la Recherche sur la Nature et les Technologies) postdoctoral fellowship. We thank Dr. Mary Ann Gawinowicz from the Protein Core Facility at Columbia University College of Physicians & Surgeons for the MALDI-TOF experiments.
Footnotes
Supporting Information Available: Synthesis procedures, pertinent spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Breslow R, editor. Artificial Enzymes. Weinheim, Germany: Wiley-VCH; 2005. [Google Scholar]
- 2.Liu L, Zhou WJ, Chruma J, Breslow R. J. Am. Chem. Soc. 2004;126:8136. doi: 10.1021/ja048671a. [DOI] [PubMed] [Google Scholar]
- 3.(a) Zhou WJ, Liu L, Breslow R. Helv. Chim. Acta. 2003;86:3560. [Google Scholar]; (b) Liu L, Breslow R. Bioorg. Med. Chem. 2004;12:3277. doi: 10.1016/j.bmc.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 4.Delaney EJ, Wood LE, Klotz IM. J. Am. Chem. Soc. 1982;104:799. [Google Scholar]
- 5.Suh J. Synlett. 2001:1343. and references therein. [Google Scholar]
- 6.Hollfelder F, Kirby AJ, Tawfik DS. J. Org. Chem. 2001;66:5866. doi: 10.1021/jo015723v. [DOI] [PubMed] [Google Scholar]
- 7.(a) Liu L, Breslow R. J. Am. Chem. Soc. 2002;124:4978. doi: 10.1021/ja025895p. [DOI] [PubMed] [Google Scholar]; (b) Liu L, Rozenman M, Breslow R. J. Am. Chem. Soc. 2002;124:12660. doi: 10.1021/ja028151k. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H, Foss FW, Breslow R. J. Am. Chem. Soc. 2008;130:12590. doi: 10.1021/ja804577q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Overberger CG, Vorcheheimer N. J. Am. Chem. Soc. 1963;85:951. [Google Scholar]; (b) Overberger CG, Corett R, Salamone JC, Yaroslavsky S. Macromolecules. 1968;1:331. [Google Scholar]; (c) Overberger CG, Shen C-M. Bioorg. Chem. 1971;1:1. [Google Scholar]; (d) Overberger CG, Glowaky RC, Vandewyer PH. J. Am. Chem. Soc. 1973;95:6008. [Google Scholar]
- 10.Breslow R, Hammond M, Lauer M. J. Am. Chem. Soc. 1980;102:421. [Google Scholar]
- 11.Bandyopadhyay S, Zhou WJ, Breslow R. Org. Lett. 2007;9:1009. doi: 10.1021/ol0630604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.