Abstract
The production of optically pure d-lactic acid via xylose fermentation was achieved by using a Lactobacillus plantarum NCIMB 8826 strain whose l-lactate dehydrogenase gene was deficient and whose phosphoketolase genes were replaced with a heterologous transketolase gene. After 60 h of fermentation, 41.2 g/liter of d-lactic acid was produced from 50 g/liter of xylose.
The production of d-lactic acid as well as l-lactic acid is of significant importance for the practical application of polylactic acid, which is an important raw material for bioplastics that can be produced from biomass. Lignocellulose, one of the most abundant forms of biomass, is inedible and contains mainly glucose that is available for microbial lactic acid fermentation and xylose that is unavailable as a nutritional resource for most microorganisms (11). Therefore, for efficient utilization of lignocellulose as a raw material for product development, it is also necessary to develop methods for the efficient utilization of xylose.
Some lactic acid bacteria (LAB) such as Lactobacillus pentosus (1), Lactobacillus brevis (2), and Leuconostoc lactis (7) are known to ferment xylose. However, without exception, all of these strains produce both lactic acid and acetic acid. In these microorganisms, xylose is converted into lactic and acetic acids by the following enzymatic reactions (11). Xylose is first isomerized by xylose isomerase (XylA) to xylulose, which is phosphorylated to xylulose-5-phosphate (X5P) by xylulokinase (XylB). Through the phosphoketolase pathway (PK pathway), X5P is finally converted to lactic acid and acetic acid in a process known as hetero-lactic acid fermentation (11).
Tanaka et al. (11) have shown that, in contrast to the above-mentioned LAB, the Lactococcus lactis IO-1 strain has a second pathway for pentose assimilation in addition to the PK pathway, namely, the pentose phosphate pathway (PP pathway). Due to the presence of the PP pathway, IO-1 produces only l-lactic acid. We had previously disrupted the PK pathway by deletion of the phosphoketolase 1 gene (xpk1) in an l-lactate dehydrogenase gene (ldhL1)-deficient L. plantarum NCIMB 8826 strain which produces optically pure d-lactic acid (10) and introduced the PP pathway by introduction of the transketolase gene from Lactococcus lactis IL 1403 (tkt) into this strain. The resulting mutant, L. plantarum ΔldhL1-xpk1::tkt, successfully produces only lactic acid (exhibiting homo-lactic acid production) from arabinose that is metabolized via an intermediate compound, X5P (3), which is also an intermediate compound in xylose conversion (9). Therefore, the goal of this study was to achieve homo-d-lactic acid production from xylose by expanding the uses of the L. plantarum ΔldhL1-xpk1::tkt strain.
The bacterial strains, oligonucleotides, and plasmids used in this study are listed in Table 1. The L. plantarum NCIMB 8826 ΔldhL1 strain (10) and its derivatives were grown at 37°C in MRS broth (Difco Laboratories, Detroit, MI) or MRS broth containing 25 μg/ml erythromycin. The plasmid used for xylose assimilation was constructed as follows. The xylAB operon that stretches from the start codon of the xylA gene to the stop codon of the xylB gene was amplified by PCR using xylA_F and xylB_R primers from the genome of L. pentosus NRIC 1069. The amplified fragment was then digested with EcoRV and XhoI and ligated into the expression vector pCU (8). The resulting plasmid for xylose assimilation was designated pCU-PXylAB and was introduced into L. plantarum ΔldhL1, L. plantarum ΔldhL1-Δxpk1 (9), and L. plantarum ΔldhL1-xpk1::tkt strains by electroporation as described previously (6). The resulting transformants were designated L. plantarum ΔldhL1/pCU-PXylAB, L. plantarum ΔldhL1-Δxpk1/pCU-PXylAB, and L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB, respectively.
TABLE 1.
Strains, plasmids, and oligonucleotide primers used in this study
| Strain, plasmid, or primer | Relevant phenotype and/or description or sequence (5′-3′)a | Source or reference |
|---|---|---|
| E. coli strains | ||
| NovaBlue | endA1 hsdR17(rK-12− mK-12+) supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB+lacIqZΔM15::Tn10 (Tetr)]; host for pCU DNA manipulation | Novagen |
| VE7108 | Carries kanamycin resistance marker; contains the wild-type repA plasmid gene (not thermosensitive); host for pG+host9 DNA manipulation | 10 |
| VE6838 | Carries kanamycin resistance marker; VE7108 carrying pG+host9 | 10 |
| L. plantarum strains | ||
| Wild type | NCIMB 8826 wild-type strain | NCIMB |
| ΔldhL1 strain | NCIMB 8826 ΔldhL1 | 16 |
| ΔldhL1-Δxpk1 strain | NCIMB 8826 ΔldhL1 Δxpk1 | 15 |
| ΔldhL1-xpk1::tkt strain | NCIMB 8826 ΔldhL1 strain with xpk1 gene replaced with tkt | 15 |
| ΔldhL1-xpk1::tkt-Δxpk2 strain | NCIMB 8826 ΔldhL1 Δxpk1::tkt Δxpk2 | This study |
| L. pentosus NRIC 1069 | NRIC 1069 wild-type strain; source of xylAB gene | NRICb |
| Plasmids | ||
| pSECE1 | Wide-host-range vector; carries erythromycin resistance marker | 19 |
| pCU | Vector under clpC UTLS promoter control; derivative of pSECE1 | 14 |
| pCU-PXy1AB | Vector for assimilation of xylose; carries erythromycin resistance marker | This study |
| pG+host9 | Thermosensitive replicon; carries erythromycin resistance marker | 9 |
| pGh9-Δxpk2 | Vector for deletion of xpk2 gene of ΔldhL1-xpk1::tkt strain; carries erythromycin resistance marker | This study |
| Oligonucleotide primers | ||
| xy1A_F | CCCGATATCATGACAAACGAATATTGGCAAGGC | |
| xy1B_R | CCGCTCGAGTTAGTGTTGTTTCCTTTGTTCCAATAG | |
| xpk2-up_F | CCGCTCGAGTACTAAAACCAGATGCAAATAAATGCAA | |
| xpk2-up_R | GGAAGATCTATATTTGTTTTATTTATCATTACGGTTATTATGATAGC | |
| xpk2-down_F | GGAAGATCTGAGCTCAGTTTTAATCAATAATTCAGCGTAAATTAAAAGC | |
| xpk2-down_R | GGACTAGTGAGACGAATCCCCATAAAAGCC | |
| xpk2-up_seq | ATTTTGGCTGATCATACCAAATTCG | |
| xpk2-down_seq | TGAAAAAGAAACGGAATTATCAACTATTATTTA | |
| xpk2_2208-2226bp_F | GCCATTCGACATGCCTGTT | |
| xpk2_2268-2288bp_R | GCATAAGCTGGCGTTGCTAAT | |
| ldhD_676-696bp_F | GATGGCGCCTACATCTTGAAC | |
| ldhD_731-753bp_R | GCCACTGTCTAAGGCTTTGATCA |
For primers, restriction enzyme sites are underlined. UTLS, untranslated leader sequence.
NRIC, Nodai Research Institute Culture Collection.
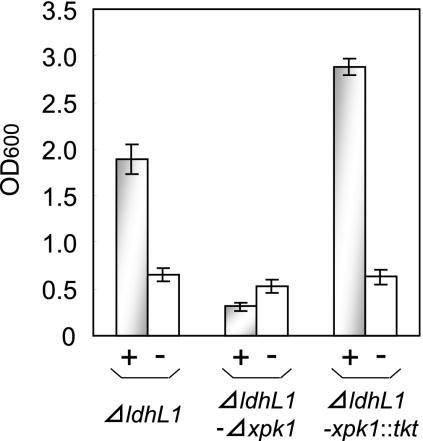
Cell growth with xylose was evaluated using modified MRS medium (MRS broth without glucose, beef extract, sodium acetate, and Tween 80) containing 2.0% (wt/vol) xylose and erythromycin at 25 μg/ml. Figure 1 shows the optical density at 600 nm (OD600) of each strain after 15 h of cultivation. The L. plantarum ΔldhL1/pCU-PXylAB strain was efficiently grown (OD600, 1.89) using xylose as the sole carbon source. In the case of control cultivation (without plasmid), growth decreased markedly, to a level (OD600, 0.66) comparable to growth without any sugar in the medium (data not shown). Alternatively, L. plantarum ΔldhL1-Δxpk1/pCU-PXylAB showed a poor level of growth (OD600, 0.31) with xylose, even lower than that of the control culture without any added sugar (OD600, 0.53). This lack of growth was restored by introducing tkt into the xpk1 locus, and the resulting strain, L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB, reached an OD600 of 2.88 with xylose. These data indicate that the PP pathway is suitable for cell growth, that Xpk1 is a crucial enzyme in the PK pathway, and that transketolase is a key enzyme for the introduction of the PP pathway, not only for arabinose metabolism (9), but also for xylose metabolism in L. plantarum.
FIG. 1.
Growth of L. plantarum NCIMB 8826 ΔldhL1, ΔldhL1-Δxpk1, and ΔldhL1-xpk1::tkt strains on xylose. Growth of the indicated strains was assayed by measurement of the OD600. +, growth of cells harboring pCU-PXylAB; −, growth of cells without pCU-PXylAB. Each bar represents the mean and standard deviation calculated from the results of three independent experiments.
Encouraged by these findings, we carried out lactic acid production via fermentation of 50 g/liter of xylose by using L. plantarum ΔldhL1/pCU-PXylAB and L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB strains in a 2.0-liter bioreactor as described previously (9), with the addition of erythromycin (final concentration, 25 μg/ml) to the fermentor. The temperature was maintained at 36°C, the agitation speed was maintained at 100 rpm, and the pH was kept at approximately 6.0 (±0.03) by the automatic addition of 10 M NH3 solution. Xylose, lactic acid, and acetic acid concentrations were measured by high-performance liquid chromatography, as described previously (9). The optical purity of lactic acid was measured using a BF-5 biosensor (Oji Scientific Instruments, Hyogo, Japan) as described previously (6). The optical purity of lactic acid was defined as follows: % optical purity = (d-lactic acid concentration − l-lactic acid concentration)/(d-lactic acid concentration + l-lactic acid concentration) × 100.
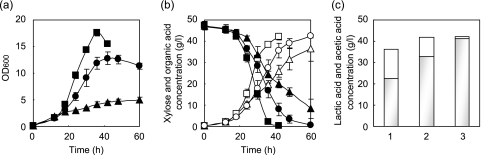
As shown in Fig. 2a and b, L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB exhibited significantly higher growth, sugar consumption, and acid production rates than L. plantarum ΔldhL1. The OD600 of the L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB culture increased with fermentation time and reached 17.6 after 36 h of fermentation, while that of the L. plantarum ΔldhL1 culture was significantly lower (OD600, 0.5), even after 36 h of fermentation (Fig. 2a). Differences in sugar consumption and acid production levels between the strains were also observed; L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB consumed all sugar within 36 h of fermentation and produced 41.9 g/liter organic acid (Fig. 2b), while fermentation by L. plantarum ΔldhL1/pCU-PXylAB continued after 36 h, with 39.0 g/liter xylose being consumed and 36.3 g/liter organic acid being produced (Fig. 2b).
FIG. 2.
Xylose fermentation by the mutant strains. Fermentation experiments with xylose were carried out using the L. plantarum NCIMB 8826 ΔldhL1 (triangles; lane 1), ΔldhL1-xpk1::tkt (squares; lane 2), and ΔldhL1-xpk1::tkt-Δxpk2 (circles; lane 3) strains harboring pCU-PXylAB. (a) OD600s of the cultures; (b) concentrations of xylose (closed symbols) and organic acid (representing the sum of lactic and acetic acids; open symbols); (c) concentrations of lactic acid (gray bars) and acetic acid (white bars). Data points represent the means and standard deviations of results from three independent experiments.
As shown in Fig. 2c and Table 2, L. plantarum ΔldhL1/pCU-PXylAB produced both lactic acid and acetic acid (22.5 and 13.8 g/liter, respectively). Unexpectedly, the ΔldhL1-xpk1::tkt/pCU-PXylAB strain produced 32.7 g/liter of lactic acid and 9.2 g/liter of acetic acid. This result differed from the results observed previously for arabinose fermentation (9) and suggests that, in the case of xylose fermentation, the replacement of xpk1 with tkt did not induce the complete shift from the PK pathway to the PP pathway that had been observed when arabinose was used as the carbon source.
TABLE 2.
Various parameters of xylose fermentationa
| Strain | Amt (g/liter) of: |
Yield (g lactic acid produced per g xylose consumed) | Optical purity of lactic acid (%) | ||
|---|---|---|---|---|---|
| Lactic acid produced | Acetic acid produced | Xylose consumed | |||
| ΔldhL1 strainb | 22.5 ± 4.6 | 13.8 ± 1.6 | 39.0 ± 3.8 | 0.57 ± 0.08 | 99.9 ± 0.04 |
| ΔldhL1-xpk1::tkt strainc | 32.7 ± 1.4 | 9.2 ± 0.2 | 47.4 ± 2.0 | 0.69 ± 0.01 | 99.2 ± 0.07 |
| ΔldhL1-xpk1::tkt-Δxpk2 strainb | 41.2 ± 0.7 | 1.0 ± 0.0 | 46.4 ± 0.6 | 0.89 ± 0.01 | 99.2 ± 0.34 |
Values are means ± standard deviations of results from three independent experiments.
Values for this strain were obtained after 60 h of cultivation.
Values for this strain were obtained after 42 h of cultivation.
Since L. plantarum has two phosphoketolase genes, xpk1 and xpk2 (4), we constructed a Δxpk2 mutant of L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB. The plasmid for disruption of the phosphoketolase 2 gene (xpk2) was constructed as follows. The 1,000-bp region upstream from the start codon of xpk2 and the 1,000-bp region downstream from the stop codon of xpk2 were amplified from the genome of L. plantarum NCIMB 8826 by PCR using the forward and reverse oligonucleotide primers xpk2-up_F with xpk2-up_R and xpk2-down_F with xpk2-down_R, respectively. The resulting fragments were digested with BglII and ligated. By using the ligated fragment (2,000 bp) as a template, the same fragment was amplified by PCR using the oligonucleotide primers xpk2-up_F and xpk2-down_R. The amplified fragment was digested with XhoI and SpeI and was subsequently inserted into the XhoI and SpeI sites of the plasmid pG+host9 (5). The resulting plasmid was designated pGh9-Δxpk2. Disruption of the xpk2 gene of L. plantarum ΔldhL1-xpk1::tkt (9) by using pGh9-Δxpk2 was carried out by pG+host plasmid-based double-crossover homologous integration, as described previously (10). The resulting xpk2 disruption strain of L. plantarum ΔldhL1-xpk1::tkt was designated L. plantarum ΔldhL1-xpk1::tkt-Δxpk2. Deletion of xpk2 was confirmed by PCR using the forward and reverse primers xpk2-up_seq and xpk2-down_seq, which anneal to the upstream region (bp 276 to 300) and the downstream region (bp 268 to 300) of xpk2, respectively (data not shown).
We then tested the effect of additional mutation of the xpk2 gene on xylose fermentation. As shown in Fig. 2, disruption of xpk2 caused a slight decrease in cell growth and fermentation rates. However, L. plantarum ΔldhL1-xpk1::tkt-Δxpk2/pCU-PXylAB maintained a higher rate of fermentation than the L. plantarum ΔldhL1/pCU-PXylAB transformant of the parental strain. The L. plantarum ΔldhL1-xpk1::tkt-Δxpk2/pCU-PXylAB strain produced a favorable ratio of lactic acid to acetic acid, generating predominantly lactic acid (41.2 g/liter) and very little acetic acid (1.0 g/liter). As a result, the ΔldhL1-xpk1::tkt-Δxpk2/pCU-PXylAB strain achieved a high lactic acid yield, 0.89 g per g of xylose consumed, compared with those achieved by L. plantarum ΔldhL1/pCU-PXylAB and L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB (0.57 and 0.69 g per g of xylose consumed, respectively) (Table 2). Moreover, the optical purity of the lactic acid produced was very high, at 99.2% (Table 2). These results strongly indicated that xpk2, in addition to xpk1, plays a crucial role in acetic acid production from xylose via the PK pathway. However, these results conflicted with those of our previous study (9), in which homo-lactic acid fermentation could be achieved by only xpk1 deletion when L. plantarum ΔldhL1-xpk1::tkt was cultivated with arabinose (9). To better understand the relative levels of xpk2 transcripts in the two above-mentioned cases, the transcription of xpk2 in xylose-fermenting L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB was quantified by real-time reverse transcription-PCR. L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB was cultured with xylose for 22 h. As a control, L. plantarum ΔldhL1-xpk1::tkt was cultured with arabinose for 22 h. Then the total RNA from each strain was purified with a RiboPure bacterial kit (ABI), and cDNA was made using a ReverTra Ace kit (TOYOBO). PCRs were run on the ABI PRISM system using the primers xpk2_2208-2226bp_F and xpk2_2208-2226bp_F. A standard curve was obtained with 10-fold serial dilutions of cDNA samples from 270 to 0.02 ng/μl by using the primers ldhD_676-696bp_F and ldhD_731-735bp_R. As expected, xpk2 transcription in xylose-fermenting L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB was about 100-fold higher than that in arabinose-fermenting L. plantarum ΔldhL1-xpk1::tkt. This result clearly shows that xpk2 expression was strongly induced when L. plantarum ΔldhL1-xpk1::tkt/pCU-PXylAB was cultivated with xylose.
In conclusion, we have successfully demonstrated homo-d-lactic acid production from xylose by disruption of xpk1 and xpk2 and by introduction of the xylAB operon and the tkt gene in an ldhL1-deficient L. plantarum strain. Our findings show that complete abolition of enzymes that have phosphoketolase activity, combined with introduction of enzymes that function as a bridge to the PP pathway, enables homo-lactic acid production by pentose sugar fermentation. This approach is applicable to other LAB, and this is the first report of homo-d-lactic acid production via xylose fermentation.
Acknowledgments
We are grateful to Emmanuelle Maguin for supplying the E. coli strains VE7108 and VE6838 (VE7108 containing the pG+host9 plasmid) and to the Meiji Dairies Corporation for supplying the pSECE1 plasmid.
This work was supported in part by a grant-in-aid for JSPS Fellows (20000860) from the Tokyo and Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe), MEXT, Japan.
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Bustos, G., A. B. Moldes, J. M. Cruz, and J. M. Domínguez. 2005. Influence of the metabolism pathway on lactic acid production from hemicellulosic trimming vine shoots hydrolyzates using Lactobacillus pentosus. Biotechnol. Prog. 21:793-798. [DOI] [PubMed] [Google Scholar]
- 2.Chaillou, S., Y.-C. Bor, C. A. Batt, P. W. Postma, and P. H. Pouwels. 1998. Molecular cloning and functional expression in Lactobacillus plantarum 80 of xylT, encoding the d-xylose-H+ symporter of Lactobacillus brevis. Appl. Environ. Microbiol. 64:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helanto, M., K. Kiviharju, M. Leisola, and A. Nyyssölä. 2007. Metabolic engineering of Lactobacillus plantarum for production of l-ribulose. Appl. Environ. Microbiol. 73:7083-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narita, J., K. Okano, T. Kitao, S. Ishida, T. Sewaki, M.-H. Sung, H. Fukuda, and A. Kondo. 2006. Display of α-amylase on the surface of Lactobacillus casei cells by use of the PgsA anchor protein, and production of lactic acid from starch. Appl. Environ. Microbiol. 72:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohara, H., M. Owaki, and K. Sonomoto. 2006. Xylooligosaccharide fermentation with Leuconostoc lactis. J. Biosci. Bioeng. 101:415-420. [DOI] [PubMed] [Google Scholar]
- 8.Okano, K., S. Kimura, J. Narita, H. Fukuda, and A. Kondo. 2007. Improvement in lactic acid production from starch using α-amylase-secreting Lactococcus lactis cells adapted to maltose or starch. Appl. Microbiol. Biotechnol. 75:1007-1013. [DOI] [PubMed] [Google Scholar]
- 9.Okano, K., S. Yoshida, T. Tanaka, H. Fukuda, and A. Kondo. 2009. Homo-d-lactic acid fermentation from arabinose by redirection of the phosphoketolase pathway to the pentose phosphate pathway in l-lactate dehydrogenase gene-deficient Lactobacillus plantarum. Appl. Environ. Microbiol. 75:5175-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okano, K., Q. Zhang, S. Shinkawa, S. Yoshida, T. Tanaka, H. Fukuda, and A. Kondo. 2009. Efficient production of optically pure d-lactic acid from raw corn starch using genetically modified l-lactate dehydrogenase gene-deficient and α-amylase-secreting Lactobacillus plantarum. Appl. Environ. Microbiol. 75:462-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka, K., A. Komiyama, K. Sonomoto, A. Ishizaki, S. J. Hall, and P. F. Stanbury. 2002. Two different pathways for d-xylose metabolism and the effect of xylose concentration on the yield coefficient of l-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 60:160-167. [DOI] [PubMed] [Google Scholar]