Abstract
Coproduction of DnaK/DnaJ in Escherichia coli enhances solubility but promotes proteolytic degradation of their substrates, minimizing the yield of unstable polypeptides. Higher eukaryotes have orthologs of DnaK/DnaJ but lack the linked bacterial proteolytic system. By coexpression of DnaK and DnaJ in insect cells with inherently misfolding-prone recombinant proteins, we demonstrate simultaneous improvement of soluble protein yield and quality and proteolytic stability. Thus, undesired side effects of bacterial folding modulators can be avoided by appropriate rehosting in heterologous cell expression systems.
The production of recombinant proteins is an essential instrument in biotechnology and biomedicine, but it has not been fully optimized in all the cell systems commonly used as factories. An important fraction of recombinant proteins fail to reach their native conformation, triggering diverse cell stress responses, and are often deposited as insoluble aggregates (usually referred to as inclusion bodies) or degraded by cellular proteases (11). Furthermore, the quality of soluble recombinant proteins is frequently compromised by the occurrence of soluble aggregates (5) and, in general, by their conformational heterogeneity (7, 17). In the widely used bacterium Escherichia coli, many strategies have been explored to enhance recombinant protein solubility. Among them, the coproduction of folding modulators, believed to be limiting in cells actively producing recombinant proteins, has attracted special attention (14). However, the efficacy of such an approach has been highly controversial. While some authors have reported enhancement of protein solubility by coproducing specific chaperones or chaperone sets, many others have found poor improvement or even impairment of protein solubility and yield. In particular, the chaperone DnaK and its cochaperone DnaJ have been very frequently incorporated as folding modulators, being present in essentially all the promising chaperone sets (6, 8). E. coli DnaK is the major cytosolic chaperone that exhibits folding and disaggregase activities (Table 1). Moreover, it is a negative regulator of the heat shock response in cooperation with DnaJ by promoting the conformation-dependent FtsH-mediated proteolytic inactivation of the stress-activated RNA polymerase subunit σ32 (Table 1) (22). This subunit controls the expression of the heat shock genes whose products cope with conformational stress, thus increasing cell survival. DnaK and DnaJ also deliver misfolded or conformationally abnormal proteins (including recombinant proteins) to the Lon and ClpP proteases, resulting in reduced protein yields (13, 23, 24). Misfolding-prone green fluorescent protein (GFP) fusions synthesized in DnaK knockouts (10) rendered higher protein yields but reduced recombinant protein solubility compared to those in wild-type cells. In contrast, the overexpression of dnaK and dnaJ genes in E. coli enhanced the proportion of soluble recombinant proteins by stimulating Lon- and ClpP-mediated proteolysis of aggregated proteins, reducing overall protein yields (10). Very recently, the molecular basis of the DnaKJ-mediated proteolytic enhancement has been solved by fine dissection of the interaction between DnaKJ and σ32. The binding of DnaK and DnaJ to distinct sites of the transcription factor promotes conformational modifications that expose a unique target site for the inactivating protease FtsH (22). Such conformational effect of the substrate seems to be the mechanistic platform of both foldase and disaggregase activities exhibited by DnaK/DnaJ on their substrates, including RepA and unfolded proteins (22). In recombinant E. coli cells, DnaK/DnaJ could mediate conformational perturbations of misfolded proteins at the surface of inclusion bodies, where DnaK localizes (4), exposing them to the stress proteases Lon and ClpP (10) and promoting digestion during refolding attempts. Such a dual role of DnaK/DnaJ in stimulating protein folding but also enhancing degradation of protease-sensitive protein species could be the cause of the controversial data obtained from their use as folding modulators and the reason for the only transient rise of soluble protein species refolded in vivo from bacterial inclusion bodies (3).
TABLE 1.
Biological activities of the E. coli chaperone DnaK
| Activity | Cochaperone(s) | Substrate(s) | Reference(s) | Evolutionary relationship(s) |
|---|---|---|---|---|
| Foldase | DnaJ, GrpE | Unfolded/misfolded proteins | 15 | Hsp70 family member |
| Holding chaperone | Hsp31 | Unfolded/misfolded proteins | 21 | Presumed Hsp70 family member |
| Disaggregase | ClpB, IbpAB | Protein aggregates | 20, 25 | Species specificity of Hsp104/Hsp70 and ClpB/DnaK cooperativity; a disaggregation activity has not yet been identified in mammalian cells |
| Negative regulator of the heat shock response (proteolytic enhancer) | DnaJ, FtsH | Transcription factor σ32 | 22 | Not reported |
| Proteolytic enhancer | DnaJ, Lon, ClpP | Abnormal proteins | 13 | Not reported |
| Proteolytic enhancer | DnaJ, Lon, ClpP | Recombinant proteins | 10 | Not reported |
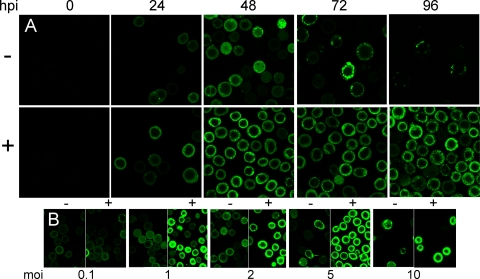
For the improved production of recombinant proteins, it would then be desirable to keep the DnaK/DnaJ foldase activity but eliminate the enhanced proteolysis indirectly promoted by these chaperones. Since DnaK/DnaJ are members of the highly conserved Hsp70 family, we anticipated that their foldase activity would be retained in organisms other than E. coli, but that was not the case for the proteolytic stimulation of recombinant proteins, which is highly dependent on the E. coli proteases Lon and ClpP (Table 1) (10). Under the assumption that insect cell proteases would not recognize the target sites exposed by the activity of DnaKJ (no Lon and ClpP orthologs have so far been identified in insects), we rehosted the E. coli DnaKJ pair for coproduction along with recombinant protein by using the baculovirus expression system. For these studies, we used a proteolytically unstable GFP (mGFP), already characterized for recombinant protein quality analysis in bacteria (10), and generated two transfer vectors designed to express mGFP either alone or together with the bacterial chaperones DnaK and DnaJ, using the vector pAcAB4 (2). mGFP and dnaJ genes were placed under the control of the p10 promoter, and dnaK was placed under the control of the polyhedrin promoter. Each transfer vector was cotransfected with Bsu36I-linearized viral DNA BAC10:KO1629 (27) into Spodoptera frugiperda Sf9 cells to obtain recombinant baculoviruses. Individual clones were plaque purified and further amplified, and the titers of the clones were determined before they were analyzed for mGFP expression. In the absence of chaperone coproduction, mGFP gene expression resulted in mild cytoplasmic fluorescence within 24 h postinfection (hpi), with punctate distribution indicative of inclusion body formation (Fig. 1A). Coexpression of mGFP along with DnaK and DnaJ rendered much higher and homogeneously distributed fluorescence. The degree of fluorescence was largely stable at least up to around 72 h and was reproducible at all the tested multiplicities of infection (MOIs) (ranging from 0.1 to 10 [Fig. 1B]). The enhancement of fluorescence in infected cells coexpressing dnaK and dnaJ was further confirmed by flow cytometry (Fig. 2A). These results were in marked contrast to those of similar experiments with E. coli, where coexpression of DnaK and DnaJ with mGFP resulted in much lower fluorescence levels than in the control cells (10).
FIG. 1.
Confocal microscopy images of baculovirus-infected Sf9 cells taken in a Leica TCS SP2 AOBS microscope. Batches of 1 × 106 cells were seeded on glass-bottom dishes, supplemented with 5% fetal calf serum, and infected at MOIs of 0.1, 1, 2, 5, and 10 with recombinant baculoviruses expressing mGFP in the presence (+) or absence (−) of DnaK and DnaJ. The time course experiment for a set MOI of 2 is shown in panel A, while cultured cells 48 h after infection at different MOIs are presented in panel B. The settings used to capture the images in panel B were maintained for direct comparison of the images.
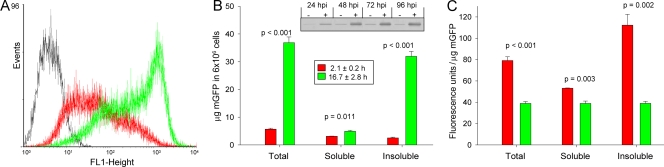
FIG. 2.
(A) Flow cytometry analysis of baculovirus-infected Sf9 cells producing mGFP in the absence (red plot) or presence (green plot) of bacterial chaperones DnaK and DnaJ. Sf9 cell cultures were set up at a density of 2 × 106 cells/ml, supplemented with 5% fetal calf serum, and infected at an MOI of 2. Cells were harvested at 72 hpi and rinsed with cold phosphate-buffered saline (PBS). Flow cytometry analyses were performed with intact cells resuspended in PBS on a FACSCalibur system (Becton Dickinson). The fluorescence emission in the FL1 channel was analyzed using WinMDI 2.9 software. Uninfected cells (black plot) were used as a negative control, and measurements were recorded for three independent replicas. (B) Protein amounts in total, soluble, and insoluble cell fractions in the absence (red bars) or presence (green bars) of bacterial chaperones DnaK and DnaJ were estimated by Western blot analysis after disruption of cells harvested at 72 hpi. Sf9 cells were disrupted in lysis buffer (extraction buffer [50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA] containing 1% Tergitol NP-9 [Sigma] and Roche's protease inhibitor cocktail [catalog no. 11836170001]) on ice for 30 min and Dounce homogenized. Cell lysates were treated with DNase (25 μg/ml) and MgSO4 (10 mM), and soluble and insoluble cell fractions were separated by centrifugation at 9,500 × g for 10 min. The insoluble cell pellet was washed with extraction buffer containing 0.5% Triton X-100 and resuspended in extraction buffer. The inset box shows the estimated half-life for mGFP in the absence (red bars) or presence (green bars) of DnaKJ after protein synthesis arrest at 24 hpi by the addition of cycloheximide at a final concentration of 100 μg/ml. Western blot analysis of a time course experiment showing mGFP production in Sf9 cells at different hpi is depicted on the inset graph. Material from the same number of cells was loaded into the gels for protein determination. (C) Specific fluorescence of mGFP produced in the absence (red bars) or presence (green bars) of bacterial chaperones DnaK and DnaJ at 72 hpi. Fluorescence emissions of lysates and soluble and insoluble cell fractions were measured in triplicate, with no further treatment, using a Cary Eclipse fluorescence spectrophotometer (Varian, Inc.). Fluorescence data were combined with protein amounts to obtain the specific fluorescence of mGFP, defined as fluorescence units per μg of mGFP. The significance of the differences between data values determined in the absence and in the presence of DnaK/DnaJ is indicated through P values from an analysis of variance test in panels B and C.
Also, the coproduction of DnaK and DnaJ resulted in a dramatic, sixfold increase of the total amount of mGFP, indicating the absence of proteolysis. Being more important in the insoluble cell fraction, this still represented an almost twofold yield enhancement for the soluble version of the protein (Fig. 2B). Consistent with previous studies, correctly folded polypeptides were deposited in inclusion bodies (9), confirming that solubility is not a good indicator of a successful protein production process (12). When the stability of mGFP produced alone or in the presence of E. coli DnaK and DnaJ was analyzed, mGFP's half-life increased from 2.1 ± 0.2 h to 16.7 ± 2.8 h (Fig. 2B, inset), indicative of an increased resistance to proteolytic degradation. This is the opposite of what has been observed in bacterial cells, where the half-life of mGFP was reduced from 5.9 ± 0.5 h to 1.9 ± 0.3 h in the presence of DnaKJ (10). This result confirms not only the lack of DnaKJ-mediated mGFP proteolysis in insect cells but also that the activity of these chaperones as folding mediators is able to protect from degradation by host cell proteases, probably by fully completing the folding process. In the absence of DnaK and DnaJ coexpression, mGFP demonstrated a clear heterogeneity in specific fluorescence when soluble and insoluble versions were compared (Fig. 2C). This is in marked contrast to what was observed when the chaperones were coexpressed, where specific fluorescence emissions of soluble and insoluble mGFPs were remarkably similar. On the other hand, the overall mGFP specific fluorescence in the presence of DnaK/DnaJ was lower than in the absence of these chaperones. This is consistent with recent observations reporting that higher protein yield in a production process necessarily results in a decrease of conformational quality (10, 16, 18).
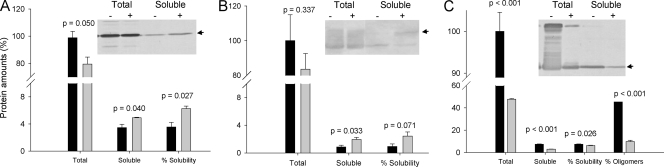
To determine whether the positive effect of DnaK and DnaJ in assisting protein folding could be extended to the expression of other recombinant proteins in insect cells, we also tested coexpression of these proteins with foot-and-mouth disease virus (FMDV) VP1 and VP2 capsid proteins and human alpha-galactosidase A. Although potential gene dosage effects due to coinfection of the virus expressing the chaperones with that expressing the recombinant protein prevented us from a fine comparative analysis of total protein amounts, unlike in our studies with mGFP, we were able to assess changes in solubility that correlated with recombinant protein expression. In general, total protein amounts were comparable when produced alone or with DnaK/DnaJ, indicating the absence of important DnaK-induced proteolysis (Fig. 3A to C). For two of the three proteins (VP1 and alpha-galactosidase), the amounts of soluble protein were significantly enhanced, reaching up to more than twofold for the enzyme, in which the solubility also improved by more than 250% (Fig. 3B). For the third protein (FMDV VP2), there was no increase in the yield of soluble protein but in the protein quality. VP2 has a tendency to spontaneously form unwanted oligomers. When coexpressed with DnaK/DnaJ, formation of these aggregates was completely cleared (Fig. 3C). The quality of soluble alpha-galactosidase, which is protease sensitive, was also enhanced, with degradation largely minimized, indicative of DnaK/DnaJ-promoted protein stabilization (Fig. 3B). At least in mGFP, chaperone coproduction did not affect the efficiency of recovery from cell extracts in further purification processes (not shown), and we do not have any experimental data suggesting that this could happen with other proteins.
FIG. 3.
Total amounts and soluble amounts of recombinant proteins in Sf9 cells coinfected at an MOI of 2 with baculoviruses encoding FMDV VP1 (A), human alpha-galactosidase (B), and FMDV VP2 (C) proteins (black bars), compared with protein data obtained during coinfection with a DnaK/DnaJ-encoding baculovirus, also at an MOI of 2 (gray bars). Data were obtained in triplicate at 72 hpi, and they were compared to the total protein amounts observed in the absence of chaperones. Percentages of solubility and VP2 oligomer occurrence are also included. Arrows indicate the positions of the full-length recombinant proteins. The significance of the differences between data values determined in the absence and in the presence of DnaK/DnaJ is indicated through P values from an analysis of variance test.
In contrast to the widespread use of folding modulators in bacteria (14), very few attempts have been made to improve cytoplasmic protein production in insect cells with the aid of chaperones. In particular, the human versions of DnaK and DnaJ, namely, Hsp70 and Hsp40, respectively, have been coproduced along with target proteins (1, 19, 26), with all cases reporting slight increases in solubility, poor gain of yield, if any, and no references to protein stability and quality. Interestingly, gain of solubility has also been generally described for bacteria when the dnaK gene is coexpressed (6). Our data demonstrate that bacterial DnaK and DnaJ do have a significant effect on protein solubility in insect cells, suggesting that these chaperones may target a broader subset of misfolded protein substrates in insect cells than human Hsp70 and Hsp40. This highlights the relevance of function selection in multifunctional folding modulators for use in heterologous hosts. The results presented here strongly support chaperone rehosting as a new concept for high-quality recombinant protein production in insect cells that permits separation of the undesirable effects observed in E. coli from the valuable foldase activity. Coexpression of DnaK and DnaJ in insect cells dramatically enhances protein yield, proteolytic stability, protein solubility, and global biological activity, with unusually mild negative effects on protein quality.
Acknowledgments
We appreciate financial support through grants BIO2007-61194 and EUI2008-03610 (MICINN) to A.V. and grant BB/C504735/1 (BBSRC) to P.R. We also acknowledge the support of the CIBER de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Spain. M.M.-A. is the recipient of a predoctoral fellowship from MEC, Spain. A.V. received an ICREA ACADEMIA award (Catalonia, Spain).
We thank Francisco Cortés, from Servei de Cultius Cel·lulars (UAB), for routine maintenance of insect cell cultures and both Servei de Microscòpia and Servei de Citometria (UAB) for helpful technical assistance.
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Ailor, E., and M. J. Betenbaugh. 1998. Overexpression of a cytosolic chaperone to improve solubility and secretion of a recombinant IgG protein in insect cells. Biotechnol. Bioeng. 58:196-203. [PubMed] [Google Scholar]
- 2.Belyaev, A. S., and P. Roy. 1993. Development of baculovirus triple and quadruple expression vectors: co-expression of three or four bluetongue virus proteins and the synthesis of bluetongue virus-like particles in insect cells. Nucleic Acids Res. 21:1219-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrio, M. M., and A. Villaverde. 2001. Protein aggregation as bacterial inclusion bodies is reversible. FEBS Lett. 489:29-33. [DOI] [PubMed] [Google Scholar]
- 4.Carrio, M. M., and A. Villaverde. 2005. Localization of chaperones DnaK and GroEL in bacterial inclusion bodies. J. Bacteriol. 187:3599-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Marco, A., and A. Schroedel. 2005. Characterization of the aggregates formed during recombinant protein expression in bacteria. BMC Biochem. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Marco, A. 2007. Protocol for preparing proteins with improved solubility by co-expressing with molecular chaperones in Escherichia coli. Nat. Protoc. 2:2632-2639. [DOI] [PubMed] [Google Scholar]
- 7.de Marco, A. 2008. Minimal information: an urgent need to assess the functional reliability of recombinant proteins used in biological experiments. Microb. Cell Fact. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Marco, A., E. Deuerling, A. Mogk, T. Tomoyasu, and B. Bukau. 2007. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Fruitos, E., N. Gonzalez-Montalban, M. Morell, A. Vera, R. M. Ferraz, A. Aris, S. Ventura, and A. Villaverde. 2005. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Fact. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Fruitos, E., M. Martinez-Alonso, N. Gonzalez-Montalban, M. Valli, D. Mattanovich, and A. Villaverde. 2007. Divergent genetic control of protein solubility and conformational quality in Escherichia coli. J. Mol. Biol. 374:195-205. [DOI] [PubMed] [Google Scholar]
- 11.Gasser, B., M. Saloheimo, U. Rinas, M. Dragosits, E. Rodriguez-Carmona, K. Baumann, M. Giuliani, E. Parrilli, P. Branduardi, C. Lang, D. Porro, P. Ferrer, M. L. Tutino, D. Mattanovich, and A. Villaverde. 2008. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb. Cell Fact. 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Montalban, N., E. Garcia-Fruitos, and A. Villaverde. 2007. Recombinant protein solubility—does more mean better? Nat. Biotechnol. 25:718-720. [DOI] [PubMed] [Google Scholar]
- 13.Jubete, Y., M. R. Maurizi, and S. Gottesman. 1996. Role of the heat shock protein DnaJ in the lon-dependent degradation of naturally unstable proteins. J. Biol. Chem. 271:30798-30803. [DOI] [PubMed] [Google Scholar]
- 14.Kolaj, O., S. Spada, S. Robin, and J. G. Wall. 2009. Use of folding modulators to improve heterologous protein production in Escherichia coli. Microb. Cell Fact. 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langer, T., C. Lu, H. Echols, J. Flanagan, M. K. Hayer, and F. U. Hartl. 1992. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356:683-689. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Alonso, M., E. Garcia-Fruitos, and A. Villaverde. 2008. Yield, solubility and conformational quality of soluble proteins are not simultaneously favored in recombinant Escherichia coli. Biotechnol. Bioeng. 101:1353-1358. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Alonso, M., N. González-Montalbán, E. García-Fruitós, and A. Villaverde. 2008. The functional quality of soluble recombinant polypeptides produced in Escherichia coli is defined by a wide conformational spectrum. Appl. Environ. Microbiol. 74:7431-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Alonso, M., N. Gonzalez-Montalban, E. Garcia-Fruitos, and A. Villaverde. 2009. Learning about protein solubility from bacterial inclusion bodies. Microb. Cell Fact. 8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Torrecuadrada, J. L., S. Romero, A. Nunez, P. Alfonso, M. Sanchez-Cespedes, and J. I. Casal. 2005. An efficient expression system for the production of functionally active human LKB1. J. Biotechnol. 115:23-34. [DOI] [PubMed] [Google Scholar]
- 20.Mogk, A., E. Deuerling, S. Vorderwulbecke, E. Vierling, and B. Bukau. 2003. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol. Microbiol. 50:585-595. [DOI] [PubMed] [Google Scholar]
- 21.Mujacic, M., M. W. Bader, and F. Baneyx. 2004. Escherichia coli Hsp31 functions as a holding chaperone that cooperates with the DnaK-DnaJ-GrpE system in the management of protein misfolding under severe stress conditions. Mol. Microbiol. 51:849-859. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez, F., F. Arsene-Ploetze, W. Rist, S. Rudiger, J. Schneider-Mergener, M. P. Mayer, and B. Bukau. 2008. Molecular basis for regulation of the heat shock transcription factor σ32 by the DnaK and DnaJ chaperones. Mol. Cell 32:347-358. [DOI] [PubMed] [Google Scholar]
- 23.Sherman, M. Y., and A. L. Goldberg. 1996. Involvement of molecular chaperones in intracellular protein breakdown. EXS 77:57-78. [DOI] [PubMed] [Google Scholar]
- 24.Sherman, M. Y., and A. L. Goldberg. 1992. Involvement of the chaperonin dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 11:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weibezahn, J., B. Bukau, and A. Mogk. 2004. Unscrambling an egg: protein disaggregation by AAA+ proteins. Microb. Cell Fact. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama, N., M. Hirata, K. Ohtsuka, Y. Nishiyama, K. Fujii, M. Fujita, K. Kuzushima, T. Kiyono, and T. Tsurumi. 2000. Co-expression of human chaperone Hsp70 and Hsdj or Hsp40 co-factor increases solubility of overexpressed target proteins in insect cells. Biochim. Biophys. Acta 1493:119-124. [DOI] [PubMed] [Google Scholar]
- 27.Zhao, Y., D. A. Chapman, and I. M. Jones. 2003. Improving baculovirus recombination. Nucleic Acids Res. 31:E6. [DOI] [PMC free article] [PubMed] [Google Scholar]