Abstract
In recent years, the genetic manipulation of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough has seen enormous progress. In spite of this progress, the current marker exchange deletion method does not allow for easy selection of multiple sequential gene deletions in a single strain because of the limited number of selectable markers available in D. vulgaris. To broaden the repertoire of genetic tools for manipulation, an in-frame, markerless deletion system has been developed. The counterselectable marker that makes this deletion system possible is the pyrimidine salvage enzyme, uracil phosphoribosyltransferase, encoded by upp. In wild-type D. vulgaris, growth was shown to be inhibited by the toxic pyrimidine analog 5-fluorouracil (5-FU), whereas a mutant bearing a deletion of the upp gene was resistant to 5-FU. When a plasmid containing the wild-type upp gene expressed constitutively from the aph(3′)-II promoter (promoter for the kanamycin resistance gene in Tn5) was introduced into the upp deletion strain, sensitivity to 5-FU was restored. This observation allowed us to develop a two-step integration and excision strategy for the deletion of genes of interest. Since this in-frame deletion strategy does not retain an antibiotic cassette, multiple deletions can be generated in a single strain without the accumulation of genes conferring antibiotic resistances. We used this strategy to generate a deletion strain lacking the endonuclease (hsdR, DVU1703) of a type I restriction-modification system that we designated JW7035. The transformation efficiency of the JW7035 strain was found to be 100 to 1,000 times greater than that of the wild-type strain when stable plasmids were introduced via electroporation.
The anaerobic sulfate-reducing bacteria (SRB) are found in a remarkable variety of habitats. These bacteria have received attention recently because they have a potential role in toxic metal bioremediation (23, 26). To fully understand the potential benefits and to maximize opportunities for successful manipulation of the SRB, it would be useful to create deletions in critically important genes. Several activities of particular interest are represented by multiple isozymes, suggesting that compensation may occur upon elimination of one or more of these genes. To fully elucidate alternative pathways, genetic approaches allowing the construction of multiple mutations are needed. The genetic manipulation of the SRB Desulfovibrio vulgaris Hildenborough has seen significant improvements in recent years (reviewed in reference 3). Chloramphenicol and kanamycin marker exchange mutagenesis methods have been developed (2, 10). Although gene deletions can be constructed, the necessary retention of antibiotic resistance limits sequential deletions, since each deletion would require an additional antibiotic cassette. To eliminate the necessity of marker retention, an in-frame markerless deletion system has been developed.
A two-step method for marker exchange/deletion that used the counterselectable marker sacB (13) was used by Fu and Voordouw (10) to generate the first deletion by marker exchange in D. vulgaris. The sacB gene from Bacillus subtilis encodes levansucrase and confers sensitivity to sucrose in many gram-negative bacteria (7, 32, 35), including D. vulgaris (8, 10, 15, 18, 19, 21). In the first step of the process, a suicide plasmid carrying DNA regions from up- and downstream of a target gene flanking a chloramphenicol resistance (Cmr) cassette was introduced into D. vulgaris by conjugation and single recombinants were selected as Cmr colonies (10). After confirmation of the integration of this plasmid, the double-recombination event was selected on medium containing chloramphenicol and sucrose. This method, with some variation, has been used to make several mutants by the Voordouw group (8, 10, 15, 18, 19, 21). One unexpected complication was the observation that 50% of sucrose resistant colonies were due to events other than the removal of the sacB gene and plasmid through a second recombination as desired (11). Also, sensitivity to sucrose is apparently strongly affected by medium composition, initial culture density, and the time of exposure (11). This method involved a large time investment, but it ultimately resulted in a marker exchange mutant (Cmr) and established the effectiveness of a two-step recombination process in D. vulgaris.
Among alternative counterselectable markers are the purine and pyrimidine salvage enzymes, phosphoribosyl transferases (PRTases). These enzymes allow the recycling of free bases from internal or environmental sources, as well as the incorporation of base analogs into nucleoside monophosphates. Importantly, the incorporation of base analogs can be lethal and are the reason these nucleotide salvage pathways have been widely used as counterselectable markers for gene knockout systems in bacteria, archaea, and eukaryotes (4, 5, 8, 9, 12, 16, 22, 24, 27, 29, 34). Specifically, the incorporation of the pyrimidine analog 5-fluorouracil (5-FU) is lethal in a number of bacteria (8, 16, 22). Mutants whose genes encoding the pertinent PRTases have been deleted are resistant to the toxic base analogs (4, 5, 8, 9, 12, 16, 22, 24, 27, 29, 34). Reintroduction of these genes restores sensitivity. In order to utilize the genes for PRTases as counterselectable markers, a deletion of the endogenous PRTase gene must be created in the host strain. We have previously shown that wild-type D. vulgaris is extremely sensitive to low levels of 5-FU, as little as 0.1 μg/ml (3). In the present study, we deleted the upp gene (DVU1025) encoding the putative uracil phosphoribosyl transferase in D. vulgaris creating strain JW710 and showed that it was resistant to 5-FU. When the upp gene was reintroduced into JW710 (Δupp), it restored sensitivity to wild-type levels of 5-FU. These phenotypic observations indicate that the loss of the upp provides a selectable marker for a two-step integration and excision strategy for the deletion of target genes without a residual marker exchange. A second advantage of using this markerless method is the facile ability to generate in-frame deletions, eliminating potential polarity.
To test the effectiveness of using the upp as a counterselectable marker in D. vulgaris, we deleted the gene encoding the endonuclease of a type I restriction-modification system, hsdR (DVU1703), and the downstream conserved hypothetical gene (CHP; DVU1702), creating strain JW7035 [Δupp Δ(hsdR-CHP)]. The type I restriction-modification system was targeted for deletion in hopes of increasing the transformation efficiency of D. vulgaris and facilitating the construction of future deletions. As anticipated, electroporation experiments with stable plasmids revealed an improvement in transformation efficiency for stable plasmids compared to wild-type D. vulgaris. Finally, Gateway Technology (Invitrogen) was applied to generate a destination vector (pMO727) containing the constitutively expressed wild-type upp gene. This vector will expedite the process of creating the required suicide deletion vectors for future markerless deletions.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Strains used in the present study are listed in Table 1. Escherichia coli strains were cultured aerobically in liquid LC medium (components per liter of medium: 10 g of tryptone, 5 g of NaCl, and 5 g of yeast extract) and recovered in SOC medium (components per liter of medium: 5 g of yeast extract, 9 g of tryptone, 0.5 g of NaCl, 0.19 g of KCl, 3.6 g of glucose, 10 ml of 1 M MgCl, and 10 ml of 1 M MgSO4) (17) after transformation. Where indicated, ampicillin, kanamycin, chloramphenicol, and spectinomycin were added to the LC medium to final concentrations of 100, 50, 30, and 100 μg/ml, respectively. When plating for blue/white selection with pBluescript plasmids (Table 1), X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was added to the LC plates at a final concentration of 80 μg/ml. Chemicals and antibiotics were obtained from Fisher Scientific (Pittsburg, PA). For plating, LC medium was solidified with 15 g of agar/liter.
TABLE 1.
Bacteria strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| GC5 competent cells | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsd R17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 tonA | Gene Choice |
| OneShot TOP10 competent cells | F− φ80lacZΔM15 ΔlacX74 recA1 endA1 mcrA Δ(mrr-hsdRMS-mcrBC) araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) nupG | Invitrogen |
| OneShot ccdB survival T1 phage-resistant cells | F−mcrA Δ(mrr-hsdRMS-mcrBC) 80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG tonA::Ptrc-ccdA | Invitrogen |
| D. vulgaris Hildenborough strains | ||
| Wild type | D. vulgaris Hildenborough, ATCC 29579 (pDV1); 5-FUs | ATCC |
| JW710 | Δupp (pDV1); 5-FUr | This study |
| JW7034 | Chromosomal insertion of pMO729 into JW710 (pDV1); Specr 5-FUs | This study |
| JW7035 | Δupp Δ(hsdR-CHP) (pDV1); 5-FUr | This study |
| JW801 | ΔpDV1; 5-FUs | Wall Laboratory |
| Plasmids | ||
| pBluescript SK(+) | Blue-white cloning vector; Ampr | Stratagene |
| pSC27 | Desulfovibrio shuttle vector; source of SRB replicon pBG1, mob; Kanr | 32 |
| pCR4Blunt-TOPO | Cloning vector; Kanr Ampr | Invitrogen |
| pCR8/GW/TOPO | Cloning vector; Specr | Invitrogen |
| pENTR/D-TOPO | Cloning vector; Kanr | Invitrogen |
| pMO711 | pBluescript containing the markerless deletion cassette for the upp gene; Ampr | This study |
| pMO715 | pCR4Blunt-TOPO containing the Paph(3′)-II-upp (1,087 bp); Kanr Ampr | This study |
| pMO719 | Desulfovibrio shuttle vector; SRB replicon pBG1 (EcoRI to EcoRI from pSC27) ligated to the EcoRI-bounded pCR8/GW/TOPO fragment; Specr | This study |
| pMO720 | Paph(3′)-II-upp inserted via PmeI/SnaBI digest from pMO715 into EcoRV site of pMO719; Specr | This study |
| pMO727 | Gateway Destination cassette [reading frame B] into AclI and BspEI of pMO720; Specr Cmr | This study |
| pMO728 | pENTR/D containing the DVU1703 (and DVU1702) markerless deletion cassette with TOPO cloning; Kanr | This study |
| pMO729 | DVU1703 (and DVU1702) markerless deletion cassette from pMO728 into pMO727 using LR clonase reaction; Specr | This study |
Strr, streptomycin resistance; Ampr, ampicillin resistance; Specr, spectinomycin resistance; Kanr, kanamycin resistance. pDV1 is an endogenous 202-kb plasmid in D. vulgaris Hildenborough. CHP, conserved hypothetical protein. The deletion in JW7035 is of both the hsdR gene (DVU1703) and the CHP gene (DVU1702); however, the strain is referred to as ΔhsdR.
D. vulgaris strains were manipulated and grown at ∼32°C in an anaerobic growth chamber (Coy Laboratory Product, Inc., Grass Lake, MI), unless indicated otherwise, in either LS4D (25) amended to contain 30 mM Tris (pH 7.4) instead of 30 mM PIPES or in Wall medium (8 mM MgCl, 20 mM NH4Cl, 0.6 mM CaCl2, 2 mM K2HPO4, 60 μM FeCl2, 120 μM EDTA, 30 mM Tris [pH 7.4], 0.1% [wt/vol] yeast extract, 1 ml of Thauers vitamin solution [6] per liter, and 6 ml of trace elements solution per liter, with the pH adjusted to 7.2). The trace elements solution contained 2.5 mM MnCl2, 1.26 mM CoCl2, 1.47 mM ZnCl2, 210 μM Na2MoO4, 320 μM H3BO3, 380 μM NiSO4, 11.7 μM CuCl2, 35 μM Na2SeO3, and 24 μM Na2WO4. For Wall medium, sodium lactate (60 mM) was used as the electron donor with either 30 mM sodium sulfate (Wall LS4) or 40 mM sodium sulfite (Wall LS3) as the terminal electron acceptor. When necessary and indicated, antibiotics were added to the Wall medium to a final concentration of 100 μg of spectinomycin/ml or 400 μg of G418/ml. G418 (RPI Corp., Mt. Prospect, IL) was used in place of kanamycin because it was found to be more effective for kanamycin resistance selection in D. vulgaris. When D. vulgaris cultures were grown in Wall medium, 5-FU (Fisher Scientific, Pittsburg, PA) was added to final concentrations of 2.5 μg/ml for liquid cultures or 40 μg/ml in solidified medium. For plating, Wall medium was solidified with 1.5% (wt/vol) agar, and two reductants were added: sodium thioglycolate (1.2 mM) and titanium citrate (380 μM). Cells were dispensed into sterile, empty petri dishes, and then molten Wall LS4 was poured over the cells and swirled. All plating steps were carried out in an anaerobic glove chamber (Coy Laboratory Products, Inc., Grass Lake, MI) in an atmosphere of ∼95% N2 and ∼5% H2. Once solidified, the plates were inverted, placed in an airtight container, and incubated at ∼32°C for 4 to 7 days. D. vulgaris cultures were routinely plated on LC plates containing 40 mM glucose and incubated in air to ensure that they were free of aerobic contaminants.
Generation of the Δupp strain, JW710.
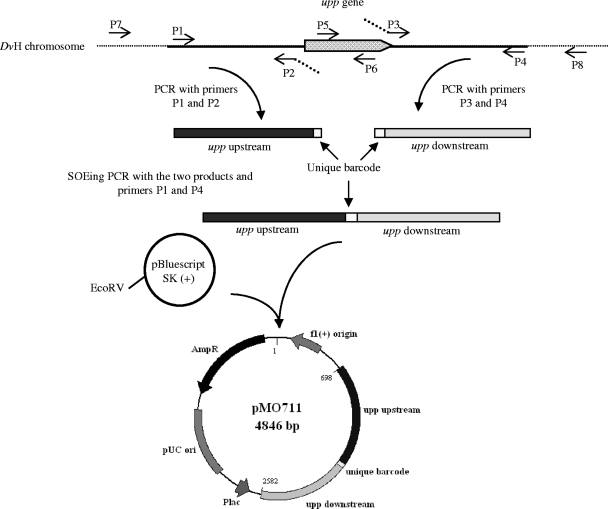
A upp deletion cassette was constructed as described by Bender et al. (2), with the differences listed below. Primers (for a listing, see Table S1 in the supplemental material), generated by IDT, Coralville, IA, were designed to amplify approximately 1,000 bp upstream (P1 and P2) and 900 bp downstream (P3 and P4) of the upp gene (DVU1025). For future deletion tracking, primers P2 and P3 were designed to contain the following unique barcode ACG TGC CTA AAG TGT ACT ACG ACA CCT CCG CGA TGA GT A, as well as five additional overlapping base pairs on each 5′ end to allow the joining of the two fragments with gene splicing by overlapping extension PCR (SOE-PCR) (20). Amplification of the two regions by PCR, the splicing together of those two products, and the capture of that deletion cassette in the EcoRV site of pBluescript SK(+) were performed as previously described (2) and illustrated in Fig. 1. The resulting pMO711 was sent to the DNA Core Facility at the University of Missouri for sequence verification before proceeding.
FIG. 1.
Construction of pMO711, the mutagenic plasmid for the deletion of upp. Regions upstream and downstream of the upp gene were amplified to contain a unique barcode (primers P2 and P3). The two products were joined together by using SOE-PCR and cloned into EcoRV site of pBluescript SK(+). The schematic is not drawn to scale.
The preparation of competent D. vulgaris and electroporation by pMO711 was performed as described previously (2), with the following modifications. Briefly, the electroporation was carried out anaerobically in a total volume of 75 μl with an ECM 630 electroporator, BTX (Genetronix, San Jose, CA) in 1-mm gapped electroporation cuvettes (Molecular BioProducts, San Diego, CA) at 1,750 V, 250 Ω, and 25 μF. The cells were allowed to recover in 1 ml of LS4D medium overnight at ∼32°C. An additional 5 ml of fresh LS4D was then added to the recovered cells, which were allowed to grow to early stationary phase. Aliquots of 1 ml were mixed with 4 ml of molten reduced top agar (1.5% [wt/vol] agar, 30 mM PIPES) and poured onto reduced LS4 (LS4D modified to contain 0.1% [wt/vol] yeast extract) solidified medium containing 40 μg of 5-FU/ml, and the plates incubated for ∼7 days at 32°C in the anaerobic chamber. 5-FUr colonies were selected, and six were grown to stationary phase in liquid LS4D medium with 40 μg of 5-FU/ml, gDNA was extracted, and the deletion of the upp gene was verified by PCRs. Size differences of the PCR amplicons indicated the presence of the wild-type target upp gene or the successful deletion (two of the six isolates). One isolate that produced no amplicon from primers P5 and P6 internal to the upp gene (Fig. 1) and a band of the predicted size for a successful deletion from primers P7 and P8 was selected and named JW710.
Colony PCR screening.
In order to screen E. coli colonies for correctly constructed plasmids, colony PCR screens were performed. Individual colonies were picked from a plate and dispersed in 50 μl of a PCR mixture in a 0.2-ml PCR tube. The remaining cells from the colony were streaked on an LC plate containing the appropriate antibiotic for plasmid maintenance. Isolates were identified that produced the expected PCR amplicons from the plasmid constructs. New plasmids were considered verified only after both DNA strands of the construct had been fully sequenced. All sequencing was performed at the DNA Core Facility at the University of Missouri.
Generation of Paph(3′)-II-upp construct.
The pMO715 plasmid (Fig. 2A) was constructed for complementation of the upp deletion. Two regions were amplified with Herc polymerase (Stratagene, La Jolla, CA). A 449-bp fragment containing the promoter for the aminoglycoside phosphotransferase II gene, aph(3′)-II from Tn5, was amplified with the primers P9 and P10, and 697 bp containing the wild-type upp gene from D. vulgaris was amplified with the primers P11 and P12. Primers P10 and P11 contained overlapping sequences to allow splicing of the two products by SOE-PCR and the primers P9 and P12 amplified the spliced product. The product was purified with the Wizard SV gel and PCR Clean-Up system (Promega) and cloned into pCR4Blunt-TOPO (Invitrogen) (see Fig. S1 in the supplemental material). The recombinant plasmid was transformed into One Shot TOP10 Competent Cells (Invitrogen) and plated on LC with 100 μg of kanamycin/ml. Individual transformants were screened for the insert by colony PCR with primers P23 and P24. After verification of the sequence, one plasmid isolate was kept as the source of the constitutively expressed upp gene and named pMO715.
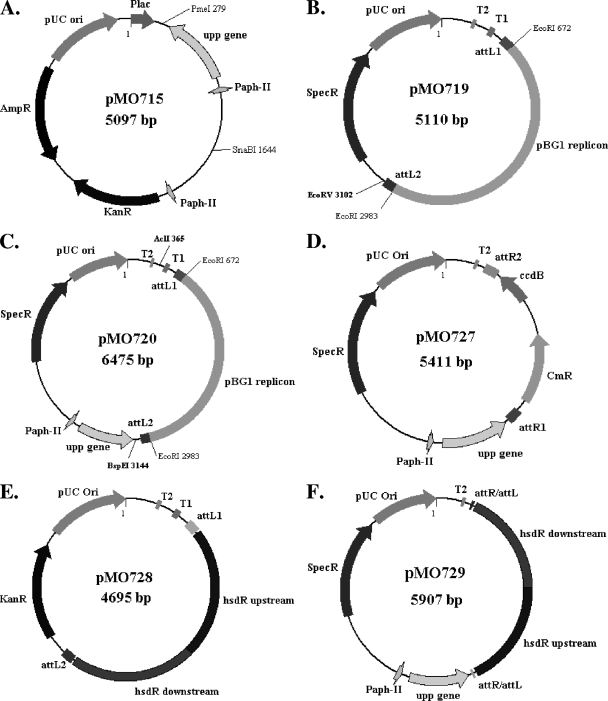
FIG. 2.
Plasmids constructed for the present study. Illustrations of various plasmids involved in the construction of the markerless deletion suicide plasmid for the type I site-specific restriction-modification system. Details of the construction of pMO715 (A), pMO719 (B), and pMO720 (C) are illustrated in Fig. S1 in the supplemental material, while the construction of pMO727 (D), pMO728 (E), and pMO729 (F) is illustrated in Fig. S2 in the supplemental material.
Generation of plasmids for deletion construction. (i) pMO719 and pMO720.
A stable plasmid conferring spectinomycin resistance to Desulfovibrio strains was constructed and designated pMO719 (Fig. 2B). The plasmid was built as a shuttle vector. The replicon for E. coli and the spectinomycin resistance were provided by the EcoRI-bounded pCR8/GW/TOPO fragment. The Desulfovibrio desulfuricans G100A replicon of the cryptic plasmid pBG1 (33) allows for replication in the SRB strains and was provided from pSC27 (31) (see Fig. S1 in the supplemental material).
The upp gene and the aph(3′)-II promoter were removed from pMO715 by digestion with the restriction endonucleases PmeI and SnaBI. The gel-purified DNA fragment was ligated into the EcoRV site of pMO719 (Fig. 2B). After verification of the sequence, one isolate was named pMO720 (Fig. 2C).
(ii) pMO727.
In order to convert pMO720 into a Gateway destination vector (Invitrogen) that would allow facile construction of mutagenic vectors for generation of markerless deletions, the Gateway vector conversion system (Invitrogen) was used. The pBG1 replicon and the attL sites from pCR8/GW/TOPO were removed from pMO720 by digestion with the restriction endonucleases AclI and BspEI (see Fig. S2 in the supplemental material). The ends of the remaining vector DNA were made blunt with DNA polymerase I, large (Klenow) fragment (NEB). After dephosphorylation of the vector by Antarctic phosphatase (NEB), the Gateway reading frame cassette B (Invitrogen) was ligated, and the recombinant plasmid transformed into One Shot ccdB survival competent cells (Invitrogen). Transformants were selected on LC medium containing chloramphenicol and screened by colony PCR with the primers P29 and P30. After verification of the sequence, one isolate was named pMO727 (Fig. 2D).
(iii) pMO728.
The plasmid pMO728 (Fig. 2E) is an intermediate in the construction of the plasmid used for the deletion of hsdR, encoding the type I restriction-modification system endonuclease (DVU1703). The deletion cassette unique to pMO728 was constructed by SOE-PCR of 1,079 bp upstream of DVU1703 (primers P13 and P14) directly to 1,040 bp of the region downstream of DVU1702 (primers P15 and P16). The amplicon was purified and cloned into pENTR/D-TOPO (Invitrogen), the sequence was verified, and the construct was designated pMO728.
(iv) pMO729.
The suicide plasmid, pMO729 (Fig. 2F), for the construction of the markerless deletion of the gene encoding the type I restriction endonuclease hsdR (DVU1703) was made from pMO727 and pMO728 with the LR Clonase Reaction as instructed by the manufacturer (Invitrogen) (see Fig. S2 in the supplemental material). After verification of the sequence, one isolate was named pMO729.
Electroporation in D. vulgaris.
For the preparation of D. vulgaris cells for electroporation, a 50-ml Wall LS3 culture was grown to an optimal optical density at 600 nm of 0.4 to 0.6. After centrifugation, cells were washed with 50 ml of chilled, sterile electroporation wash buffer (30 mM Tris-HCl buffer [pH 7.2], not anaerobic), and centrifuged again. The resulting pellet was resuspended in 0.5 ml of chilled wash buffer. To 50 μl of prepared cells, 5 μl (∼1 μg) of the plasmid was added, and the mixture was transferred to a cuvette. The electroporation parameters were the same as those described for Δupp construction, except that the electroporation was performed aerobically. The transformed cells were allowed to recover in 1 ml of Wall LS3 medium overnight at ∼32°C anaerobically. Various aliquots of the electroporated, recovered cells were plated in Wall LS4 medium with spectinomycin. The plates were incubated for 4 to 7 days at 32°C until spectinomycin-resistant transformant colonies were visible.
Southern blot analysis.
Southern blots were performed as previously described (2), with the template for the probe being the PCR amplicon of the upstream region of the hsdR gene (primers P13 and P14).
RESULTS
Construction of a Δupp host strain.
The natural resistance of the SRBs to many antibiotics (28) limits the available markers for facile selection in these bacteria. The use of a PRTase as the basis for a counterselection strategy in D. vulgaris would eliminate the necessity of a residual antibiotic resistance for each mutation constructed. A counterselection is possible if (i) the wild-type strain is sensitive to a toxic base analog, (ii) a strain deleted for the gene encoding the PRTase is resistant to the analog, and (iii) the reintroduction of the PRTase gene restores sensitivity. The PRTase deletion mutant becomes the host for the construction of further deletions without retention of antibiotic resistance markers and each deletion can be made without generating polar mutations.
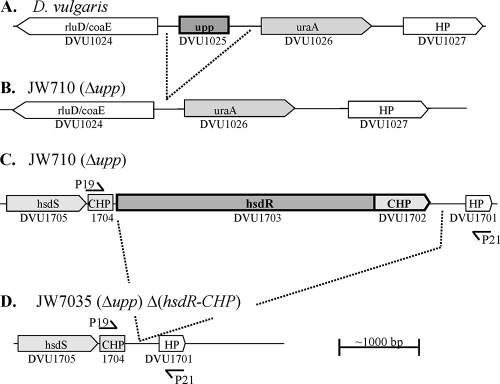
Analysis of the genome of D. vulgaris revealed a gene annotated as uracil PRTase, upp, DVU4025 (Fig. 3). Experiments to determine whether D. vulgaris was inhibited by the toxic pyrimidine analog 5-FU indicated that the bacterium was sensitive to as little as 0.1 μg of 5-FU/ml (3). In order to use 5-FU for counter selection, a host strain with the upp gene deleted (5-FUr) was constructed. An electroporation of D. vulgaris was performed with the suicide plasmid vector pMO711 containing 987 bp upstream and 858 bp downstream of the upp gene fused together. By selection for 5-FUr, transformants were obtained that were deleted for the upp gene after a double recombination with the mutagenic plasmid. After PCR verification (data not shown), one isolate was designated JW710 (Fig. 3B). The resistance of JW710 to 5-FU was confirmed (Fig. 4B), and this deletion strain serves as the host strain for the markerless deletion system in D. vulgaris.
FIG. 3.
Illustration of the upp (A and B) and hsdR (C and D) operons before (A and C) and after (B and D) the deletion of genes within the operons. Shaded boxes and boxed arrows are predicted to be open reading frames in the same operon. Arrowheads indicate direction of transcription. Dark-outlined open reading frames with bold letters were targets for deletions. Operons, gene annotation, and DVU numbers were taken from MicrobesOnline (http://www.microbesonline.org/) (1). (The primers are not drawn to scale.)
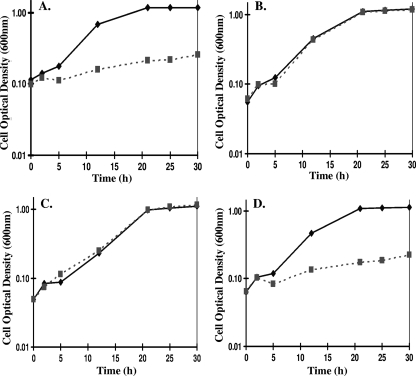
FIG. 4.
The aph(3′)-II promoted upp gene restores 5-FU sensitivity of JW710 (Δupp) to wild-type D. vulgaris levels. The growth of wild-type D. vulgaris (A), JW710 (Δupp) (B), JW710 (Δupp) carrying the SRB stable plasmid pMO719 without upp (C), and JW710 (Δupp) carrying the SRB stable plasmid pMO720 with the aph(3′)-II promoted upp gene (D) was determined. (A and B) Strains were grown in Wall LS4 medium with either 2.5 μg of 5-FU/ml (▪, dashed lines) or without (⧫, solid line). (C and D) Strains were grown in the same medium modified by the inclusion of 100 μg of spectinomycin/ml. Cultures were incubated anaerobically at 37°C for 30 h, and growth was measured as the optical density at 600 nm. Each point is the average of triplicate samples; errors bars are within data symbols and represent standard deviations of ≤0.031.
Complementation of Δupp.
To complement the upp deletion, the upp gene was cloned under the control of the aph(3′)-II promoter, which is known to be constitutively expressed in D. vulgaris. The promoter and gene were captured in the pCR4Blunt-TOPO plasmid, creating pMO715 (Fig. 2A). The restriction fragment containing the promoter and upp was then cloned into the EcoRV site of pMO719 (Fig. 2B), generating pMO720 (Fig. 2C), a stable plasmid capable of replicating in D. vulgaris. Both pMO719 and pMO720 were electroporated into JW710, the strain deleted for upp, to explore 5-FU resistance phenotypes.
D. vulgaris and JW710 (Δupp) grown in the presence or absence of 2.5 μg of 5-FU/ml showed that D. vulgaris was inhibited at this concentration of the pyrimidine analog (Fig. 4A), whereas the growth of JW710 was not affected (Fig. 4B). JW710 cells carrying pMO719, the vector without the upp gene, were resistant to 5-FU (Fig. 4C); however, JW710 cells carrying pMO720, constitutively expressing upp, were inhibited at the same levels as wild-type D. vulgaris (Fig. 4). The restoration of 5-FU sensitivity to JW710 demonstrated that the construct was fully functional. Therefore, the deletion of the upp gene in D. vulgaris created a strain, JW710, to serve as the host for the two step integration and excision markerless deletion procedure.
Generation of the destination vector, pMO727, and its use in deleting the gene for the type 1 restriction-modification endonuclease (DVU1703-1702).
Since a suicide deletion plasmid containing three features—(i) a fusion of the region ∼1 kb upstream and ∼1 kb downstream of the gene of interest, (ii) an antibiotic resistance gene (spectinomycin resistance), and (iii) a functional upp gene—is required for the generation of each markerless mutant, Gateway Technology (Invitrogen) was utilized to expedite the mutagenic vector construction process. A universal destination vector, pMO727 (Fig. 2D), was constructed containing the aph(3′)-II promoter fused to upp, a gene conferring spectinomycin resistance, and the attR1/attR2 sites. DNA fragments flanking the gene to be deleted were captured in pENTR/D-TOPO, and the movement of that cassette into pMO727 to create the specific mutagenic vectors was performed in vitro by site-specific recombination enzymes from temperate bacteriophage (Invitrogen). The effectiveness of pMO727 as a destination vector was tested by the generation of the deletion vector for a type I restriction endonuclease gene, hsdR gene (DVU1703), pMO729 (Fig. 2F). For the deletion of the hsdR, a markerless deletion cassette was generated by fusing ∼1 kb of the region upstream of hsdR (DVU1703) directly to ∼1 kb of the region downstream of DVU1702. This markerless deletion cassette was then cloned into pENTR/D-TOPO, producing the entry vector, pMO728 (Fig. 2E). An LR clonase reaction allowed for recombination between the attR1/attR2 sites in the destination vector pMO727 and the attL1/attL2 sites in the entry clone pMO728, which generated the markerless deletion suicide vector pMO729 that was then electroporated into JW710.
Southern blot analysis was performed (data not shown) on several transformants to confirm the expected integration of pMO729 into the chromosome of JW710 (Fig. 5, step 1). One spectinomycin-resistant 5-FUs isolate was confirmed for appropriate integration of this mutagenic plasmid into the JW710 chromosome and was designated JW7034 (Fig. 5).
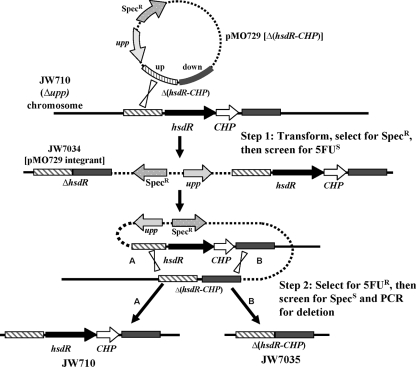
FIG. 5.
Schematic representation of the multistep process used for the construction of the markerless deletion mutant JW7035 in D. vulgaris. Step 1 shows the integration of the deletion plasmid pMO729 into the chromosome of the Δupp strain, JW710, selected as resistance to spectinomycin. A plasmid integrant achieved in step 1 (JW7034) was grown in the absence of spectinomycin and plated on Wall LS4 medium containing 40 μg of 5-FU/ml, selecting for 5-FUr transformants. Depending on the location of the second recombinational event, the 5-FUr transformants were either wild type for hsdR (A) or had hsdR deleted without retention of an antibiotic resistance gene (B). PCR screens were performed on transformants to identify possible markerless deletions. Southern blot analysis was then used to further verify the proper construction of the markerless deletion of hsdR-CHP, JW7035. The schematic is not drawn to scale.
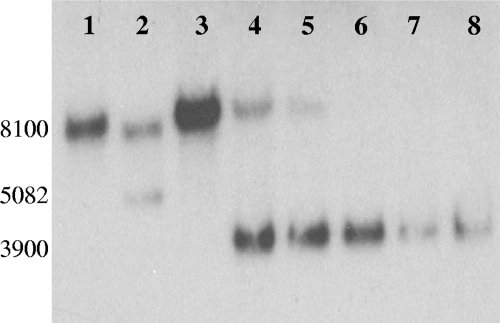
The second step for creating the ΔhsdR mutant was the selection of strains that had lost pMO729 sequences from the chromosome by a second recombination event. Such a recombinational event would render cells 5-FUr and spectinomycin sensitive by loss of the upp gene and the gene encoding the antibiotic resistance, respectively. The location of that second recombinational event determined whether the 5-FUr strain had deleted or restored the wild-type hsdR gene (Fig. 5, Step 2). JW7034 was grown in Wall LS4 medium (in the absence of spectinomycin) for 24 h to allow the second recombinational event to occur that would resolve the partial diploid (Fig. 5, step 2). Sixteen 5-FUr colonies were picked, grown, and screened by PCR for the deletion of hsdR (primer positions shown in Fig. 3). Nine of the sixteen transformants had clearly identifiable bands of the expected size for the deletion of hsdR-CHP (data not shown). Southern blot analysis was performed on six of the nine isolates with a probe corresponding to the DNA upstream of DVU1703 (Fig. 6). DNA from the host strain JW710, hsdR+, produced a single BclI band of the predicted size (8,100 bp; Fig. 6, lane 1). Strain JW7034 with the integrated mutagenic plasmid produced two bands (7,165 and 5,082 bp) as expected (Fig. 6, lane 2) because it is a merodiploid for the region probed (Fig. 5, step 1). Genomic DNA from a successful deletion of the hsdR-CHP genes was predicted to produce a single 3,900-bp BclI band. In contrast to the PCR results, one isolate clearly yielded only the wild-type hsdR BclI band of 8100 bp (Fig. 6, lane 3). Genomic DNA from five of the transformants had a 3,900-bp band (Fig. 6, lanes 4 to 8); however, two of these transformants also contained an 8,100-bp band with homology to the probe (Fig. 6, lanes 4 and 5), suggesting that mixed colonies may have been selected. Three isolates appeared to contain a genome structure consistent with a deletion of the hsdR-CHP region (Fig. 6). An isolate (Fig. 6, lane 6) was kept as the markerless deletion mutant lacking the hsdR and the downstream CHP gene and was designated JW7035.
FIG. 6.
Southern blot verification of deletion of type I restriction modification, R-subunit (hsdR). BclI digestion of gDNA—JW710 (lane 1), JW7034 (lane 2), and six putative isolates of JW7035 (lanes 3 to 6)—was performed. Homology was predicted to be found in a 8,100-bp band for the wild-type hsdR gene, two bands at 7,165 and 5,082 bp for the merodiploid produced by integration of pMO729 into the chromosome, and a 3,900-bp fragment for the successful markerless deletion the hsdR gene. DNA fragment sizes were determined by comparison with the migration of the GeneRuler 1-kb Plus DNA ladder (Fermentas [data not shown]).
Characterization of transformation efficiency of JW7035.
To determine whether the deletion of the type I restriction endonuclease hsdR gene had an effect on the transformation efficiency, transformation experiments with plasmids that replicate in D. vulgaris (containing the endogenous SRB cryptic plasmid pBG1) were performed. Competent cells of wild-type D. vulgaris and mutant strains JW710, JW801, and JW7035 were prepared and electroporations performed with either pSC27 or pMO719. Competent cells subjected to electroporation without plasmid DNA served as controls to determine the number of spontaneously antibiotic resistant colonies. Three separate electroporation experiments were performed for each strain, and the number of transformant CFU were counted and compared. Although the number of transformants recovered varied among the electroporation experiments, the differences in transformation efficiencies between the strains were consistent (Table 2).
TABLE 2.
Characterization of transformation efficienciesa
| D. vulgaris strainb | No. of transformants/μg of plasmid DNA |
|
|---|---|---|
| pSC27 (G418r) | pMO719 (Specr) | |
| Wild type | 2.1 × 100 | 4.4 × 100 |
| JW801 | 2.3 × 102 | 6.3 × 103 |
| JW710 (Δupp) | 3.1 × 10−1c | 3.2 × 102 |
| JW7035 (Δupp ΔhsdR) | 2.4 × 103 | 2.8 × 103 |
Data are averages from two separate electroporation experiments, with multiple plates counted per experiment.
Spontaneous resistance to G418 or spectinomycin was not detected in the sample sizes of the recipient strains plated. Therefore, fewer than 1 in 109 cells of each recipient strain were spontaneously resistant.
The low transformation efficiency for JW710 resulted from a lack of transformation in one of the two experiments used for the averages. Additional experiments showed that 2 to 10 transformants per μg of plasmid DNA was a more typical result.
Transformation of wild-type D. vulgaris by pSC27, a 9.1-kb plasmid containing a kanamycin resistance determinant, or pMO719, a 5.1-kb plasmid conferring spectinomycin resistance, resulted in low, but similar efficiencies of two to five transformants per μg of plasmid DNA. Loss of the upp gene did not improve transformation by pSC27 but, unexpectedly, did appear to increase the transformation efficiency of pMO719. In contrast, loss of the hsdR gene resulted in 102- to 103-fold increases in the numbers of transformants obtained for these plasmids compared to the wild type. The loss of the native plasmid, pDV1, from wild-type D. vulgaris that generated JW801 also improved transformation efficiencies by increases similar to JW7035. Annotation of pDV1 revealed a type II restriction modification system, and its loss by plasmid curing is a possible cause of the increased transformation observed for JW801. Experiments to confirm this hypothesis are under way.
To verify that the increase in transformation efficiency of JW7035 was due to the deletion of hsdR and not due to the loss of the native pDV1 plasmid, PCR experiments were performed to amplify genes annotated on either pDV1 or the chromosome. DVUA0015 encoding the nifH gene was targeted as a reporter for the presence of pDV1. Chromosomal genes, DVU3152 (encoding a histidine kinase) and DVU0942 (encoding a fur homolog), were amplified to confirm the quality of the gDNA templates. The PCR products confirmed that pDV1 was present in wild-type D. vulgaris, JW710, and JW7035 (see Fig. S3, lanes 2 to 4, in the supplemental material) and absent in JW801 (see Fig. S3, lanes 1, in the supplemental material). These data were consistent with our interpretation that loss of the type I restriction endonuclease caused by deletion of the hsdR produced the increase in transformation efficiency in JW7035.
DISCUSSION
Much is still to be learned about the metabolism and electron flow of the SRB, through studies of the model strain D. vulgaris Hildenborough. Although advances have been made in the ability to generate marker exchange deletion mutants (2), the introduction of multiple deletions into a single strain has been limited (28). In a recent article, Giaever and Nislow (14) discuss the importance of obtaining multiple mutations and knockouts to fully dissect and understand the architecture of cellular pathways, including regulatory networks. As more SRBs are sequenced and genomes annotated, the need to make multiple deletions to test metabolic pathways is ever increasing.
In the present study, we have described the development of a new markerless deletion system in D. vulgaris Hildenborough. We have shown that the upp gene, in combination with the toxic pyrimidine base analog 5-FU, is an effective counterselectable marker in D. vulgaris. The uracil PRTase salvages pyrimidine bases during nucleotide turnover in the bacterium and permits incorporation of the toxic analog 5-FU. In comparison with most antibiotic resistance determinants whose loss can be detected only by screening a rather small number of cells, the loss of the upp gene can be selected as the rare acquisition of resistance to 5-FU. This provides a powerful counterselection. The upp gene itself was successfully deleted from wild-type D. vulgaris by direct selection of a rare double recombination event, generating the 5-FUr JW710 (Δupp). JW710 then provided the background for the markerless deletion system (Fig. 5).
To test the mutagenesis system, an unmarked deletion of the gene for a type 1 restriction endonuclease was created. In the final step, two genotypes with identical 5-FU resistance were predicted from the resolution of the partial diploid state. One recombinational event (Fig. 5, step 2B) generates the desired mutation, while the other restores the wild-type gene and chromosomal structure (Fig. 5, step 2A). The screening process to distinguish these events can become tedious, especially if the wild-type has any growth advantage over the mutant. However, compared to the widely used counterselectable sacB gene, which confers sucrose sensitivity (8, 10, 15, 18, 19, 21), the selection for 5-FU resistance is more straight forward and cleaner in our hands. From the resolution of the merodiploid, one would expect the theoretical ratio of wild-type gene to desired mutant to be equal, 50% wild type and 50% mutant, if the relative sizes of homologous DNA available for the recombination events up and downstream of the target gene are equal. In our initial PCR screening for the hsdR deletion, we were able to obtain these theoretical levels, since ∼56% of the transformants screened were consistent with the desired gene deletion.
Preliminary data indicate JW7035 not only shows an increase in the efficiency of transformation of stable plasmids but also shows an increase in the number of transformants when the selecting events depend on both transformation and recombination (e.g., Fig. 5, step 1). By using JW7035 as the host strain for the markerless deletion system for D. vulgaris, instead of JW710, we should be able to increase the efficiency of construction of markerless deletion mutants because of the required plasmid integration.
JW801 also had increased transformation efficiency of plasmids compared to wild-type D. vulgaris that may be due to the loss of a type II restriction system encoded on pDV1. We have provided data showing that the increase in efficiency of JW7035 is not due to the loss of pDV1. We are currently in the process of making a markerless deletion of the type II restriction system to test the role of these genes in the transformation efficiency of D. vulgaris.
As we begin to make multiple deletions to test the metabolic pathways in D. vulgaris, the wild type may quickly gain a growth advantage over multiple gene deletion mutants. A growth advantage of wild type over a mutant would make the screening process for a markerless deletion very long and tedious. We are working to modify the procedure described here to eliminate the resolution of the merodiploid resulting in the wild-type gene and drastically reduce screening by a procedure adapted from B. subtilis (9) and in Methanosarcina acetivorans (30) incorporating the use of the Flp recombinase.
Overall, we have demonstrated the functionality of the upp counterselection strategy for markerless deletion in D. vulgaris by construction of an in-frame deletion in the hsdR gene. Since the upp counterselectable marker is recyclable, many genes can be sequentially deleted in the same strain for study of metabolic pathways. In addition, these markerless deletions can be designed to remain “in-frame” for the deletion of the genes within an operon without having polar effects on the downstream genes. This genetic exchange system can also be used to modify genes for the production of tagged proteins expressed from their native promoters. In addition, site-directed mutagenesis of target genes should be possible. This procedure and future improvements will greatly improve the genetic accessibility of this important group of environmental anaerobes.
Supplementary Material
Acknowledgments
This study was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through the grants ESPP-ESPP2 (http://VIMSS.lbl.gov) from the DOE Office of Science; Genomics: GTL through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy; and by the Office of Science (BER), U.S Department of Energy, grant OBER BioHydrogen Production and BioEthanol DE-FG02-083464691.
Footnotes
Published ahead of print on 16 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res. 15:1015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, K. S., H.-C. B. Yen, C. L. Hemme, Z. Yang, Z. He, Q. He, J. Zhou, K. H. Huang, E. J. Alm, T. C. Hazen, A. P. Arkin, and J. D. Wall. 2007. Analysis of a ferric uptake regulator (fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 73:5389-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, K. S., H.-C. Yen, and J. D. Wall. 2006. Analysing the metabolic capabilities of Desulfovibrio species through genetic manipulation. Biotechnol. Genet. Eng. Rev. 23:157-174. [DOI] [PubMed] [Google Scholar]
- 4.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 6.Brandis, A., and R. K. Thauer. 1981. Growth of Desulfovibrio species on hydrogen and sulfate as sole energy source. J. Gen. Microbiol. 126:249-252. [Google Scholar]
- 7.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovely. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolla, A., B. K. J. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol. 174:143-151. [DOI] [PubMed] [Google Scholar]
- 9.Fabret, C., S. D. Ehrlich, and P. Noirot. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 10.Fu, R., and G. Voordouw. 1997. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 11.Fu, R., and G. Voordouw. 1998. ISDI, an insertion element from the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough: structure, transposition, and distribution. Appl. Environ. Microbiol. 64:53-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukagawa, T., N. Hayward, J. Yang, C. Azzalin, D. Griffin, A. F. Stewart, and W. Brown. 1999. The chicken HPRT gene: a counter selectable marker for the DT40 cell line. Nucleic Acids Res. 27:1966-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gay, P., D. Le Coq, M. Steinmetz, E. Ferrari, and J. A. Hoch. 1983. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J. Bacteriol. 153:1424-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giaever, G., and C. Nislow. 2009. Knocking sense into regulatory pathways. Nat. Biotechnol. 27:149-150. [DOI] [PubMed] [Google Scholar]
- 15.Goenka, A., J. K. Voordouw, W. Lubitz, W. Gaertner, and G. Voordouw. 2005. Construction of a [NiFe]-hydrogenase deletion mutant of Desulfovibrio vulgaris Hildenborough. Biochem. Soc. Trans. 33:59-60. [DOI] [PubMed] [Google Scholar]
- 16.Goh, Y. J., M. A. Azcaŕate-Peril, S. O'Flaherty, E. Durmaz, F. Valence, J. Jardin, S. Lortal, and T. R. Klaenhammer. 2009. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 75:3093-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Haveman, S. A., V. Brunelle, J. K. Voordouw, G. Voordouw, J. F. Heidelberg, and R. Rabus. 2003. Gene expression analysis of energy metabolism mutants of Desulfovibrio vulgaris Hildenborough indicates an important role for alcohol dehydrogenase. J. Bacteriol. 185:4345-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haveman, S. A., E. A. Greene, C. P. Stilwell, J. K. Voordouw, and G. Voordouw. 2004. Physiological and gene expression analysis of inhibition of Desulfovibrio vulgaris Hildenborough by nitrite. J. Bacteriol. 186:7944-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, R. M., Z. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-8535. [PubMed] [Google Scholar]
- 21.Keon, R. G., F. Rongdian, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 22.Kristich, C. J., D. A. Manias, and G. M. Dunny. 2005. Development of a method for markerless genetic exchange in Enterococcus faecalis and its use in construction of a srtA mutant. Appl. Environ. Microbiol. 71:5837-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel, C., M. Brugna, C. Aubert, A. Bernadac, and M. Bruschi. 2001. Enzymatic reduction of chromate: comparative studies using sulfate-reducing bacteria. Key role of polyheme cytochromes c and hydrogenases. Appl. Microbiol. Biotechnol. 55:95-100. [DOI] [PubMed] [Google Scholar]
- 24.Moore, B. C., and J. A. Leigh. 2005. Markerless mutagenesis in Methanococcus maripaludis demonstrates roles for alanine dehydrogenase, alanine racemase, and alanine permease. J. Bacteriol. 187:972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay, A., Z. He, E. J. Alm, A. P. Arkin, E. E. Baidoo, S. C. Borglin, W. Chen, T. C. Hazen, Q. He, H.-Y. Holman, K. Huang, R. Huang, D. C. Joyner, N. Katz, M. Keller, P. Oeller, A. Redding, J. Sun, J. Wall, J. Wei, Z. Yang, H.-C. Yen, J. Zhou, and J. D. Keasling. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne, R. B., D. M. Gentry, B. J. Rapp-Giles, L. Casalot, and J. D. Wall. 2002. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl. Environ. Microbiol. 68:3129-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peck, R. F., S. Dassarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 28.Postgate, J. R. 1984. The sulphate-reducing bacteria. Cambridge University Press, London, United Kingdom.
- 29.Pritchett, M. A., J. K. Zhang, and W. W. Metcalf. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rother, M., and W. W. Metcalf. 2005. Genetic technologies for Archaea. Curr. Opin. Microbiol. 8:745-751. [DOI] [PubMed] [Google Scholar]
- 31.Rousset, M., L. Casalot, B. J. Rapp-Giles, Z. Dermoun, P. de Philip, J. P. Bélaich, and J. D. Wall. 1998. New shuttle vectors for the introduction of cloned DNA in Desulfovibrio. Plasmid 39:114-122. [DOI] [PubMed] [Google Scholar]
- 32.Steyert, S. R., and S. A. Pineiro. 2007. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 73:4717-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wall, J. D., B. J. Rapp-Giles, and M. Rousset. 1993. Characterization of a small plasmid from Desulfovibrio desulfuricans and its use for shuttle vector construction. J. Bacteriol. 175:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, G., S. P. Kennedy, S. Fasiludeen, C. Rensing, and S. DasSarma. 2004. Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J. Bacteriol. 186:3187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, S. S., and D. Kaiser. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178:5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.