Abstract
Several microorganisms are known for their efficient anaerobic conversion of glycerol to 1,3-propanediol, with Clostridium diolis DSM 15410 as one of the better performers in terms of molar yield and volumetric productivity. However, this performance is still insufficient to compete with established chemical processes. Previous studies have shown that high concentrations of 1,3-propanediol, glycerol, and fermentation side products can limit the productivity of C. diolis DSM 15410. Here, we describe the use of genome shuffling for improved 1,3-propanediol fermentation by the strict anaerobe C. diolis DSM 15410. By using chemical mutagenesis, strains with superior substrate and product tolerance levels were isolated and higher product yields were obtained. These superior strains were then used for genome shuffling and selection for 1,3-propanediol and organic acid tolerance. After four rounds of genome shuffling and selection, significant improvements were observed, with one strain attaining a 1,3-propanediol volumetric yield of 85 g/liter. This result represents an 80% improvement compared to the yield from the parental wild-type strain.
The use of biomass instead of petrochemical feedstock could facilitate the sustainable production of many chemicals, but this approach has proven economically feasible in only a few cases (4, 17, 20, 21). The microbial production of 1,3-propanediol (1,3-PD) provides an interesting case study because this monomer is used to produce several plastics, including the relatively new and highly versatile polytrimethylene terephthalate, which has significantly increased demand for 1,3-PD (12, 23, 24). Polytrimethylene terephthalate is currently produced from petrochemical feedstock in a process that involves the conversion of ethylene oxide into 3-hydroxypropionaldehyde by hydroformylation under high pressure and then further into 1,3-PD by hydrogenation using a nickel or rubidium catalyst (21).
Several companies have investigated the sustainable production of 1,3-PD from biomass. For example, DuPont and Genencor transferred the relevant 1,3-PD biosynthetic genes from Klebsiella pneumonia into Escherichia coli and further modified carbohydrate metabolism and transport so that 1,3-PD could be synthesized from glucose (10, 11; M. Emptage, S. L. Haynie, L. A. Laffend, J. Pucci, and G. M. Whited, 2001, Patent Cooperation Treaty international application WO 2001/01/12833). Large-scale production of 1,3-PD by this approach is likely to be too expensive due to the high input costs of vitamin B12 and antibiotics, so the use of glycerol as an alternative feedstock has been investigated, although this strategy requires additional enzymes and a shift from aerobic to anaerobic conditions (15, 25, 30).
Clostridium diolis DSM 15410 (formerly C. butyricum DSM 5431) can produce 1,3-PD from glycerol under anaerobic conditions and is therefore a desirable alternative to E. coli given the relative costs of industrial aerobic and anaerobic fermentation (6, 8, 9, 22). However, the efficiency of conversion is not yet high enough for an industrial process. The production of 1,3-PD by C. diolis is limited by inhibition from both substrates and products, as well as organic acids produced as fermentation by-products (13).
Classical strain improvement has significantly increased 1,3-PD production, but this is a slow process and the mutations are predominantly neutral or detrimental (2). We have therefore approached the problem using genome shuffling, which is more efficient and reliable for engineering complex phenotypes, as demonstrated in several other examples of microbial strain development (16, 26, 31). Genome shuffling offers the advantages of accumulated beneficial mutations and the removal of unnecessary mutations due to simultaneous changes at different positions throughout the genome and, therefore, yields microbes of superior fitness (29). We applied both the classical approach and genome shuffling to C. diolis DSM 15410 to improve the production of 1,3-PD, which is a necessary prerequisite for the fermentation process. To our knowledge, this study is the first example of genome shuffling in a strictly anaerobic microorganism.
MATERIALS AND METHODS
Bacteria and media.
C. diolis DSM 15410 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany). Spores were stored at 4°C in reinforced clostridial medium (RCM; Carl-Roth, Germany). Standard cultivation was carried out in a Bactron IV anaerobic chamber (Omni Life Sciences, Hamburg) at 34°C. The preculture medium (improved minimal medium [IMM]) contained the following components per liter of distilled water: 18 g glycerol, 13.6 g KH2PO4, 17.4 g K2HPO4, 2 g (NH4)SO4, 0.2 g MgSO4, 20 mg CaCl2, 5 mg FeSO4, 1 g yeast extract, and 2 ml trace element solution SL7 (7). For high-throughput screening in 96-deep-well plates, IMM was used with the following additions or changes: 60 g/liter glycerol, 0.1 g/liter K2HPO4, 100 mM MES [2-(N-morpholino)ethanesulfonic acid], 1 g/liter CaCO3, 5 g/liter yeast extract, and bromothymol blue as a pH indicator. Cultivation in 96-deep-well plates was carried out for 3 days. The medium for bioreactor fermentation was the same as IMM except for the use of 1 g/liter K2HPO4, 0.5 g/liter KH2PO4, and 5 g/liter yeast extract.
An exponential-phase culture grown in preculture medium was used as the inoculum at 6% (vol/vol) for 96-deep-well plate fermentation and 5% (vol/vol) for batch or fed-batch fermentation in a 250-ml bioreactor vessel (Ochs GmbH, Bovenden/Lenglern, Germany). The pH and feed were coupled by using a ratio of 1.5 M KOH to 3.75 M glycerol. The pH was monitored and maintained at 7.0 with a T50 titrator (Mettler Toledo GmbH, Giessen, Germany). Feed consisting of 20 g/liter yeast extract, 6 g/liter NH4SO2, 60 mg/liter CaCl2, 20 mg/liter FeSO4, 6 ml/liter trace element solution SL7, and 8 M glycerol was manually added during the fermentation to maintain the glycerol concentration between 20 and 25 g/liter and to refresh the medium composition. The growth temperature for 96-deep-well plate fermentations was 34°C, whereas that for fed-batch fermentations was 28°C. The working volume for deep-well fermentations was 1 ml, and that for fed-batch fermentations was 220 ml.
Crude glycerol.
Crude glycerol was kindly provided by M. Menner (Fraunhofer Institute IVV, Freising, Germany) from a biodiesel production plant in Mainburg (Germany). It was at pH 11 and contained the following components: glycerol, 70% (wt/vol); potash, 15%; fatty acids, 5 to 10%; and methanol, <1%. For the fed-batch fermentations, the fatty acids were removed by acidification and subsequent extraction. Glycerol was concentrated by a rotary evaporator.
Mutagenesis and selection.
N-Methyl-N″-nitro-N-nitrosoguanidine (NTG) was used for mutagenesis in C. diolis at the initial exponential phase: 10-ml aliquots of cells were treated with different concentrations of NTG for at least 1 h at 34°C. The cells were then centrifuged for 10 min at 4,000 × g, washed twice with anaerobic potassium phosphate buffer, resuspended in IMM, and regenerated for at least 1 h. Cells were spread onto agar plates containing different concentrations of 1,3-PD or glycerol. For high-throughput screening, the mutated cells were spread onto IMM agar plates. Colonies were picked from 96-deep-well plates.
Analytical methods.
The cell concentrations were estimated as the dry weight of cells by using a predetermined correlation between A600 (measured with a BioPhotometer [Eppendorf, Hamburg, Germany]) and the dry weight of cells. For the analysis of fermentation metabolites, either high-performance liquid chromatography (HPLC) or gas chromatography-mass spectrometry (GC-MS) was applied. For HPLC, we used a Shimadzu Prominence HPLC system equipped with a refractive index detector. For the HPLC separation, we used a Rezex RFQ-Fast Acid column (100 by 7.8 mm) from Phenomenex (Aschaffenburg, Germany), and metabolites were detected by a refractive index detector. The following operating conditions were employed: oven temperature, 60°C; mobile phase, 5 mM sulfuric acid; and flow rate, 1 ml/min. As an internal standard, 48 mM 1,5-pentanediol was used. For GC-MS analysis, a Shimadzu GCMS-QP2010S system was used. For sample preparation prior to analyses of 1,3-PD and glycerol concentrations, deep-well plates were centrifuged for 10 min at 3,000 × g and 25 μl of biomass-free supernatant was transferred into 975 μl ethanol containing 11 mM 1,4-butanediol as an internal standard. Additional centrifugation was carried out for 10 min at 3,000 × g. A total of 600 μl of the sample was transferred into a new deep-well plate. A Shimadzu GCMS-QP2010S system equipped with an Rtx Wax GC column (internal diameter, 0.18 mm; length, 10 m [Restek, Bad Homburg, Germany]) was used for metabolite analysis. For separation of the compounds, the following temperature program was used: a split injection temperature of 250°C and a temperature gradient program starting at 130°C for 1 min, with heating of the column at 40°C per min to 250°C. Mass detection was done with electric ionization at 1 keV by using the selected-ion monitoring/scan mode for masses of m/z 50 to 100. Standards for the fermentation broth products were purchased from Sigma-Aldrich.
Genome shuffling and selection.
Cells were cultivated in RCM until they reached mid-log phase and harvested by centrifugation at 3,000 × g for 10 min. For protoplast formation, 500 μl of the culture was mixed with clostridium protoplasting buffer (CPB), which consisted of RCM with 500 mM sucrose, 25 mM MgCl2, 25 mM CaCl2, and 20 mg/liter lysozyme. Enzyme treatment was continued for 1 h at 35°C. The appearance of spherical cells as judged by light microscopy was used as an indicator of protoplast formation. Protoplasts were centrifuged twice for 5 min at 2,000 × g and washed with CPB lacking lysozyme. To generate fusions between protoplast preparations, approximately equal numbers of protoplasts from different populations were mixed and the mixture was centrifuged and resuspended in 100 μl CPB lacking lysozyme. Nine volumes of 30 to 60% polyethylene glycol 6000 in CPB were added, and the protoplast mix was incubated for 15 to 30 min at room temperature. For regeneration of protoplasts, 5 ml CPB lacking lysozyme was added and the protoplasts were centrifuged, washed, and resuspended in CPB containing 100 mM N-acetyl-glucosamine. Protoplasts were incubated for 5 h at 34°C and spread onto regeneration plates consisting of RCM plus 12.5 mM MgCl2, 12.5 mM CaCl2, 10 g/liter casein hydrosylate, and 40 g/liter gelatin. The plates were scraped, and the cells were spread onto selection medium consisting of IMM and different concentrations of glycerol, 1,3-PD, and butyric and acetic acids.
The formation of protoplasts and protoplast fusion, subsequent regeneration, and selection were repeated four times, with the pooled regenerated cells from one fusion serving as the inoculum for the subsequent protoplast culture. Controls were prepared by protoplast formation and regeneration without exposure to polyethylene glycol.
RESULTS
Improvement of glycerol tolerance through chemical mutagenesis.
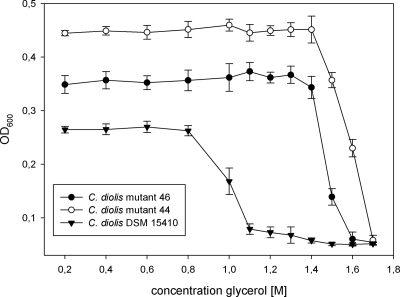
C. diolis DSM 15410 cannot grow on agar plates containing concentrations of glycerol greater than 1 M. In order to obtain mutants with the ability to grow at higher concentrations of glycerol, C. diolis DSM 15410 was subjected to treatment with NTG, followed by selection on agar plates with increasing concentrations of glycerol. We used a concentration of NTG that allowed 1% of cells to survive, and this protocol has turned out to be effective for strain improvement. The starting concentration for the following rounds of mutagenesis was 900 mM glycerol. The isolated mutants were used for subsequent mutagenesis at higher concentrations. In total, five rounds of mutagenesis and selection were carried out for the improvement of glycerol tolerance. Five mutants with enhanced abilities to grow on glycerol concentrations of up to 1.5 M were isolated (Fig. 1). All mutants produced more biomass than the wild type and showed similar behaviors with regard to glycerol tolerance.
FIG. 1.
Comparison of levels of inhibition of C. diolis DSM 15410 and mutants grown for 3 days at 35°C in media with increasing glycerol concentrations. Standard deviations were calculated from the results of at least six independent experiments. OD600, optical density at 600 nm.
Improvement of 1,3-PD tolerance through chemical mutagenesis.
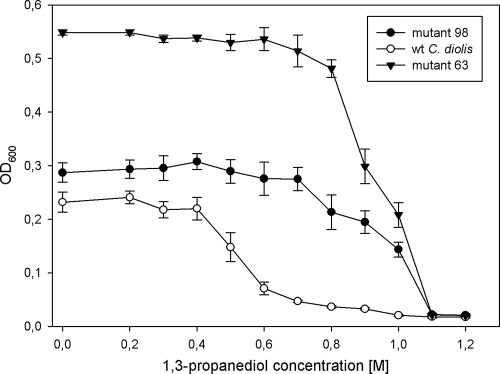
Growth of C. diolis is hampered by inhibition from 1,3-PD at concentrations greater than 600 mM. Chemical mutagenesis was employed to obtain a mutant that can tolerate more than 1 M 1,3-PD. The mutagenesis protocol was equivalent to that of the glycerol tolerance improvement approach. After four rounds of mutagenesis, four mutants that were able to grow on 1,3-PD at concentrations of up to 1.2 M were isolated. Figure 2 shows a representative characterization of the two best selected mutants. All mutants yielded significantly more biomass than the wild-type strain.
FIG. 2.
Comparison of levels of inhibition of C. diolis wild-type (wt) strain DSM 15410 and mutants grown for 3 days at 35°C in media with increasing 1,3-PD concentrations. Standard deviations were calculated from the results of at least six independent experiments. OD600, optical density at 600 nm.
Batch fermentations with C. diolis DSM 15410 and selected glycerol mutants.
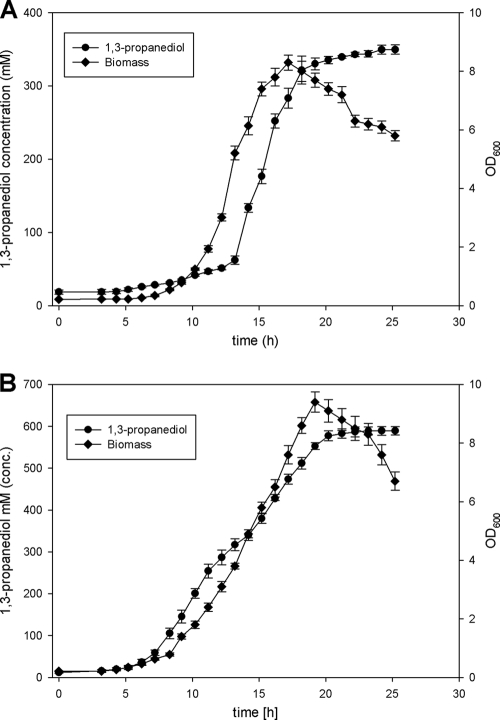
In order to test the performances of the C. diolis wild type and the selected glycerol-tolerant mutants in 1,3-PD formation, batch fermentations with these strains were carried out. Glycerol concentrations were chosen according to the highest concentrations tolerated by the wild type and mutant 98. Starting glycerol concentrations of 673 mM for the wild-type strain and 1.19 M for the mutant were used. Figure 3 shows the levels of 1,3-PD production by the wild-type strain and mutant 98. Mutant 98 produced 597 mM 1,3-PD, and the wild-type strain produced 328 mM 1,3-PD. For mutant 98, the ratio of moles of 1,3-PD produced to moles of glycerol consumed (Y1,3-PD) was 0.50, and the Y1,3-PD for the wild-type strain was 0.48.
FIG. 3.
(A) Results from batch fermentation with C. diolis DSM 15410. For the wild type, 62 g/liter glycerol was used as a starting concentration. Standard deviations were calculated from the results of at least four independent experiments. (B) Results from batch fermentation with the glycerol-tolerant mutant 98. A glycerol concentration of 112 g/liter was used as the starting concentration. Standard deviations were calculated from the results of at least four independent experiments. OD600, optical density at 600 nm; conc., concentration.
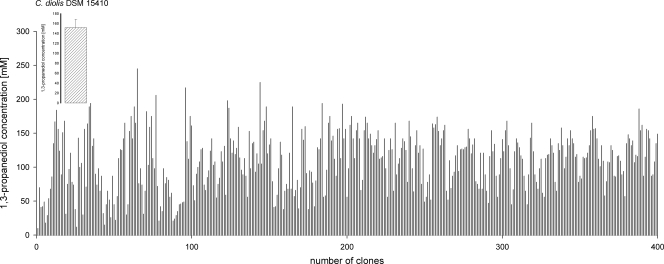
High-throughput screening for a superior 1,3-PD producer.
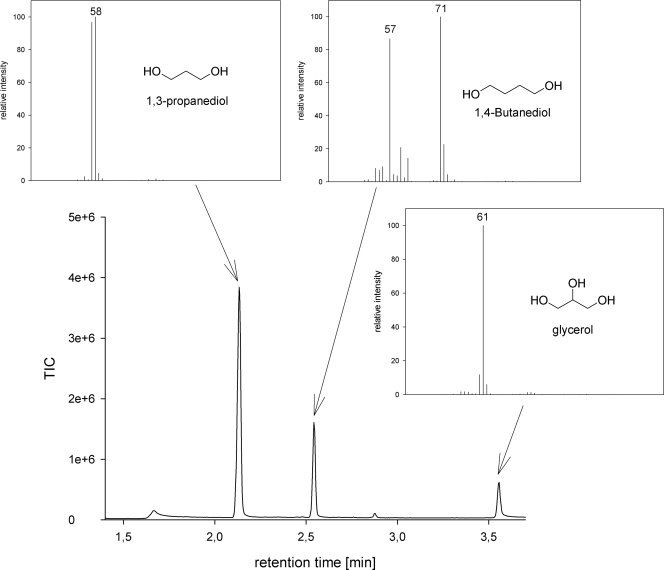
Due to the lack of direct selection methods for improved productivity, a mutagenesis approach with subsequent screening was performed. For this purpose, different concentrations of NTG were applied to the wild-type strain. Mutants were randomly picked and then tested for their productivity in a 96-deep-well small-scale fermentation format. A severe problem that was partially overcome was rapid acidification of the medium and subsequent growth inhibition due to the generation of organic acids as metabolic side products. We have tested various buffer conditions and found that a dynamic buffering system using MES and calcium carbonate attenuated the fast acidification. For analysis of product concentrations, 1,3-PD was extracted from the fermentation broth and analyzed by a fast yet accurate GC-MS method. In Fig. 4, a GC chromatogram and the corresponding MS spectra from the high-throughput screening approach are shown. Because of different growth conditions in the deep-well plates, mutants that showed improved productivity were retested for superior 1,3-PD production. This retesting was carried out by statistical analysis of the mutants and the wild type. At least eight independent small-scale fermentations were performed. This procedure was done in iterative rounds, and mutants with a modest but reproducible improvement in 1,3-PD production were obtained. In Fig. 5, a representative result for the final 1,3-PD yield from a small-scale fermentation is shown. Differences in product formation are clearly demonstrated. The wild-type strain C. diolis DSM 15410 produced up to 150 mM 1,3-PD. Among the screened mutants, PSM 7 produced up to 242 mM 1,3-PD under the applied screening conditions. In total, more than 4,000 mutants were screened for superior 1,3-PD production. Altogether, four mutants with superior 1,3-PD production in the high-throughput screening were isolated, with PSM 7 as the most productive one.
FIG. 4.
GC-MS analysis of fermentation broth showing the total ion chromatogram (TIC) together with the mass spectra of 1,3-PD, 1,4-butanediol, and glycerol.
FIG. 5.
Overview of 1,3-PD product formation after mutagenesis. Screening for a mutant with increased 1,3-PD production was performed in small-scale fermentations using 96-deep-well plates. The standard deviation in levels of 1,3-PD production by C. diolis DSM 15410 was calculated from the results of least eight independent experiments.
Genome shuffling for improvement of 1,3-PD production.
Successful formation of protoplasts from several different Clostridium spp. has been demonstrated previously (28), and the approach was slightly modified. Genome shuffling in mutants possessing enhanced tolerance for 1,3-PD and glycerol, as well as mutants with improved 1,3-PD productivity, was performed. The colonies that appeared after regeneration and subsequent selection were scraped from agar plates, pooled, and used for the next iterative cycle of genome shuffling with higher concentrations of 1,3-PD and butyric and acetic acids. A total of four rounds of genome shuffling were performed. Two strains, GSHM 2 and GSHM 4, that were able to grow at high 1,3-PD concentrations (1.05 M) and organic acid concentrations greater than 150 mM were isolated. These strains were further tested in fed-batch fermentations for their productivity using pharmaceutical or crude glycerol as the carbon source.
Comparative fed-batch fermentations with C. diolis DSM 15410 and strains obtained by high-throughput screening and genome shuffling.
In order to test the performances of the strains obtained from the high-throughput screening campaign and genome shuffling, fed-batch fermentations using pharmaceutical glycerol as the carbon source were conducted. Here, the wild-type strain produced a mean ± standard deviation of 624 ± 38 mM 1,3-PD, and the mutant PSM 7 produced 746 ± 55 mM 1,3-PD (Table 1). PSM 7, which was isolated from a high-throughput screening for elevated 1,3-PD production, offered a better yield than the wild type and a mildly improved specific production rate. The strains derived from genome shuffling showed better volumetric productivity and specific production rates and reached higher yields than the wild type and PSM 7. In total, strain GSHM 2 produced 1,032 ± 79 mM 1,3-PD and strain GSHM 4 produced 1,114 ± 86 mM 1,3-PD. These values represent an 80% improvement in the yield of 1,3-PD. Moreover, the strains showed a different acid profile from that of the wild type. GSHM 2 produced more acetic acid than butyric acid, whereas strain GSHM 4 generated more butyric acid. Both strains generated more biomass than the wild type.
TABLE 1.
Fermentation data for C. diolis DSM 15410 and mutants grown in fed-batch cultures
| Strain | Time (h) | Biomass (g liter−1) | Concn (mM) of glycerol useda | Concn (mM) of producta: |
Y1,3-PD | Q1,3-PDb | q1,3-PDc | ||
|---|---|---|---|---|---|---|---|---|---|
| Acetate | Butyrate | 1,3-PD | |||||||
| Wild type | 22 | 1.84 | 989 (56) | 63 (4) | 96 (4) | 624 (38) | 0.586 | 2.147 | 1.17 |
| PSM 7 | 27 | 1.76 | 1,204 (75) | 73 (5) | 92 (5) | 746 (55) | 0.614 | 2.099 | 1.19 |
| GSHM 2 | 28 | 1.95 | 1,695 (95) | 160 (14) | 50 (2) | 1,032 (79) | 0.642 | 2.821 | 1.44 |
| GSHM 4 | 31 | 2.10 | 1,815 (114) | 58 (3) | 240 (18) | 1,114 (86) | 0.632 | 2.745 | 1.30 |
Values are the averages of four independent determinations. Standard deviations are given in parentheses.
Q1,3-PD, 1,3-PD volumetric productivity (g liter−1 h−1).
q1,3-PD, 1,3-PD specific formation rate (g g of cells [dry weight]−1 h−1).
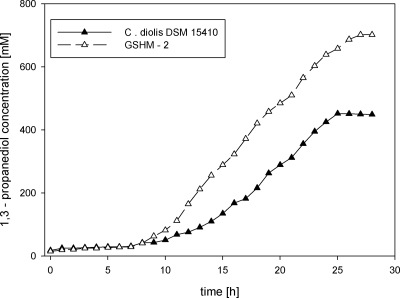
Comparative fed-batch fermentations with C. diolis DSM 15410 and GSHM 2 using crude glycerol as a carbon source.
Crude glycerol obtained as a side product in biodiesel production is a more attractive carbon source than pharmaceutical glycerol due to significant cost reduction and the recirculation of a waste product from biodiesel production. However, crude glycerol products often differ greatly in terms of quality, the glycerol amount, and possible components that may inhibit the growth of C. diolis. Crude glycerol was kindly provided by M. Menner (Fraunhofer Institute IVV) and contained large amounts of fatty acids that may hamper the growth of C. diolis; these were therefore removed by liquid extraction. This crude glycerol was used for comparative fed-batch fermentations with the wild-type strain and the generated strain GSHM 2.
Both C. diolis DSM 15410 and GSHM 2 produced less 1,3-PD when crude glycerol was used than when pharmaceutical glycerol was used as a carbon source. The wild-type strain reached a final yield of 448 mM, and GSHM 2 yielded 706 mM 1,3-PD (Fig. 6). Both strains displayed a longer lag phase with crude glycerol than with pharmaceutical glycerol. The molar yields of 1,3-PD were 0.46 for the wild-type strain and 0.49 for GSHM 2, lower than those obtained by fed-batch fermentation with pharmaceutical glycerol.
FIG. 6.
Results from fed-batch fermentations with C. diolis and GSHM 2 using crude glycerol as a carbon source. Levels of 1,3-PD product formation by the different strains are shown.
DISCUSSION
Although the classical strain improvement approach has been successful in the development of industrial fermentation strains, it is a time-consuming process that often generates only incremental improvements, as seen in our attempts to select for both glycerol and PD tolerance (Fig. 1 and 2). This characteristic reflects practical limitations such as the simultaneous accumulation of desirable and undesirable mutations and the inability to introduce heterologous genes (29). Classical strain improvement competes with current rational metabolic engineering approaches, which can specifically address shortcomings in the natural producer or even establish entire pathways in a better-suited heterologous host (3). However, such rational methods rely on biochemical models of the relevant metabolic pathways and a detailed understanding of the genes involved and their regulation. For complex phenotypes such as tolerances to certain inhibitory molecules, multiple genes distributed throughout the genome are often involved (26). This pattern raises serious obstacles for the application of genetic engineering to complex strain improvement and favors the employment of combinatorial approaches such as genome shuffling.
The factors limiting 1,3-PD production by C. diolis are not clearly defined, and a multitude of factors related to membrane effects, metabolic feedback regulation, transport, and competing metabolic pathways seem to be involved (1, 5, 14). Recursive protoplast fusion of improved strains leads to homologous recombination and allows the generation of a combinatorial library. This approach provides for simultaneous changes at different positions throughout the genome, even in the absence of detailed genetic information. Beneficial mutations can accumulate, whereas neutral or detrimental mutations are weeded out, thus increasing fitness rapidly (29).
In the present study, we demonstrated the application of genome shuffling to the strict anaerobic microorganism C. diolis for high-volume 1,3-PD production. Industrial 1,3-PD production by C. diolis is hampered primarily by inhibition from fermentation products (14). By using a classical strain improvement approach, mutants that showed an 80% improvement in terms of 1,3-PD tolerance and a 15% increase in the product yield were generated. For generating genetic diversity in the genome shuffling approach, natural 1,3-PD producers isolated from other sources, e.g., environmental samples, could have been used instead of a well-characterized 1,3-PD producer derived from the classical approach. However, isolation of 1,3-PD producers from environmental samples can result in the coisolation of potentially pathogenic organisms (19). Genome shuffling in the mutants derived from chemical mutagenesis led to a dramatic increase in the volumetric productivity and specific production rate, demonstrating the power of applying of these methods to a strict anaerobe. These results are consistent with those of previous studies in which notable improvements in Lactobacillus and Streptomyces species were shown (26, 31). An indication of the mechanism underlying the improved productivity is provided by the data in Table 1, which show that the mutants produced more biomass than the wild type and that higher biomass may confer tolerance. The enhanced growth rate may reflect mutations that allow the cells to use nutrients in the medium more effectively or to export inhibitory molecules more rapidly, as suggested in previous studies (2).
Crude glycerol, which is an abundant by-product from biodiesel production, is an attractive substrate for the production of high-value chemicals such as 1,3-PD. Previous investigations showed that crude glycerol can be used as the sole carbon source for 1,3-PD production by C. butyricum (18, 27). The results of our fed-batch fermentation with crude glycerol showed reduced productivity and a prolonged lag phase for both the mutants and the wild type, thus indicating inhibitory or missing compounds in the fermentation medium. However, the quality and composition of crude glycerol from biodiesel production plants vary. Additional rounds of genome shuffling may be required to further improve the utilization of crude glycerol. For an industrial strain under process conditions, tolerance of substrates and fermentation products is desired, in addition to high-volume productivity. The medium composition must also be taken into account. A major cost factor is the use of yeast extract, which should be replaced with a synthetic medium if possible. A previous study gave some indications that p-aminobenzoic acid and biotin are important vitamins for C. butyricum (18). Based on the characteristics of the mutants isolated in the present study, further improvements with regard to volumetric productivity, fermentation cost, and the use of crude glycerol can be expected to be realized by further optimization of the fermentation process.
Acknowledgments
This work was financially supported by the German Federal Ministry of Education and Research (BMBF; through grant no. 0315046A).
We thank N. Koch for excellent technical assistance.
Footnotes
Published ahead of print on 23 October 2009.
REFERENCES
- 1.Abbad-Andaloussi, S., E. Guedon, E. Spiesser, and H. Petitdemange. 1996. Glycerol dehydratase activity: the limiting step for 1,3-propanediol production by Clostridium butyricum DSM 5431. Lett. Appl. Microbiol. 22:311-314. [Google Scholar]
- 2.Abbad-Andaloussi, S., C. Manginot-Durr, J. Amine, E. Petitdemange, and H. Petitdemange. 1995. Isolation and characterization of Clostridium butyricum DSM 5431 and mutants with increased resistance to 1,3-propanediol and altered production of acids. Appl. Environ. Microbiol. 61:4413-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alper, H., K. Miyaoku, and G. Stephanopoulos. 2005. Construction of lycopene-overproducing E. coli strains by combining systematic and combinatorial gene knockout targets. Nat. Biotechnol. 23:612-616. [DOI] [PubMed] [Google Scholar]
- 4.Berovic, M., and M. Legisa. 2007. Citric acid production. Biotechnol. Annu. Rev. 13:303-343. [DOI] [PubMed] [Google Scholar]
- 5.Biebl, H. 1991. Glycerol fermentation of 1,3-propanediol by Clostridium butyricum. Measurement of product inhibition by use of a pH-auxostat. Appl. Microbiol. Biotechnol. 35:701-705. [Google Scholar]
- 6.Biebl, H., S. Marten, H. Hippe, and W. D. Deckwer. 1992. Glycerol conversion to 1,3-propanediol by newly isolated clostridia. Appl. Microbiol. Biotechnol. 36:592-597. [Google Scholar]
- 7.Biebl, H., and N. Pfenning. 1981. Isolation of members of the family Rhodospirillaceae, p. 267-273. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes. Springer-Verlag KG, Berlin, Germany.
- 8.Biebl, H., and C. Sproer. 2002. Taxonomy of the glycerol fermenting clostridia and description of Clostridium diolis sp. nov. Syst. Appl. Microbiol. 25:491-497. [DOI] [PubMed] [Google Scholar]
- 9.Biebl, H., A. P. Zeng, K. Menzel, and W. D. Deckwer. 1998. Fermentation of glycerol to 1,3-propanediol and 2,3-butanediol by Klebsiella pneumoniae. Appl. Microbiol. Biotechnol. 50:24-29. [DOI] [PubMed] [Google Scholar]
- 10.Cameron, D. C., N. E. Altaras, M. L. Hoffmann, and A. J. Shaw. 1998. Metabolic engineering of propanediol pathways. Biotechnol. Prog. 14:116-125. [DOI] [PubMed] [Google Scholar]
- 11.Christopher, A. T., S. B. Tummala, and E. T. Papoutsakis. 2005. Metabolic engineering of solventogenic clostridia, p. 814. In P. Dürre (ed.), Handbook on clostridia. CRC Press, Boca Raton, FL.
- 12.Chuah, H. 1996. Corterra poly(trimethylene terephthalate)—new polymeric fiber for carpets. Chem. Fibers Int. 46:424-428. [Google Scholar]
- 13.Colin, T., A. Bories, C. Lavigne, and G. Moulin. 2001. Effects of acetate and butyrate during glycerol fermentation by Clostridium butyricum. Curr. Microbiol. 43:238-243. [DOI] [PubMed] [Google Scholar]
- 14.Colin, T., A. Bories, and G. Moulin. 2000. Inhibition of Clostridium butyricum by 1,3-propanediol and diols during glycerol fermentation. Appl. Microbiol. Biotechnol. 54:201-205. [DOI] [PubMed] [Google Scholar]
- 15.Coombs, A. 2007. Glycerin bioprocessing goes green. Nat. Biotechnol. 25:953-954. [DOI] [PubMed] [Google Scholar]
- 16.Dai, M., and S. D. Copley. 2004. Genome shuffling improves degradation of the anthropogenic pesticide pentchlorphenol by Shingobium chlorophenolicum ATCC 39723. Appl. Environ. Microbiol. 70:2391-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datta, R., S. P. Tsai, P. Bonsignore, S. H. Moon, and J. R. Frank. 1995. Technological and economical potential of poly(lactic acid) and lactic acid derivates. FEMS Microbiol. Rev. 16:221-231. [Google Scholar]
- 18.Gonzalez-Pajuelo, M., J. C. Andrade, and I. Vasconcelos. 2004. Production of 1,3-propanediol by Clostridium butyricum VPI 3266 using a synthetic medium and raw glycerol. J. Ind. Microbiol. Biotechnol. 31:442-446. [DOI] [PubMed] [Google Scholar]
- 19.Hao, J., R. Lin, Z. Zheng, H. Liu, and D. Liu. 2008. Isolation and characterization of microorganisms able to produce 1,3-propanediol under aerobic conditions. World J. Microbiol. Biotechnol. 24:1731-1740. [Google Scholar]
- 20.Hermann, B. G., K. Blok, and M. K. Patel. 2007. Producing bio-based bulk chemicals using industrial biotechnology saves energy and combats climate change. Environ. Sci. Technol. 41:7915-7921. [DOI] [PubMed] [Google Scholar]
- 21.Hermann, B. G., and M. Patel. 2007. Today's and tomorrow's bio-based bulk chemicals from white biotechnology. Appl. Biochem. Biotechnol. 136:361-388. [DOI] [PubMed] [Google Scholar]
- 22.Homann, T., C. Tag, H. Biebl, W. D. Deckwer, and B. Schick. 1990. Fermentation of glycerol to 1,3-propanediol by Klebsiella and Citrobacter strains. Appl. Microbiol. Biotechnol. 33:121-126. [Google Scholar]
- 23.Kurian, J. V. 2005. A new polymer platform for the future—Sorona from corn derived 1,3-propanediol. J. Polym. Environ. 13:159-167. [Google Scholar]
- 24.Lligadas, G., J. C. Ronda, M. Galia, and V. Cadiz. 2007. Poly(ether urethane) networks from renewable resources as candidate biomaterials: synthesis and characterization. Biomacromolecules 8:686-692. [DOI] [PubMed] [Google Scholar]
- 25.O'Brien, J. R., C. Raynaud, C. Croux, L. Girbal, P. Soucaille, and W. N. Lanzilotta. 2004. Insights into the mechanism of the B12 independent glycerol dehydratase from Clostridium butyricum: preliminary biochemical and structural characterization. Biochemistry 43:4635-4645. [DOI] [PubMed] [Google Scholar]
- 26.Patnaik, R., S. Louie, V. Gavrilovic, K. Perry, W. P. Stemmer, C. M. Ryan, and S. B. del Cardayre. 2002. Genome shuffling of Lactobacillus for improved acid tolerance. Nat. Biotechnol. 20:707-712. [DOI] [PubMed] [Google Scholar]
- 27.Petitdemange, E., C. Durr, S. A. Andaloussi, and G. Raval. 1995. Fermentation of raw glycerol to 1,3-propanediol by new strains of Clostridium butyricum. J. Ind. Microbiol. Biotechnol. 15:498-502. [Google Scholar]
- 28.Stal, M. H., and H. P. Blaschek. 1985. Protoplast formation and cell wall regeneration of Clostridium perfringens. Appl. Environ. Microbiol. 50:1097-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stephanopoulos, G. 2002. Metabolic engineering by genome shuffling. Nat. Biotechnol. 20:666-668. [DOI] [PubMed] [Google Scholar]
- 30.Tang, X., Y. Tan, H. Zhu, K. Zhao, and W. Shen. 2009. Microbial conversion of glycerol to 1,3-propanediol by an engineered strain of Escherichia coli. Appl. Environ. Microbiol. 75:1628-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, Y. X., K. Perry, V. A. Vinci, K. Powell, W. P. Stemmer, and S. B. del Cardayre. 2002. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:664-666. [DOI] [PubMed] [Google Scholar]