Abstract
Species of bacteria associated with Stylophora pistillata were determined by analyses of 16S ribosomal genes. Coral samples were taken from two distinct sites at Kenting, in the far south of Taiwan; three coral colonies at each site were tagged and sampled in the winter and summer of 2007. Six hundred 16S rRNA gene clones were selected and sequenced for diversity analysis and community comparison. LIBSHUFF and nonparametric multiple dimensional scaling analyses showed variations in the composition of the coral-associated bacteria in the different samples, suggesting that seasonal and geographic factors and variations in individual coral colonies were all vital drivers of the structure of the S. pistillata-associated bacterial community. To examine the association between species specificity and environmental impacts on the structure of the coral-associated bacterial community, we conducted an integrated, comparative analysis of 44 coral-associated bacterial data sets, including the present study's data. The clustering analysis suggests that the influence of spatial and temporal factors on the coral-associated bacteria population structure is considerable; nonetheless, the effect of species specificity is still detectable in some coral species, especially those from the Caribbean Sea.
Microbes are abundant in the ocean and thrive around corals. In earlier investigations over the past decades, microbes have been detected in coral mucus (8), in coral tissues (5), and in the surrounding reef waters (25). Although we understand little about the real interactions between these coral-associated microbes and the coral itself, or their mutual roles, much indirect evidence suggests that these microbes may play an important role in the coral holobiont, with respect to coral nutrition, health, and disease (14, 18, 30).
It is now known that most microbes are uncultivable by present laboratory methods (1, 28, 10). To understand more about coral-associated microbial communities, to identify the diversity of microbes associated with particular corals, and to assess whether these microbes are indeed species specific or represent only opportunistic interactions with the coral animal, some relatively comprehensive studies have been carried out in recent years based on culture-independent molecular techniques, e.g., construction of 16S rRNA gene clone libraries or denaturing gradient gel electrophoresis (3, 6, 9, 11, 13, 16, 17, 19, 20, 21, 23, 30, 31, 33). Consequently, coral-associated microbes are now known to be not only highly diverse and dynamic but also substantially coral specific.
Recently, the specificity of association between coral and bacterial species has been a topic of much discussion. Earlier reports suggested that similar microbial communities were specifically associated with identical coral species, regardless of whether they were isolated from distinct geographic regions or at different times (3, 9, 20, 21); however, environmental factors have also recently been found to significantly influence the specificity of bacteria-coral associations (2, 17). Such inconsistency might reasonably be expected in light of the complexity of interaction in the coral holobiont, which includes coral, algae, fungi, bacteria, archaea, and other biotic and environmental factors (30). Nonetheless, more-detailed studies are needed for a better comprehension of species-specific bacterial associations.
In this study, we selected Stylophora pistillata, a widely distributed coral in western Pacific reefs (26), to study the diversity and composition of the coral-associated bacteria and the effects of spatial and temporal differences on such population structures. We also examined the species specificity of such coral-bacteria associations by comparing our data with another 44 coral-associated bacterial data sets. This biodiversity analysis shows the presence of a large variation in the composition of S. pistillata-associated bacterial communities, suggesting that specificity between S. pistillata and associated bacteria is significantly influenced by geographic and seasonal factors. Furthermore, a comparison with 10 previous studies of coral-associated microbes showed that spatial and temporal factors play a role in affecting the population structure of coral-associated bacteria, though distinct species-specific bacterial profiles are detectable in some corals of the Caribbean Sea.
MATERIALS AND METHODS
Sample collection and processing.
Coral and bacterial samples were taken from two sampling sites near Kenting, at the southern tip of Taiwan (Fig. 1), namely, the Third Nuclear Power Plant (21°55.0′N, 120°44.0′E) and Tiaoshi (21°56.9′N, 120°48.5′E). Three S. pistillata colonies at a depth of approximately 7 m were tagged at each site; they were marked as N61, N63, and N68 and T36, T37, and T40, respectively. Selected coral colonies were more than 10 m apart. Samples were collected in January 2007 and June 2007 (before bleaching season). The average water temperatures were 24.4°C in January and 28.0°C in June. Coral samples approximately 1.5 cm in length were broken off with a hammer and transferred to sterile bottles. The samples were washed twice with sterile synthetic seawater and quickly sprayed by using a sprayer with TE buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA [pH 8.0]) in a seaside laboratory close to the sampling sites and stored in liquid nitrogen after processing. The entire procedure was completed in less than 1 h.
FIG. 1.
Two sampling sites, Tiaoshi (21°56.9′N, 120°48.5′E) and the Third Nuclear Power Plant (21°55.0′N, 120°44.0′E), at the southern tip of Taiwan. The map was constructed using Generic Mapping Tools ver. 3.4.2 (http://gmt.soest.hawaii.edu).
DNA extraction and purification.
Total DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB)-mediated method (32). Coral samples frozen in liquid nitrogen were homogenized to a fine powder using a sterile mortar and pestle. The samples were moved to clean microtubes and centrifuged at 12,000 × g. The pellet was resuspended in TE buffer, centrifuged again at 12,000 × g, and then resuspended in 567 μl of TE buffer, 30 μl of 10% sodium dodecyl sulfate, and 3 μl of 20 mg/ml proteinase K. After vigorous vortexing, the samples were incubated at 37°C for 1 h before 100 μl of 5 M NaCl and 80 μl of CTAB-NaCl solution (4.1 g NaCl and 10 g of CTAB dissolved in 100 ml H2O) were added with vortexing, and they were then incubated at 65°C for a further 10 min. A two-thirds volume of chloroform-isoamyl alcohol (24:1) was added, and samples were mixed and centrifuged for 5 min at 12,000 × g at 4°C. The top aqueous layer was collected in a clean microtube, an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added, and samples were mixed and centrifuged for 5 min at 12,000 × g at 4°C. The top aqueous layer was collected in a clean tube containing a two-thirds volume of isopropanol, gently mixed, and centrifuged at 12,000 × g for 7 min. The supernatant was then discarded, and the pellet was washed with 70% prechilled ethanol. The DNA pellet was air dried and resuspended in sterile Milli-Q water. After the DNA concentration was determined, aliquots of the DNA samples were placed in individual PCR microtubes to avoid DNA degradation by repeated freezing and thawing in subsequent processes.
Amplification of bacterial 16S rRNA genes.
The universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) were used to amplify 16S rRNA genes. The 16S rRNA genes were amplified in a 50-μl reaction mixture consisting of 5 μl of 10× PCR buffer, 1 μl of a 2.5 mM total deoxynucleoside triphosphate mixture, 2.5 μl of a 10 μM concentration of each primer, 1 μl of template DNA, and 1.2 U of DyNAzyme EXT DNA polymerase (Finnzymes, Finland), according to the conditions recommended by the manufacturer. The amplification conditions for the bacterial library PCR consisted of an initial denaturation step of 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min, with a final step of 72°C for 10 min and then cooling at 4°C, using a PxE thermal cycler (Thermo Electron Corporation). These low-stringency, loose PCR conditions were adopted to minimize primer bias in the PCR. After the PCR, the quality of the product was confirmed by DNA agarose electrophoresis visualized using an ImageQuant 300 UV transilluminator (GE Healthcare).
Clone library construction.
The PCR products of bacterial 16S rRNA were purified using a QIAquick gel extraction kit (Qiagen) for cloning, according to the manufacturer's instructions. Cloned DNA was recovered in 20 μl of sterile Milli-Q water (Q-Gard 1 purification pack; Millipore). The purified PCR products were ligated into TOPO-TA cloning vector (TOP10 genotypes of Escherichia coli) by following the manufacturer's instructions (Invitrogen). Positive colonies were selected by blue and white screening on Luria-Bertani agar plates with ampicillin (50 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM) and 5-bromo-4-chloro-3-indoyl-β-d-galactopyranoside (X-Gal; 50 μg/ml). To confirm the successful clones, each positive colony was examined by PCR with M13 forward (5′-GTAAAACGACGGCCA G-3′) and reverse (5′-CAGGAAACAGCTATGAC-3′) primers. The amplification conditions were the same as the bacteria universal primer PCR conditions (see the previous section). Reaction products were checked by DNA agarose electrophoresis, as described above.
Clone-picking bias was avoided by the following process. In each transformation plate, the number of positive colonies (i.e., white colonies) needed to be more than 1,000 (10- to 20-μl transformation solutions for each plate) to guarantee sufficient transformation efficiency. One hundred positive colonies were picked from a one-eighth area of a plate for further streaking; there was no preference for large or small colonies when picking colonies. According to a random number table created by Microsoft Excel, we then randomly picked 50 white colonies from these 100 numbered white colonies for sequencing.
Taxonomic assessment.
The sequences of the selected colonies were submitted to RDP (http://rdp.cme.msu.edu) to identify their taxonomic identities based on the best hit with SeqMatch. We organized these RDP search results into class-level group lists.
Diversity analysis and related statistics.
DOTUR software was used to estimate the biodiversity of coral-associated microbes in the winter and summer coral samples (22). The diversity indices determined were the Shannon-Weaver diversity index, Chao1 estimator, and evenness index. The furthest neighbor grouping method was used for DOTUR in this study, with the precision option set to 0.01 and a 3% cutoff value to demark an operational taxonomic unit (OTU).
Comparison of microbial communities.
Two methods were used to compare microbial communities, LIBSHUFF (http://libshuff.mib.uga.edu) (24) and nonmetric multidimensional scaling ordination (nMDS) (12). Both statistical methods allow determinations of the likelihood that two communities are different or not. The clone libraries were compared using LIBSHUFF with Bonferroni correction of confidence levels; nMDS then provided a visual map of the relationship between different microbial communities.
Phylogenetic analysis.
We analyzed the phylogenetic relationships of the 16S rRNA sequences detected in the present study. Thirty reference sequences (including an outgroup sequence; see Table S1 in the supplemental material) were retrieved from the RDP website and used for tree construction by the neighbor-joining method using MEGA software, version 4 (27). Multiple bootstrapped datasets (1,000 samplings) of the alignment data were used to determine the significance of the classification. The Kumura-2-parametric mutation model was used. The analyzed nucleotides were 541 bases long on average, with a total of 116 taxa analyzed, including 30 reference sequences and 86 OTUs (one sequence was omitted because of insufficient length).
Hierarchical cluster analysis.
To compare the coral-associated microbial profiles, we used Gene Cluster 3.0 (7) to perform a clustering analysis of the results of this study and data from another 10 reports (3, 4, 13, 15, 16, 17, 20, 21, 29, 33), for a total of 44 coral samples. The coral data sets came from a diversity of seawaters, including those from the Kenting (Taiwan; West Pacific Ocean), Palau (West Pacific Ocean), Davis, Magnetic, and Orpheus Island Reefs (Great Barrier Reef, Australia; South Pacific Ocean); Cape Armitage, Scott Base, and McMurdo Station (McMurdo Sound, Ross Sea, Antarctica); Whalebone Bay, Hog Breaker Reef, and Bocas del Toro (Bermuda; Caribbean Sea); the Jim Atria wreck sites off Pompano Beach, FL (Caribbean Sea); Playa Hundu (Curaçao, Netherlands Antilles; Caribbean Sea); Sedot Yam (Israel; Mediterranean Sea); and the Gulf of Eilat (Israel; Red Sea).
The gene similarity and array similarity matrices were constructed and clustered by Euclidean distance and average linkage. Various weights were adopted to discriminate among major groups, minor groups, and unclassified bacteria. Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria were commonly detected groups and given a weight value of 0.3; other groups were weighted at 0.1, and the unclassified bacteria were given a weight value of 0.05. However, other weight value schemes, such as setting all weights equal to 0.1 or setting the weight value for the major groups to 0.1 were also tested, and the results were consistent (data not shown). The program Maple Tree (http://sourceforge.net/projects/mapletree/?abmode=1) was used to view and adjust the map, with the negative RGB (red green blue) color set at 1.102.0 and the contrast value set at 13.0.
RESULTS
Biodiversity analysis of S. pistillata-associated bacteria.
Six hundred positive colonies were selected from the 16S rRNA gene libraries of S. pistillata samples collected from Tiaoshi and the Third Nuclear Power Plant at Kenting, Taiwan, in the winter and summer (Fig. 1; Table 1). The diversity of the microbial communities in the winter was very low, with the most diverse community being found at site N68 (Shannon's index, 1.3; Chao1, 11). The diversity of the microbial communities in summer was much higher, with the greatest diversity again seen at N68 (Shannon's index, 2.63; Chao1, 33).
TABLE 1.
Diversity indices for the bacterial communities as represented in the 16S rRNA gene librariesa
| Season and site | Results for indicated index |
|||||
|---|---|---|---|---|---|---|
| S | N | Evenness | Richness | Shannon's | Chao1 | |
| Winter | ||||||
| Tiaoshi | ||||||
| T36 | 4 | 50 | 0.62 | 1.77 | 0.86 | 3 (3-3) |
| T37 | 7 | 50 | 0.57 | 3.53 | 1.1 | 8.5 (7-22) |
| T40 | 4 | 50 | 0.62 | 1.77 | 0.86 | 3 (3-3) |
| Nuclear power plant | ||||||
| N62 | 6 | 50 | 0.51 | 2.94 | 0.92 | 9 (6-31) |
| N63 | 7 | 50 | 0.53 | 3.53 | 1.03 | 9 (7-22) |
| N68 | 8 | 50 | 0.63 | 4.12 | 1.3 | 11 (8-31) |
| Summer | ||||||
| Tiaoshi | ||||||
| T36 | 10 | 50 | 0.82 | 5.30 | 1.89 | 11 (10-21) |
| T37 | 13 | 50 | 0.88 | 7.06 | 2.25 | 18 (14-45) |
| T40 | 14 | 50 | 0.92 | 7.65 | 2.44 | 14 (15-22) |
| Nuclear power plant | ||||||
| N62 | 9 | 50 | 0.77 | 4.71 | 1.69 | 9 (9-14) |
| N63 | 15 | 50 | 0.79 | 8.24 | 2.13 | 17 (15-28) |
| N68 | 22 | 50 | 0.87 | 12.36 | 2.68 | 33 (25-64) |
Calculations are based on OTUs formed at an evolutionary distance of <0.03. S, number of OTUs; N, number of sequences; Evenness, Shannon's index/ln(number of OTUs); Richness, (number of singleton OTUs − 1)/log N. The maximum richness value is (N − 1)/log N. Confidence intervals for the Chao1 estimator are shown in parentheses.
Comparison of microbial communities.
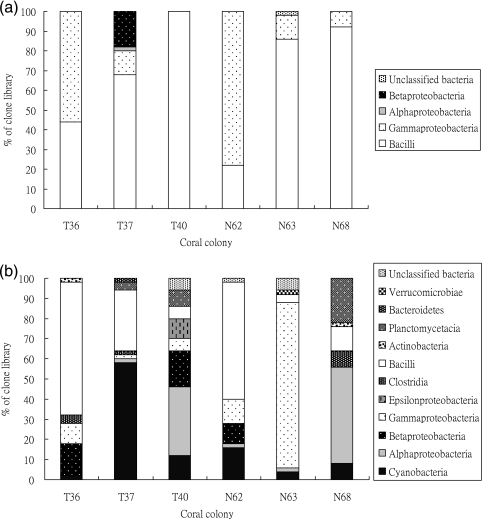
By looking at the composition of each 16S rRNA gene library based on sequence similarity, the difference between the winter and summer samples is noticeable (Fig. 2). In the winter, corals at all sampling sites housed very similar microbe profiles, with the Gammaproteobacteria and Bacilli being the two major groups detected. However, there was much more variation in coral-associated bacteria in the summer samples; in addition, the microbial communities in the coral colonies were more distinct from each other. Cyanobacteria and Bacilli were observed in all clone libraries, though the dominant microbes varied in different coral samples. Bacilli was the major group in T36 and N36. Corals T40 and N68 hosted more Alphaproteobacteria, while Cyanobacteria and Gammaproteobacteria dominated in the T37 and N63 profiles.
FIG. 2.
Distribution of the bacterial 16S rRNA gene sequences from clone libraries. Histograms of the samples isolated in winter (a) and summer (b). Corals T36, T37, and T40 were from Tiaoshi, and corals N62, N63, and N68 were from the Third Nuclear Power Plant.
To examine whether any microbial group may be present for a longer term in corals, we conducted a distribution analysis of the largest microbial groups; OTUs were used to define a species with a cutoff value of 3% (Table 2). The largest OTUs in winter were dissimilar to those in summer, suggesting that the coral-associated microbial community changed greatly between the seasons. Bacillus cereus or Bacillus thuringiensis-like bacteria was the largest OTU, appearing mainly in winter samples. The next two largest OTUs corresponded to Paenibacillus algionlyticus and Gammaproteobacteria (Enterobacteria, Shigella sp.). The former appeared in both winter and summer samples, but the latter was detected only in winter; however, most of the larger OTUs in the summer samples were not detected in winter, including cyanobacteria, marine Alphaproteobacteria, Bacilli, and Gammaproteobacteria species. Such great differences are likely affected by local climate and environment and differences between individual coral colonies (see Table S2 in the supplemental material).
TABLE 2.
Distribution of the largest OTUs
| Group | Total no. of samples | Clone name (OTU) | Closest relative (description) [% identity]a | Distribution |
|||
|---|---|---|---|---|---|---|---|
| Winter |
Summer |
||||||
| T | N | T | N | ||||
| Bacilli | 90 | SW44 | B. cereus or B. thuringiensis (soil bacteria) [99] | 22 | 67 | 1 | 0 |
| Bacilli | 75 | SW7 | P. alginolyticus or P. chondroitinus (soil bacteria) [97] | 39 | 22 | 14 | 0 |
| Gammaproteobacteria | 72 | W71 | Uncultured gammaproteobacterium (Shigella sp.) [97] | 28 | 44 | 0 | 0 |
| Bacilli | 35 | SW1 | Bacillus simplex (soil bacteria) [99] | 1 | 0 | 8 | 26 |
| Bacilli | 33 | W75 | Staphylococcus pasteuri (animal- or food-associated bacteria) [98] | 33 | 0 | 0 | 0 |
| Betaproteobacteria | 25 | SW3 | Delftia acidovorans (synonym, Comamonas acidovorans; rhizosphere bacteria) [99] | 6 | 0 | 17 | 2 |
| Bacilli | 22 | S2 | Uncultured Firmicutes bacterium (soil bacteria) [98] | 0 | 0 | 18 | 3 |
| Bacilli | 19 | SW4 | Paenibacillus wynnii (soil bacteria) [97] | 0 | 2 | 9 | 8 |
| Gammaproteobacteria | 15 | S5 | Uncultured gammaproteobacterium (Oceanospirillale; coral-associated bacteria) [96] | 0 | 0 | 0 | 15 |
| Gammaproteobacteria | 14 | S8 | Alteromonas macleodii (marine bacteria) [96] | 0 | 0 | 0 | 14 |
| Alphaproteobacteria | 13 | S6 | Marine alphaproteobacterium (uncultured coral-associated bacteria) [94] | 0 | 0 | 0 | 13 |
| Cyanobacteria | 11 | S9 | Synechococcus sp. (marine bacteria) [97] | 0 | 0 | 11 | 0 |
The similarity search was done on 23 April 2009 using the RDP database.
Phylogenetic analysis.
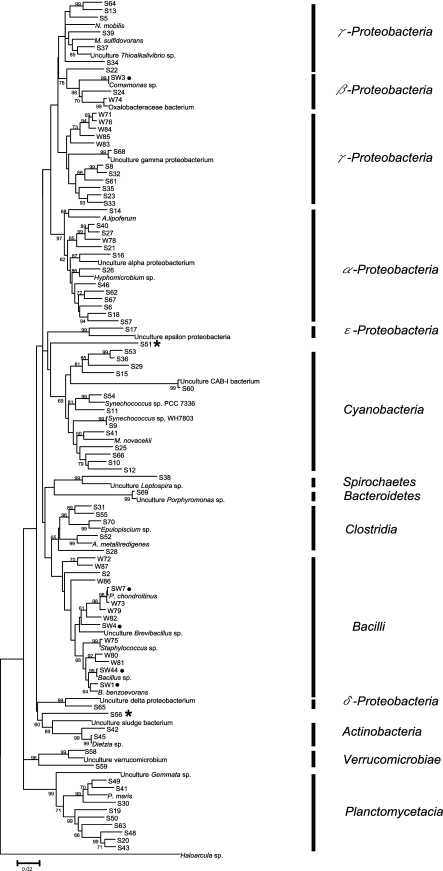
To identify the taxonomic relations between these microbes, we conducted a phylogenetic analysis. All the sequences were classified with Clustal W and DOTUR and categorized into different OTUs, and one species from each OTU was selected as a representative species. We added 30 reference sequences to the analysis, including an outgroup sequence from the archaeon Haloarcula sp. These closely related sequences were retrieved from the RDP website.
In the 16S rRNA gene tree (Fig. 3), a number of novel microbes remained alone in a subclade, such as S8, S32, S61, S23, S33, and S35 in the Gammaproteobacteria clade and S19, S50, S63, S48, S20 and S43 in the Planctomycetacia clade. In addition, two OTUs, S51 and S56, could not be placed into any bacterial group. We found only five OTU groups (four of them in the bacilli clade), i.e., SW1, SW3, SW4, SW7, and SW44, that appeared in both summer and winter samples. SW7 was dominant in both locations in winter but only one location in summer; whereas SW3 was detected in both locations in summer but one in winter. These microbes likely to be longer-term inhabitants of S. pistillata and are thus less influenced by changes of season.
FIG. 3.
Neighbor-joining tree of S. pistellata-associated bacteria based on 16S rRNA gene sequences. Bootstrapping was performed with 1,000 replications. Selected OTUs from all 16S rRNA gene libraries are presented as S numbers (e.g., S30, a sample isolated in summer), W numbers (e.g., W80, a sample isolated in winter), and SW numbers (e.g., SW7, a sample from both winter and summer). ✶, groups not determined. •, groups detected in both seasons. Reference bacterial sequences selected from the NCBI database were those for Nitrococcus mobilis (L35510), Methylophaga sulfidovorans (X95461), uncultured Thioalkalivibrio sp. (AB189351), Comamonas sp. (AJ002803), uncultured Gammaproteobacteria (AJ969451), Azospirillum lipoferum (Z29619), uncultured Alphaproteobacteria (DQ289899), Hyphomicrobium sp. (AY934488), uncultured Epsilonproteobacteria (AJ515715), uncultured candidate division CAB-I bacterium (DQ200412.1), Synechococcus sp. PCC 7336 (AF448078), Synechococcus sp. WH 7803 (AF311291), Microcystis novacekii (AB035551), uncultured Leptospira sp. (AY995721), Porphyromonas sp. (AF385560), Epulopiscium sp. (M99574), Alkaliphilus metalliredigens (AY137848), Paenibacillus chondroitinus (AB073206), uncultured Brevibacillus sp. (EU647528), Staphylococcus sp. (AJ276810), Bacillus sp. (AF290558), Bacillus benzoevorans (Y14693), Oxalobacteraceae sp. (EU057875), uncultured Deltaproteobacteria (EF076071), uncultured sludge bacterium (AF234754), uncultured Verrucomicrobium (AJ401107), uncultured Gemmata sp. (EU341290), Planctomyces maris (AJ231184), Dietzia sp. (Y17518), and Haloarcula sp. (EU708436).
Comparison of S. pistillata-associated microbial communities.
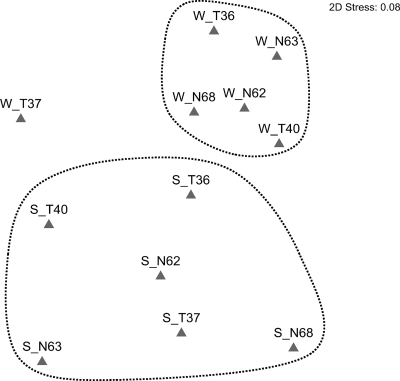
To describe the similarity of these coral-associated microbial communities, two statistical comparative analyses, LIBSHUFF and nMDS, were employed to compare the OTU compositions of each clone library. We pooled three libraries from each sampling site at a particular season into a combination library, giving four combination libraries, designated ST (summer, Tiaoshi), SN (summer, Third Nuclear Power Plant), WT (winter, Tiaoshi), and WN (winter, Third Nuclear Power Plant), for LIBSHUFF analysis.
In the LIBSHUFF analysis, each library was significantly different from the others with Bonferroni correction (P < 0.0042) (Table 3). Similarly, in the nMDS map, all samples were distant from each individual clone library. These analyses showed no significant similarity based on location (Fig. 4), although the winter 16S rRNA gene libraries were relatively more related to each other were than those from the summer (see Table S3 in the supplemental material), suggesting that season is a considerable influencing factor on the coral-associated bacterial population structure.
TABLE 3.
Community comparison with LIBSHUFFa
| Sample description | P value for indicated comparison |
|||
|---|---|---|---|---|
| ST | SN | WT | WN | |
| ST | — | 0.001* | 0.001* | 0.001* |
| SN | 0.001* | — | 0.001* | 0.001* |
| WT | 0.001* | 0.001* | — | 0.001* |
| WN | 0.001* | 0.001* | 0.006 | — |
The values above and to the right of the dashes are for the Y library, and those below and to the left of the dashes are for the X library. *, significant difference (P value [Bonferroni correction], 0.0042); —, not compared.
FIG. 4.
Nonmetric multidimensional scaling analysis. In the map, the abbreviations are formulated as season_location tag number (for example, S_T36 is the sample collected from the coral tagged as number 36 from Tiaoshi in the summer). The parameters in plotting the map were 25 for restarts, 0.01 for minimum stress, and Kruskal fit scheme set 1. 2D Stress, two-dimensional stress.
Factors determining specificity of coral-bacteria association.
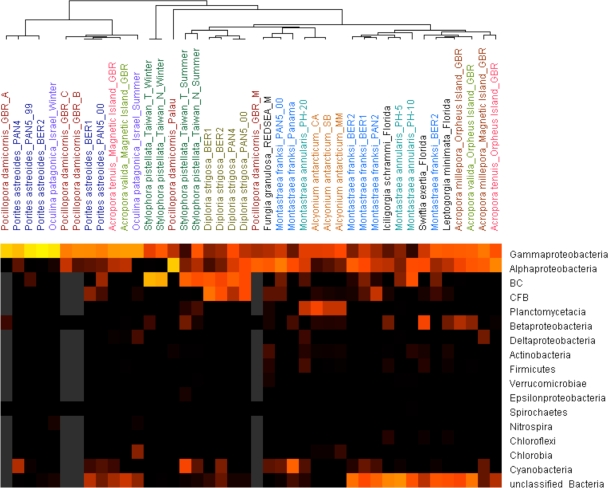
Although corals may harbor their own specific microbial community, as suggested by several reports, the results of this study show that specificity of association between bacteria and S. pistillata is weak. To assess how specific the coral-bacteria association is, we undertook an integrated comparison of our data with other coral-associated microbial communities from the literature (Fig. 5.). This included 44 samples from 15 species of corals, viz., Acropora millepora, Acropora valida, Acropora tenuis, Alcyonium antacticum, Diploria strigosa, Fungia granulosa, Iciligorgia schrammi, Leptogorigia minimata, Montastraea annularis, Montastraea franksi, Oculina patagonica, Pocillopora damicornis, Porites astreoides, S. pistillata, and Swiftia exertia (see references in Materials and Methods). By two-way clustering, the bacterial communities of these several corals can be seen to be quite distinct from each other. In considering whether spatial factors may influence the bacterial composition of coral holobiont, three corals, Alcyonium antacticum, D. strigosa, and Porites astreoides, from different locations nonetheless maintained similar microbial communities. However, other examples of the same coral species from different habitats showed different bacterial associations, such as in Acropora spp. and Pocillopora damicornis. For example, P. damicornis-associated microbes were quite different in the samples isolated from different parts of the Great Barrier Reef. Similarly, various Acropora spp. from the Great Barrier Reef clearly showed that similar bacterial profiles can be seen in the same habitat (Magnetic Island) although the corals in question comprised different species. The influence of differences in habitat seemed very considerable in that example. However, the coral-associated microbes isolated from three different corals, I. schrammi, L. minimata, and Swiftia exertia, in Florida were quite dissimilar, suggesting strong coral specificity. Last, considering the influence of temporal factors, changes in season may alter the composition of the bacterial associates dramatically, according to the clustering results of Oculina- and Stylophora-associated bacteria.
FIG. 5.
Two-way clustering analysis of various coral-associated bacterial communities. Forty-four coral samples were included in a cluster analysis using Gene Cluster 3.0. The bacterial community is divided into 17 taxonomic groups. The Gammaproteobacteria were the most dominant bacterial group and the Alphaproteobacteria were the second. Lighter colors (e.g., yellow) mean more members of the bacterial group detected, darker colors mean fewer were detected. The gray color represents undetected. The coral samples studied here originated from different seas (see Materials and Methods). BC, Bacillus/Clostridium; CFB, Cytophaga-Flavobacterium/Flexibacter-Bacteroides; GBR_A, tissue slurry colony A, Great Barrier Reef; GBR_B, tissue slurry colony B, Great Barrier Reef; GBR_ C, tissue slurry colony C, Great Barrier Reef; GBR_M, coral mucus, Great Barrier Reef; PAN2, Smithsonian Tropical Research Institute's field station, Bocas Del Toro, Panama, sampling site 2; PAN3, Smithsonian Tropical Research Institute's field station, Bocas Del Toro, Panama, sampling site 3; PAN4, Smithsonian Tropical Research Institute's field station, Bocas Del Toro, Panama, sampling site 4; PAN5_99, Smithsonian Tropical Research Institute's field station, Bocas Del Toro, Panama, sampling site 5, sampled in 1999; PAN5_00, Smithsonian Tropical Research Institute's field station, Bocas Del Toro, Panama, sampling site 5, sampled in 2000; BER1, Whalebone Bay, Bermuda; BER2, Hog Breaker Reef, Bermuda; REDSEA_M, coral mucus, Red Sea; PH-5, Playa Hundu, Curaçao, 5-m depth; PH-10, Playa Hundu, 10-m depth; PH-20, Playa Hundu, 20-m depth; CA, Cape Armitage, Antarctica; SB, Scott Base, Antarctica; and MM, McMurdo Station intake jetty, Antarctica.
DISCUSSION
This study provides the first analysis and comparison of microbial communities from a coral from Taiwan's reefs. S. pistillata-associated bacterial profiles changed rapidly by season or geography; moreover, the associated bacterial populations were highly variable among individual coral colonies. This result suggests that seasonal, geographical, and individual coral physiological factors are critical to the composition of S. pistillata-associated bacteria, and these factors ultimately drive coral species specificity. Moreover, a comparative analysis of 44 coral-associated bacteria profiles demonstrates that environmental impacts (including the season) have a large influence on the composition of bacterial associates, especially for the corals in the Great Barrier Reef, whereas a greater species specificity of coral-associated bacteria was seen in corals inhabiting the Caribbean Sea (21).
Where are the soilborne bacterial associates from?
A considerable number of soilborne bacteria were detected in microbial communities in S. pistillata, particularly in those samples collected in the winter. This phenomenon has not been reported for other coral-associated bacterial communities. We carefully examined if there was any contamination during sample processing; however, two repeat examples showed similar results (data not shown). Although there is no direct evidence to elucidate this result, we speculate that there are two climate-related activities that may be relevant to the phenomenon. In the winter (i.e., from January to March each year), strong winds regularly carry a great volume of dust or sand from afar that falls on the coast at Kenting, sufficient to maintain the beach area and to form large sand hills several meters in height (locally known as the “sea-entrance desert”). Although there are no figures available for how much dust or sand is moved and how much falls onto the reef nearby, the influence is likely significant. The dust would move a considerable number of soilborne bacteria into the reef, particularly in the shallower waters. In addition, the higher rainfall in summer than in winter (an average of 371 mm in June versus an average of 25 mm in January) often causes floods that flush out a great amount of terrestrial sediment, sufficient to render coast water turbid for 10 days. These two local climate activities are likely the cause of the differences of the soilborne bacterial associates in the coral.
The difference between the average seawater temperatures at the times of sampling in the winter (24.4°C) and summer (28°C) was only 3.6°C; therefore, temperature is not likely to be a key factor in the big dissimilarity between winter and summer clone libraries.
Species specificity plays weak role in structuring the S. pistillata-associated bacterial community.
Each tagged coral colony actually housed different bacterial associates (Fig. 2 and 3) (see Tables S2 and S3 in the supplemental material), regardless of location; particularly, no correlation was detected among bacterial communities in those corals isolated from the same location. These results suggest that the physiological difference between coral colonies is likely to be a bigger factor underlying differences in the dominant bacteria in each colony. The sensitive influence of coral physiology has been seen before, where differential bacterial communities were detected from separate parts of the same coral colony (3, 21).
We detected very little species specificity for S. pistillata-associated bacteria, with a high degree of variation in the makeup of bacterial communities among the samples. It is, however, difficult to assert that such differences were caused only by S. pistillata naturally possessing weak species specificity, as interference from other factors, such as the environment and variations in colony physiology may be masking the effect of species specificity. Indeed, the taxonomic analysis of the clone libraries of winter samples (Fig. 2 and Table 1) strongly suggested a prevalent environmental effect because of the large number of soilborne bacteria detected and the low biodiversity of the coral-associated bacterial community. On the contrary, many typical coral-associated, sponge-associated bacteria and marine bacteria appeared in the clone libraries of the summer samples (Table 2), indicating that the coral still maintained certain selective pressures on its bacterial associates. In addition, another interpretation for the weak species specificity for S. pistillata-associated bacteria would be that this coral is able to harbor a broad range of bacteria, diluting the effect of species specificity. Unfortunately, no other data on S. pistillata-associated microbes are available for comparison.
The studies of Acropora species-associated microbes in the Great Barrier Reef show that species specificity is weak compared to the influence of environmental effects (17). The result is likely due both to environmental effects and to the fact that Acropora species can support a broad range of bacteria, making the influence of species specificity unclear. Similarly, there are no similar studies of Acropora species from other places for further comparative analysis. We speculate that the species specificity of different kinds of corals may be variable, regardless of external influencing factors, though this hypothesis doubtlessly requires further experiments to validate.
Anthropogenic effects may be influencing microbial community.
In the table showing the most abundant bacterial groups (Table 2), two groups (SW3 and S2) were affiliated with the bacteria isolated from petroleum or oil. Although there is no direct evidence to prove it, these bacteria may be thriving due to the oil spill from the Greek-registered vessel MV Amorgos which, in 2001, collided with a reef and sank off Kenting. More than 95% of the ship residues remain at the incident location and pollution likely still exists (as reported by local media). In addition, small recreational watercraft and motorboat activity has been increasing around the sampling sites since 2001. It is therefore hard to tell whether these bacteria are the result of the oil spill incident or of other human activity.
Temporal factors have a considerable impact on coral-associated microbial community.
Temporal factors have been demonstrated to have a clear impact on the microbial associates in this study (Fig. 2 and Table 2). Similar observations were also reported in recent years. For instance, denatured gradient gel electrophoresis showed that Acropora-associated bacteria varied in different seasons in the Great Barrier Reef (17). O. patagonica-associated bacterial communities differed greatly in the winter and summer in the Mediterranean Sea, a fact that was also shown in our cluster map (Fig. 5). However, Rohwer and his colleagues found little difference in the community structures of Porites astreoides-associated bacteria isolated yearly (21). Their result is dissimilar to our cluster analysis, which showed a difference between two Porites astreoides samples (PAN5_99 and PAN5_00). This inconsistency may exist for two reasons. (i) The actual temporal factors studied differed between the experiments. The former three studies including ours did actually detect the community changes between winter and summer and indicated the effect of seasonal fluctuation, while the study of Rohwer et al. (21) looked at the bacterial community in the same season but in different years. (ii) The clustering result was more sensitive than the analysis method used in the report (21). For example, in the comparison between the two bacterial communities PAN5_99 and PAN5_00, cyanobacteria were detected in large numbers in PNA5_99 (i.e., 17 of a total of 88 sequences), but only a few were seen in PAN5_00 (2 of a total of 55 sequences) This difference was distinguishable in our cluster analysis, but not theirs, suggesting that temporal influences may still have existed in their sample but were not detected.
The coral-associated bacterial community is affected by multiple, dynamic factors.
In this study, multiple factors seem to have been involved in driving the coral-associated bacterial community population structure. Because coral species specificity could not be observed clearly in single coral species samples, we made a more-comprehensive comparison with another 10 reported coral holobiont profiles. In that analysis, S. pistillata-associated bacteria isolated from two different locations were tightly and independently clustered together, presenting much clearer evidence of species specificity; on the same clustering map, the Acropora spp. and Pocillopora damicornis still remain separated as per the same observations in the reports of Littman et al. (17) and Bourne and Munn (3), which confirms the accuracy of our analysis. Overall, the clustering results suggest that the impact of species specificity and environmental factors in different corals may be differential. The bacteria harbored by corals isolated from the Great Barrier Reef were more influenced by environmental parameters than by coral species specificity; on the other hand, some of the coral-associated bacteria from the Caribbean Sea show stronger species specificity, particularly with Porites astreoides-associated bacteria. This observation echoes the analysis results of those previous reports.
The association between coral and bacteria has been convincingly shown to be highly specific, according to the significant difference of the bacterial profiles between coral and nearby water samples in many reports (3, 6, 9, 11, 13, 16, 17, 19, 20, 21, 23, 30, 31, 33). There are several studies demonstrating species specificity of coral bacteria, but a few other reports suggest that environmental factors (including seasonal factors) have more significant impact and that there is little evidence for species-specific bacterial association. Based on the clustering comparison we conducted here, we suggest a multifactor interaction model for this issue. Environmental factors, coral species specificity, differences in physiology between and within individual coral colonies, differences of bacterial population composition in situ, and temporal factors should all be considered to have an integrated, compound effect on the association between coral and bacteria. Each factor may become a major influence under particular conditions. The effects on different coral species in the same reef may also differ. Some corals, such as Porites astreoides, might maintain consistently high selection against local environmental factors; however, other corals, such as Acropora or Pocillopora spp., might be rather more flexible and more easily influenced by environmental factors (3, 17).
More experiments to address the issue of species specificity are needed. We suggest choosing a few corals that are dispersed worldwide as model organisms to study. The results of such a future study would simplify interpretation, remove species inconsistency, and provide clearer evidence of the influence of various factors on coral-associated bacterial population structures.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support of this study by thematic project funding (AS-97-TP-B01) of Academia Sinica and the Biodiversity Research Center.
We also thank Hsiao-Chi Chen and Harry Wilson for English proofreading and suggestions and Garmin Hsu and the colleagues of Allen C. L. Chen for assistance with sample collection.
Footnotes
Published ahead of print on 23 October 2009.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne, D., Y. Iida, S. Uthicke, and C. Smith-Keune. 2008. Changes in coral-associated microbial communities during a bleaching event. Isme J. 2:350-363. [DOI] [PubMed] [Google Scholar]
- 3.Bourne, D. G., and C. B. Munn. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7:1162-1174. [DOI] [PubMed] [Google Scholar]
- 4.Bruck, T. B., W. M. Bruck, L. Z. Santiago-Vazquez, P. J. McCarthy, and R. G. Kerr. 2007. Diversity of the bacterial communities associated with the azooxanthellate deep water octocorals Leptogorgia minimata, Iciligorgia schrammi, and Swiftia exertia. Mar. Biotechnol. (NY) 9:561-576. [DOI] [PubMed] [Google Scholar]
- 5.Bunkley-Williams, L., J. Morelock, and E. H. Williams, Jr. 1991. Lingering effects of the 1987 mass bleaching of Puerto Rican coral reefs in mid to late 1988. J. Aquat. Anim. Health 3:242-247. [Google Scholar]
- 6.Cooney, R. P., O. Pantos, M. D. Le Tissier, M. R. Barer, A. G. O'Donnell, and J. C. Bythell. 2002. Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4:401-413. [DOI] [PubMed] [Google Scholar]
- 7.de Hoon, M. J., S. Imoto, J. Nolan, and S. Miyano. 2004. Open source clustering software. Bioinformatics 20:1453-1454. [DOI] [PubMed] [Google Scholar]
- 8.Ducklow, H. W., and R. Mitchell. 1979. Composition of mucus released by coral reef coelenterates. Limnol. Oceanog. 24:706-714. [Google Scholar]
- 9.Frias-Lopez, J., A. L. Zerkle, G. T. Bonheyo, and B. W. Fouke. 2002. Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68:2214-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gewin, V. 2006. Genomics: discovery in the dirt. Nature 439:384-386. [DOI] [PubMed] [Google Scholar]
- 11.Kellogg, C. A. 2004. Tropical Archaea: diversity associated with the surface microlayer of corals. Mar. Ecol. Prog. Ser. 273:81-88. [Google Scholar]
- 12.Kenkel, N. C., and L. Orlóci. 1986. Applying metric and nonmetric multidimensional scaling to ecological studies: some new results. Ecology 67:919-928. [Google Scholar]
- 13.Klaus, J. S., I. Janse, J. M. Heikoop, R. A. Sanford, and B. W. Fouke. 2007. Coral microbial communities, zooxanthellae and mucus along gradients of seawater depth and coastal pollution. Environ. Microbiol. 9:1291-1305. [DOI] [PubMed] [Google Scholar]
- 14.Knowlton, N., and F. Rohwer. 2003. Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am. Nat. 162:S51-62. [DOI] [PubMed] [Google Scholar]
- 15.Kooperman, N., E. Ben-Dov, E. Kramarsky-Winter, Z. Barak, and A. Kushmaro. 2007. Coral mucus-associated bacterial communities from natural and aquarium environments. FEMS Microbiol. Lett. 276:106-113. [DOI] [PubMed] [Google Scholar]
- 16.Koren, O., and E. Rosenberg. 2006. Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl. Environ. Microbiol. 72:5254-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littman, R. A., B. L. Willis, C. Pfeffer, and D. G. Bourne. 2009. Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68:152-163. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie, K. B., and G. W. Smith. 2004. Microbial communities of coral surface mucopolysaccharide layers, p. 259-263. In E. Rosenberg and Y. Loya (ed.), Coral health and disease. Springer-Verlag, Berlin, Germany.
- 19.Ritchie, K. B. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322:1-14. [Google Scholar]
- 20.Rohwer, F., M. Breitbart, J. Jara, F. Azam, and N. Knowlton. 2001. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20:7. [Google Scholar]
- 21.Rohwer, F., V. Seguritan, F. Azam, and N. Knowlton. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1-10. [Google Scholar]
- 22.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekar, R., D. K. Mills, E. R. Remily, J. D. Voss, and L. L. Richardson. 2006. Microbial communities in the surface mucopolysaccharide layer and the black band microbial mat of black band-diseased Siderastrea siderea. Appl. Environ. Microbiol. 72:5963-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorokin, Y. I. 1973. Trophical role of bacteria in the ecosystem of the coral reef. Nature 242:415-417. [Google Scholar]
- 26.Takabayashi, M., D. A. Carter, J. V. Lopez, and O. Hoegh-Guldberg. 2003. Genetic variation of the scleractinian coral Stylophora pistillata, from western Pacific reefs. Coral Reefs 22:6. [Google Scholar]
- 27.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 28.Wagner, M., R. Amann, H. Lemmer, and K. H. Schleifer. 1993. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 59:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webster, N. S., and D. Bourne. 2007. Bacterial community structure associated with the Antarctic soft coral, Alcyonium antarcticum. FEMS Microbiol. Ecol. 59:81-94. [DOI] [PubMed] [Google Scholar]
- 30.Wegley, L., R. Edwards, B. Rodriguez-Brito, H. Liu, and F. Rohwer. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9:2707-2719. [DOI] [PubMed] [Google Scholar]
- 31.Wegley, L., Y. Yu, M. Breitbart, V. Casas, D. I. Kline, and F. Rohwer. 2004. Coral-associated Archaea. Mar. Ecol. Prog. Ser. 273:89-96. [Google Scholar]
- 32.Wilson, K. 2001. Preparation of genomic DNA from bacteria, Chapter 2, Unit 2-4. Curr. Protocols Mol. Biol., John Wiley and Sons, New York, NY. [DOI] [PubMed]
- 33.Yokouchi, H., Y. Fukuoka, D. Mukoyama, R. Calugay, H. Takeyama, and T. Matsunaga. 2006. Whole-metagenome amplification of a microbial community associated with scleractinian coral by multiple displacement amplification using phi29 polymerase. Environ. Microbiol. 8:1155-1163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.