Abstract
Magnetosomes are unique bacterial organelles comprising membrane-enveloped magnetic crystals produced by magnetotactic bacteria. Because of several desirable chemical and physical properties, magnetosomes would be ideal scaffolds on which to display highly complicated biological complexes artificially. As a model experiment for the functional expression of a multisubunit complex on magnetosomes, we examined the display of a chimeric bacterial RNase P enzyme composed of the protein subunit (C5) of Escherichia coli RNase P and the endogenous RNA subunit by expressing a translational fusion of C5 with MamC, a known magnetosome protein, in the magnetotactic bacterium Magnetospirillum gryphiswaldense. As intended, the purified C5 fusion magnetosomes, but not wild-type magnetosomes, showed apparent RNase P activity and the association of a typical bacterial RNase P RNA. Our results demonstrate for the first time that magnetosomes can be employed as scaffolds for the display of multisubunit complexes.
Magnetosomes are unique organelles comprising membrane-enveloped magnetic crystals of iron minerals (Fe3O4 or Fe3S4) produced by magnetotactic bacteria (1, 11). The bacteria employ magnetosomes to sense the environmental magnetic field, probably in order to recognize their favorite environments. Compared with chemically or physically synthesized magnetic nanoparticles, magnetosomes have a variety of desirable features, including their genetically controlled uniform size and morphology, characteristic crystal habits, and their coverage by a biological membrane that can be addressed by functionalization (1, 4, 11). Based on these features, magnetosomes would be ideal scaffolds on which to display biological molecules artificially.
Until now, several heterologous target proteins have been examined for artificial display on magnetosomes (1, 11). For example, reporter proteins such as luciferase and green fluorescent protein were employed to analyze the targeting, expression, and stability of chimeric proteins displayed on magnetosomes (14, 18, 23, 30, 41). For more-practical applications, general antibody-binding proteins (protein A and protein G) were displayed to capture desired antibodies (16, 17, 25, 33, 34, 37, 41). Such antibody-captured magnetosomes are applicable for the magnetic separation of target molecules and cells. Displays of G protein-coupled receptors (the D1 dopamine receptor and the ligand binding domain of the estrogen receptor) were also examined for screening of drugs targeting these receptors (38, 39, 40).
There are two major strategies for the construction of functionalized magnetosomes: subsequent chemical modifications of purified magnetosomes (3) and in vivo expression of modified magnetosome proteins (1, 19). The latter approach is confined to biological molecules that can be expressed as a genetic fusion with a magnetosome protein inside a magnetotactic bacterium. By this approach, the target-displaying magnetosomes can be constructed inside cells or under physiological conditions in the presence of a variety of chaperons, are recoverable under mild conditions employing a magnetic field, and provide control by genetic means. Thus, the approach is highly promising for the display of a naïve target such as a multisubunit complex. To date, however, experimental evidence that magnetosomes can be employed as scaffolds for the display of such targets is still lacking. In order to demonstrate this potential of magnetosomes, here, we examined the display of a holoenzyme of bacterial RNase P, one of the simplest complexes composed of a single RNA and a single protein subunit (10, 12), by expressing a fusion of a protein component of the RNase P and a magnetosome membrane protein.
MATERIALS AND METHODS
Construction of a strain expressing the MamC-C5 fusion protein.
The coding region of the protein subunit of the Escherichia coli C5 protein was amplified from a C5 protein expression vector, pET-3a-C5 (a gift from H. Shiraishi), by PCR using ExTaq polymerase (Takara Bio, Japan) with primers C5-F {5′-CATATGGGTG GCAGCGGCGG TTCTGGTGGT AGCGGCGGCA GCGGTGTTAA GCTCGCATTT CCCAGGG-3′; a restriction enzyme [NdeI] site is italicized, and a flexible linker [(GlyGlySer)4Gly] sequence is underlined} and C5-R (5′-GGATCCTCAT TAGGACCCGC GAGCCAGGCG ACAG-3′; a restriction enzyme [BamHI] site is italicized, and stop codons are underlined). The amplified fragment was cloned into the pGEM-T Easy vector (Promega), and its sequence was confirmed from both directions. The fragment with the correct sequence was recovered by digestion with NdeI and BamHI and was then inserted downstream of the mamC gene of pBBR-gpx1-Cstrep (C. Lang and D. Schüler, unpublished data) digested with the same enzymes, resulting in an expression vector, pPmamg-mamC-C5, that expresses the MamC-C5 fusion protein under the control of the PmamDC promoter. The plasmid was transferred into M. gryphiswaldense (MSR1 derivative strain, R3/S1) by conjugation from E. coli S17-1 as described previously (31). The expression of the fusion protein was analyzed by Western blotting using an anti-MamC antibody as described previously (6, 18).
Preparation of magnetosomes.
Bacterial culture and production of magnetosomes were carried out essentially as described previously (18). Briefly, 4 liters of stationary-phase cultures was harvested by centrifugation, washed with 20 mM HEPES-HCl (pH 7.5)-10 mM MgCl2, and resuspended in buffer A (50 mM HEPES-HCl [pH 7.5], 10 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol [DTT], 5% glycerol, 0.1 mM phenylmethylsulfonyl fluoride, 10 U/ml RiboLock RNase inhibitor [Fermentas, Germany]). The cells were disrupted by two passages through a benchtop constant cell disruptor (Constant Systems) at 1.35 × 108 Pa, followed by centrifugation at 800 × g for 5 min to remove cell debris. The magnetosome-containing supernatant fluid was then passed through a MACS magnetic separation column (Miltenyi Biotech, Germany) placed between Sm-Co magnets (Miltenyi, Bergisch Gladbach, Germany), washed with 20 column volumes (100 ml) of buffer B (buffer A plus 100 mM NaCl [final concentration, 200 mM]), and washed with 5 column volumes (25-ml) of buffer A again. The washed magnetosomes were eluted with buffer C (10 mM HEPES-HCl [pH 7.5], 10 mM MgCl2) by removing the magnetic field. Until use, the recovered magnetosomes were stored in buffer C supplemented with DTT (final concentration, 1 mM), phenylmethylsulfonyl fluoride (0.1 mM), and RiboLock RNase inhibitor (1 U/ml) at 4°C. After cell harvest, all manipulations were carried out under RNase-free conditions in a cold room (4°C). Typically, magnetosomes containing ∼1 mmol of Fe were recovered from a 4-liter culture, as determined by the colorimetric ferrozine method (5). Two independent preparations of the C5 fusion magnetosomes were examined for all the following experiments.
RNase P reaction.
A precursor tRNA (pre-tRNA) used as a substrate for the RNase P reaction was prepared by in vitro transcription using SP6 RNA polymerase (Invitrogen) with MvaI-digested p67-YFO as the template DNA in the presence of [α-32P]GTP (GE Healthcare) as described previously (28). The internally labeled transcript was fractionated by 10% denaturing polyacrylamide gel electrophoresis and was recovered by a crash-and-soak method. RNase P reactions were carried out with the equivalent of 1 mM magnetosome-bound iron in buffer P (10 mM HEPES-HCl [pH 7.5], 7 mM MgCl2, 100 mM NH4Cl, 1 mM DTT, 0.2 U/μl RNase inhibitor) at 37°C. The reactions were started and stopped by adding pre-tRNA (final concentration, 1 nM) and 2 volumes of the stop solution (90% formamide, 10 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol), respectively. The reaction products were separated by 10% denaturing polyacrylamide gel electrophoresis, and the RNA bands were quantified with a Storm phosphorimager (Amersham Biosciences).
Cloning and quantification of magnetosome-associated RNase P RNA.
Magnetosome-associated RNAs were eluted from the purified magnetosomes by addition of EDTA (final concentration, 50 mM) and phenol-chloroform extraction, followed by ethanol precipitation. The recovered RNAs were cloned by a 5′ adaptor ligation/3′ polyadenylation method as described previously (32). Poly(A) polymerase and reverse transcriptase (ReverTra Ace) were purchased from Takara Bio and Toyobo (Japan), respectively. A synthetic oligo-RNA, S-1 (26), and pGEM-T Easy were used as the 5′ adaptor and cloning vector, respectively.
The magnetosome-associated RNase P RNA was quantified by Northern blotting using a DIG Northern starter kit (Roche) with sense and antisense strands of an RNase P RNA fragment from clone F08 (see Fig. S1 in the supplemental material) as the positive control and the probe, respectively. The fragment, containing the RNase P RNA fragment and pGEM-T Easy vector-derived T7 and SP6 promoters, was amplified by PCR from the plasmid DNA of clone F08 with M13 primers M4 and RV (Takara Bio). Using the PCR product as the template, the probe and control RNAs were prepared by in vitro transcription with T7 RNA polymerase and SP6 RNA polymerase in the presence and absence of digoxigenin-11-UTP, respectively. Northern blotting was carried out according to the manufacturer's protocol, and the RNA bands were quantified by the LAS-1000plus luminescent image analysis system (Fujifilm).
RESULTS
Construction of RNase P-displaying magnetosomes.
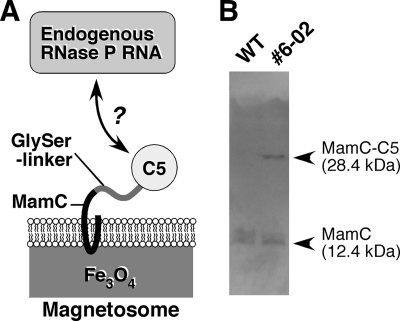
RNase P is a highly conserved ribonucleoprotein complex that catalyzes the hydrolytic cleavage of the 5′ leader of the tRNA precursor to generate mature tRNA (10, 12). Bacterial RNase P is one of the simplest multisubunit enzymes, composed of a single RNA and a single protein subunit. Because the protein subunit (C5) of Escherichia coli RNase P is known to form the active RNase P complex with RNA subunits from a wide variety of bacterial strains (8, 35, 36), it is expected that the expression of C5 protein on magnetosomes would lead to the display of the active heterogenic RNase P complex composed of E. coli C5 and the endogenous RNA subunit (Fig. 1A). To achieve this, a major magnetosome protein, MamC, was chosen as a fusion partner. In previous studies it was shown that MamC is the most abundant magnetosome protein (7, 29) and can be used as an efficient anchor for the ectopic display of target proteins on magnetosomes (18). C5 protein was fused to the C-terminal end of MamC via 13 residues of a flexible linker to avoid steric hindrance, which could inhibit complex formation (Fig. 1A), and this fusion protein was expressed in the magnetotactic bacterium Magnetospirillum gryphiswaldense (31) under the control of the strong, endogenous PmamDC promoter (17). One transconjugant (clone 6-02), which expressed the C5 fusion MamC as efficiently as the endogenous MamC, was chosen for the preparation of the RNase P-displaying magnetosomes (referred to below as the C5 fusion magnetosomes) (Fig. 1B). As a control, magnetosomes were also prepared from the parental, nontransconjugant bacteria (referred to as “wild type” [“WT”] in this article).
FIG. 1.
(A) Representation of the strategy for the display of RNase P on magnetosomes. (B) Expression of the MamC-C5 fusion protein in M. gryphiswaldense. The expression of the fusion protein was analyzed by Western blotting using an anti-MamC antibody. The transconjugant strain (clone 6-02) (right) expressed the fusion protein as efficiently as endogenous MamC. As a control, a cell lysate of the WT strain was loaded in the left lane.
Analysis of the RNase P activity of C5 fusion magnetosomes.
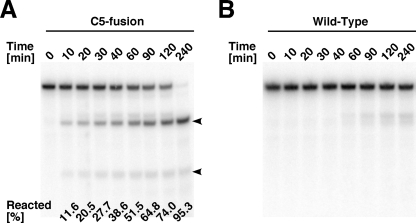
The RNase P activity of the C5 fusion or WT magnetosomes was confirmed by analysis of the cleavage of pre-tRNA under physiological conditions (in the presence of 7 mM MgCl2 and 100 mM NH4Cl at a neutral pH at 37°C). As intended, the C5 fusion magnetosomes showed apparent RNase P activity (Fig. 2A). Although the WT magnetosomes also showed some degradation of pre-tRNA, the sizes of the cleavage products were heterogeneous, and they were slightly longer than the expected RNase P cleavage product (Fig. 2B). In addition, no 5′ leader fragment was produced by the WT magnetosomes, suggesting that degradation of the fragile 5′ leader in the control was caused by contaminating nonspecific nucleases rather than by RNase P.
FIG. 2.
RNase P reaction assay of the C5 fusion (A) and WT (B) magnetosomes. The reactions were carried out under the physiological buffer condition (10 mM HEPES-HCl [pH 7.5], 7 mM MgCl2, 100 mM NH4Cl, 1 mM DTT) at 37°C. Arrowheads indicate the cleavage products (the 5′ leader fragment and mature tRNA). Average percentages (from three independent experiments with the two independent preparations) of fractions reacted by the C5 fusion magnetosomes are given in panel A.
Isolation of RNase P RNA associated with C5 fusion magnetosomes.
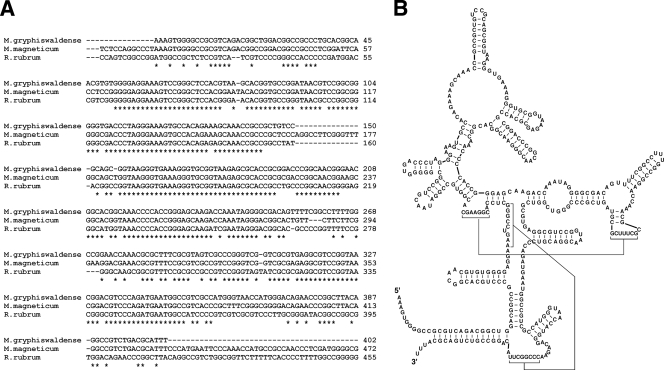
If the C5 fusion magnetosomes displayed the chimeric RNase P as intended, the endogenous RNase P RNA subunit should associate with the C5 fusion magnetosomes. To confirm this, the RNAs associated with the magnetosomes were recovered and cloned by the 5′ adaptor ligation/3′ polyadenylation method (32). As expected, fragments of a typical type A RNA subunit of bacterial RNase P were cloned (Fig. 3; see also Fig. S1 in the supplemental material). The reconstituted sequence of the RNA subunit is expected to form a secondary structure highly similar to the reported secondary structure of the closely related organism Rhodospirillum rubrum (9) (Fig. 3). A highly similar sequence is also found in the genome of another magnetotactic species, Magnetospirillum magneticum AMB1 (24) (Fig. 3).
FIG. 3.
(A) Alignment of RNase P RNAs. The isolated RNase P RNA of M. gryphiswaldense was aligned with the homologous sequence in the M. magneticum genome (genome location, 4237281 to 4237836) (24) and the RNase P RNA of Rhodospirillum rubrum (9) by ClustalW2 (20). (B) Predicted secondary structure of the isolated RNase P RNA. The secondary structure was predicted by comparison with that of the RNase P RNA of R. rubrum; the structure prediction program based on energy minimization, Mfold (22, 42), was used.
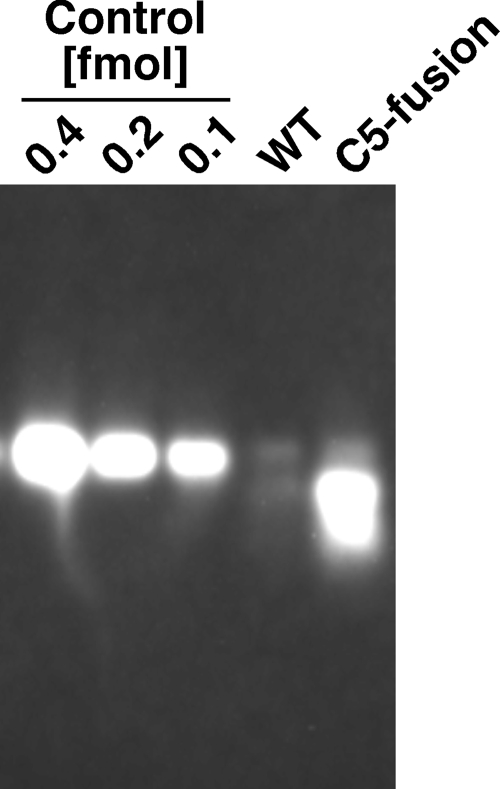
Next, the level of association of the RNase P RNA with the magnetosomes was confirmed by Northern blotting. As expected, the C5 fusion magnetosomes clearly showed an association with the RNA subunit (Fig. 4; note that the control RNA carries not only a fragment of the RNase P RNA but also the tag sequences for cloning and a portion of the vector sequence). In contrast, WT magnetosomes showed only faint signals, which could be caused by nonspecific hybridization or by slight contamination with the cytosolic RNase P RNA. According to the intensity of the band, the level of association is calculated as 10.0 (±0.2) nmol/mol of Fe contained in the magnetosomes. Assuming that the whole amount of the associated RNA subunit formed the active holoenzyme complex, its initial reaction rate is calculated as 1.1 min−1 in the presence of 1 nM pre-tRNA under the physiological condition. This value is comparable to the values reported for the E. coli RNase P holoenzyme (2, 21, 27).
FIG. 4.
Northern blotting of the RNase P RNA associated with the magnetosomes. RNAs eluted from 200 nmol of Fe contained in the magnetosomes were loaded in each lane. As a control for the quantification, a transcript of the cloned fragment of the RNase P RNA (clone F08) (see Fig. S1 in the supplemental material) was also loaded.
DISCUSSION
In this study, we examined the display of a multisubunit enzyme complex, bacterial RNase P, on magnetosomes. Our results clearly show that the chimeric RNase P holoenzyme was successfully displayed on magnetosomes, as intended.
Although the displayed RNase P holoenzyme has a heterologous pairing of the RNA and protein subunits, this chimeric RNase P showed activity comparable to, or even higher than, that reported for the natural E. coli holoenzyme (2, 21, 27). This observation may be due to the preferable environment on magnetosomes, additional association of enhancer factors such as molecular chaperons, and/or complete formation of an active holoenzyme complex under the intracellular condition (the reported holoenzymes might be incompletely reconstituted). However, the possibility that the chimeric holoenzyme has intrinsically higher activity or that slight differences in the assay conditions might unexpectedly affect the activities cannot be excluded and should be investigated by further analyses.
It is known that bacterial RNase P RNAs are especially stable in a complex with the protein subunit (13). However, we failed to isolate the full-length RNA and cloned only parts of the RNA (see Fig. S1 in the supplemental material). One possible explanation is that the strong secondary structures of the RNA might interfere with the extension in the reverse transcription. Because the cloning procedure employed in this study requires an uncapped 5′ terminus, there is another possibility, that our inability to isolate full-length RNA may be caused by the 5′ capping of the RNase P RNA. Recently, mass spectrometry-based analysis of chemical modifications of intracellular RNAs revealed several previously unidentified 5′ cap structures of bacterial RNAs (15).
According to the Western blotting result (Fig. 1B), the expression level of the MamC-C5 fusion protein was comparable to that of endogenous MamC. Assuming that the display of the fusion on the magnetosomes was as efficient as the reported display of other proteins employing an anchor homologous to MamC (41), it is calculated that only a portion of the displayed C5 (less than 10%) formed a complex with the RNase P RNA. This is probably because most of the endogenous RNase P RNA forms a complex with the endogenous protein subunit. In order to achieve a higher efficiency of complex formation, knockdown/knockout of the endogenous protein or overexpression of the RNA is required.
Here we report the first example of successful construction of magnetosomes displaying the active RNase P holoenzyme complex, and we demonstrate that magnetosomes can be employed for the generation of a magnetic nanoparticle displaying a multisubunit complex. The display could be achieved with only a single fusion protein, and a similar strategy would be applicable for the display of far more complicated complexes, such as ribosomes, by expressing only one of the ribosomal proteins anchored by a magnetosome protein. In addition, it has been demonstrated that several other magnetosome proteins are also applicable as anchors (18). Thus, not only biological complexes with a single anchored protein, but also complexes with multiple anchored proteins, could be displayed on magnetosomes. Such a multiple anchoring strategy would increase local concentrations of the displayed proteins and may be used for the generation of an artificial multienzyme-like complex for novel, completely artificial reaction pathways with high efficiency and accuracy. The results of this study open the door for future technology for the production of elaborate, highly functionalized magnetic nanoparticles as nanosized reactors and sensors via simple fermentation processes.
Supplementary Material
Acknowledgments
We thank all the members of the Schüler lab, especially Claus Lang and Manuela Löhr, for their kind help with the preparation of magnetosomes. We also thank Hideaki Shiraishi of Kyoto University (Japan) for the plasmids carrying the C5 protein and pre-tRNA genes and for helpful comments on the manuscript.
This work was sponsored by the LMU-Todai Cooperation Program.
Footnotes
Published ahead of print on 16 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Arakaki, A., H. Nakazawa, M. Nemoto, T. Mori, and T. Matsunaga. 2008. Formation of magnetite by bacteria and its application. J. R. Soc. Interface 5:977-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baer, M. F., D. Wesolowski, and S. Altman. 1989. Characterization in vitro of the defect in a temperature-sensitive mutant of the protein subunit of RNase P from Escherichia coli. J. Bacteriol. 171:6862-6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ceyhan, B., P. Alhorn, C. Lang, D. Schüler, and C. M. Niemeyer. 2006. Semisynthetic biogenic magnetosome nanoparticles for the detection of proteins and nucleic acids. Small 2:1251-1255. [DOI] [PubMed] [Google Scholar]
- 4.Faivre, D., and D. Schüler. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875-4898. [DOI] [PubMed] [Google Scholar]
- 5.Fish, W. W. 1988. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158:357-364. [DOI] [PubMed] [Google Scholar]
- 6.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrier-Takada, C., K. Gardiner, T. Marsh, N. Pace, and S. Altman. 1983. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849-857. [DOI] [PubMed] [Google Scholar]
- 9.Harris, J. K., E. S. Haas, D. Williams, D. N. Frank, and J. W. Brown. 2001. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA 7:220-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsieh, J., A. J. Andrews, and C. A. Fierke. 2004. Roles of protein subunits in RNA-protein complexes: lessons from ribonuclease P. Biopolymers 73:79-89. [DOI] [PubMed] [Google Scholar]
- 11.Jogler, C., and D. Schüler. 2009. Genetics, genomics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501-521. [DOI] [PubMed] [Google Scholar]
- 12.Kazantsev, A. V., and N. R. Pace. 2006. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 4:729-740. [DOI] [PubMed] [Google Scholar]
- 13.Kim, Y., and Y. Lee. 2009. Novel function of C5 protein as a metabolic stabilizer of M1 RNA. FEBS Lett. 583:419-424. [DOI] [PubMed] [Google Scholar]
- 14.Komeili, A., H. Vali, T. J. Beveridge, and D. K. Newman. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc. Natl. Acad. Sci. USA 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowtoniuk, W. E., Y. Shen, J. M. Heemstra, I. Agarwal, and D. R. Liu. 2009. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl. Acad. Sci. USA 106:7768-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhara, M., H. Takeyama, T. Tanaka, and T. Matsunaga. 2004. Magnetic cell separation using antibody binding with protein A expressed on bacterial magnetic particles. Anal. Chem. 76:6207-6213. [DOI] [PubMed] [Google Scholar]
- 17.Lang, C., A. Pollithy, and D. Schüler. 2009. Identification of promoters for efficient gene expression in. Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 75:4206-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang, C., and D. Schüler. 2008. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl. Environ. Microbiol. 74:4944-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang, C., D. Schüler, and D. Faivre. 2007. Synthesis of magnetite nanoparticles for bio- and nanotechnology: genetic engineering and biomimetics of bacterial magnetosomes. Macromol. Biosci. 7:144-151. [DOI] [PubMed] [Google Scholar]
- 20.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 21.Lazard, M., and T. Meinnel. 1998. Role of base G-2 of pre-tRNAfMet in cleavage site selection by Escherichia coli RNase P in vitro. Biochemistry 37:6041-6049. [DOI] [PubMed] [Google Scholar]
- 22.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 23.Matsunaga, T., A. Arakaki, and M. Takahoko. 2002. Preparation of luciferase-bacterial magnetic particle complex by artificial integration of MagA-luciferase fusion protein into the bacterial magnetic particle membrane. Biotechnol. Bioeng. 77:614-618. [DOI] [PubMed] [Google Scholar]
- 24.Matsunaga, T., Y. Okamura, Y. Fukuda, A. T. Wahyudi, Y. Murase, and H. Takeyama. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12:157-166. [DOI] [PubMed] [Google Scholar]
- 25.Matsunaga, T., M. Takahashi, T. Yoshino, M. Kuhara, and H. Takeyama. 2006. Magnetic separation of CD14+ cells using antibody binding with protein A expressed on bacterial magnetic particles for generating dendritic cells. Biochem. Biophys. Res. Commun. 350:1019-1025. [DOI] [PubMed] [Google Scholar]
- 26.Ohuchi, S. P., Y. Ikawa, and Y. Nakamura. 2008. Selection of a novel class of RNA-RNA interaction motifs based on the ligase ribozyme with defined modular architecture. Nucleic Acids Res. 36:3600-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pomeranz Krummel, D. A., and S. Altman. 1999. Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA. Proc. Natl. Acad. Sci. USA 96:11200-11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampson, J. R., and O. C. Uhlenbeck. 1988. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA 85:1033-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffel, A., A. Gärdes, K. Grünberg, G. Wanner, and D. Schüler. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 190:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultheiss, D., R. Handrick, D. Jendrossek, M. Hanzlik, and D. Schüler. 2005. The presumptive magnetosome protein Mms16 is a poly(3-hydroxybutyrate) granule-bound protein (phasin) in Magnetospirillum gryphiswaldense. J. Bacteriol. 187:2416-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 32.Sun, G., H. Li, and J. J. Rossi. 2007. Cloning and detecting signature microRNAs from mammalian cells. Methods Enzymol. 427:123-138. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi, M., Y. Akiyama, J. Ikezumi, T. Nagata, T. Yoshino, A. Iizuka, K. Yamaguchi, and T. Matsunaga. 2009. Magnetic separation of melanoma-specific cytotoxic T lymphocytes from a vaccinated melanoma patient's blood using MHC/peptide complex-conjugated bacterial magnetic particles. Bioconjugate Chem. 20:304-309. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi, M., T. Yoshino, H. Takeyama, and T. Matsunaga. 2009. Direct magnetic separation of immune cells from whole blood using bacterial magnetic particles displaying protein G. Biotechnol. Prog. 25:219-226. [DOI] [PubMed] [Google Scholar]
- 35.Waugh, D. S., and N. R. Pace. 1990. Complementation of an RNase P RNA (rnpB) gene deletion in Escherichia coli by homologous genes from distantly related eubacteria. J. Bacteriol. 172:6316-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wegscheid, B., C. Condon, and R. K. Hartmann. 2006. Type A and B RNase P RNAs are interchangeable in vivo despite substantial biophysical differences. EMBO Rep. 7:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshino, T., H. Hirabe, M. Takahashi, M. Kuhara, H. Takeyama, and T. Matsunaga. 2008. Magnetic cell separation using nano-sized bacterial magnetic particles with reconstructed magnetosome membrane. Biotechnol. Bioeng. 101:470-477. [DOI] [PubMed] [Google Scholar]
- 38.Yoshino, T., C. Kaji, M. Nakai, F. Saito, H. Takeyama, and T. Matsunaga. 2008. Novel method for evaluation of chemicals based on ligand-dependent recruitment of GFP labeled coactivator to estrogen receptor displayed on bacterial magnetic particles. Anal. Chim. Acta 626:71-77. [DOI] [PubMed] [Google Scholar]
- 39.Yoshino, T., F. Kato, H. Takeyama, M. Nakai, Y. Yakabe, and T. Matsunaga. 2005. Development of a novel method for screening of estrogenic compounds using nano-sized bacterial magnetic particles displaying estrogen receptor. Anal. Chim. Acta 532:105-111. [Google Scholar]
- 40.Yoshino, T., M. Takahashi, H. Takeyama, Y. Okamura, F. Kato, and T. Matsunaga. 2004. Assembly of G protein-coupled receptors onto nanosized bacterial magnetic particles using Mms16 as an anchor molecule. Appl. Environ. Microbiol. 70:2880-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshino, T., and T. Matsunaga. 2006. Efficient and stable display of functional proteins on bacterial magnetic particles using Mms13 as a novel anchor molecule. Appl. Environ. Microbiol. 72:465-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.