Abstract
Two strains, identified as Rhodococcus wratislaviensis IFP 2016 and Rhodococcus aetherivorans IFP 2017, were isolated from a microbial consortium that degraded 15 petroleum compounds or additives when provided in a mixture containing 16 compounds (benzene, toluene, ethylbenzene, m-xylene, p-xylene, o-xylene, octane, hexadecane, 2,2,4-trimethylpentane [isooctane], cyclohexane, cyclohexanol, naphthalene, methyl tert-butyl ether [MTBE], ethyl tert-butyl ether [ETBE], tert-butyl alcohol [TBA], and 2-ethylhexyl nitrate [2-EHN]). The strains had broad degradation capacities toward the compounds, including the more recalcitrant ones, MTBE, ETBE, isooctane, cyclohexane, and 2-EHN. R. wratislaviensis IFP 2016 degraded and mineralized to different extents 11 of the compounds when provided individually, sometimes requiring 2,2,4,4,6,8,8-heptamethylnonane (HMN) as a cosolvent. R. aetherivorans IFP 2017 degraded a reduced spectrum of substrates. The coculture of the two strains degraded completely 13 compounds, isooctane and 2-EHN were partially degraded (30% and 73%, respectively), and only TBA was not degraded. Significant MTBE and ETBE degradation rates, 14.3 and 116.1 μmol of ether degraded h−1 g−1 (dry weight), respectively, were measured for R. aetherivorans IFP 2017. The presence of benzene, toluene, ethylbenzene, and xylenes (BTEXs) had a detrimental effect on ETBE and MTBE biodegradation, whereas octane had a positive effect on the MTBE biodegradation by R. wratislaviensis IFP 2016. BTEXs had either beneficial or detrimental effects on their own degradation by R. wratislaviensis IFP 2016. Potential genes involved in hydrocarbon degradation in the two strains were identified and partially sequenced.
The pollution of soils by petroleum compounds is of great concern mainly because of the solubilities of the different molecules in water, which can endanger aquifers in contact with polluted zones. Petroleum storage facilities are frequently the source of pollution due to leaks and spills during fuel transfer and storage. For example, in the United States in 2007, the EPA indicated that nearly 110,000 old leaks have not yet been cleaned up, and there are an unknown number of petroleum brownfield sites (estimated to be over 200,000) that are predominately old abandoned gas stations (http://www.epa.gov/OUST/pubs/OUST_FY07_Annual_Report-_Final_4-08.pdf). In these locations, the contamination can be generated by diesel oil and/or gasoline leaks from storage tanks, resulting in a complex mixture of compounds with different water solubilities and different biodegradabilities. Among all the phenomena occurring at polluted sites, (i) the interactions between the different compounds can result in enhanced solubility for low-solubility compounds, (ii) the differences in biodegradability levels between the dissolved molecules can lead to dispersion of the poorly biodegradable or nonbiodegradable compounds, and (iii) in the presence of mixtures of compounds, interactions between some of them can lead to detrimental or beneficial effects. For example, methyl tert-butyl ether [MTBE] could enhance the mobility of dissolved benzene, toluene, ethylbenzene, and xylenes [BTEXs] by exerting a cosolvent effect that decreases sorption-related retardation (30). The impact of additive use after petroleum refining to meet specific requirements is a point that deserves more study. MTBE was used extensively in the United States and elsewhere in the world. Several states banned the use of MTBE because of numerous reports of groundwater pollution, but this compound is still used in Europe. Although its use is decreasing, it still remains high, and by the end of 2007, global MTBE production was about 15 million tons. Ethyl tert-butyl ether (ETBE) is used in Europe (France, Spain, Belgium, and Germany), with European production reaching 626,300 tons in 2004 (http://www.agriculture.total.fr). MTBE and ETBE can be added at up to 15% to gasoline in order to reach the octane index requirement; their use was shown to limit noncombusted hydrocarbon release in exhaust pipe fumes. 2-Ethylhexyl nitrate (2-EHN) is the nitric ester of 2-ethyl-1-hexanol, and it is added to diesel formulations at up to 0.4%. The 2-EHN market is about 100,000 tons/year. Alkylates are native components of petroleum products, but in view of the use of ethanol in gasoline, alkylates, like 2,2,4-trimethylpentane (isooctane), are among the few high-octane alternatives that have been proposed, if only the minimal volume of ethanol required to meet oxygenate requirements are used in reformulated gasoline. In this case, other fuel constituents would be needed to make up the resulting 5% gap and the octane shortfall of about 1.5 octane points. Isooctane has an octane rating of 100 and would be attractive for refiners as an octane enhancer since it can be produced by former MTBE production plants (35).
Data concerning the use of additives have to be taken into account to assess the impact of petroleum products on polluted sites, such as gas stations, where leaks from different storage tanks can occur, leading to contamination by complex mixtures of petroleum products. The biodegradation of monoaromatic (BTEX) compounds and alkanes has been studied extensively, and both are generally quite biodegradable under aerobic conditions (44, 46). Regarding the biodegradability of MTBE, several microorganisms have been isolated with specific degradation capacities, and some of the genes involved in the biodegradation pathway have been characterized (28). The first-order attenuation rates for MTBE in the plumes in which biodegradation occurred varied from 0.56 to 4.3 year−1, a rate of biodegradation not sufficient to contain the plume (7). In addition, there are numerous sites for which no biodegradation was observed (3, 10), and the presence of BTEXs and MTBE has been shown in the case of Methylibium petroleiphilum PM1 to delay the onset of MTBE biodegradation (13).
The behavior of ETBE when spilled in the environment has not been as well studied as that of MTBE, and the extent of contamination has not been documented sufficiently. Similar to that of MTBE, the biodegradation of ETBE is not always observed in microcosms with soils or aquifers derived from contaminated sites (3). Microorganisms able to grow on ETBE have been isolated, and the first monooxygenase system able to degrade ETBE was identified as a cytochrome P450 monooxygenase (encoded by the ethRABCD genes) in Rhodococcus ruber IFP 2001 (6, 21). Highly similar eth gene clusters were also isolated from Rhodococcus zopfii IFP 2005 and Gordonia sp. strain IFP 2009 (4, 16). R. ruber IFP 2001, R. zopfii IFP 2005, and Gordonia sp. strain IFP 2009 were able to grow on ETBE at the expense of the C2 moiety being released by the cleavage of the ether bond with the accumulation of tert-butyl alcohol (TBA) in the growth culture (C. Malandain, F. Fayolle-Guichard, and T. Vogel, submitted for publication). Interestingly, other microorganisms belonging to the genus Rhodococcus were reported to have biodegradation capacities toward ether fuels. Mo et al. (31) isolated a Rhodococcus sp. strain able to degrade MTBE to a low extent; R. aetherivorans, a new species that belongs to MTBE-degrading actinomycetes (20) was characterized, but the enzymatic system responsible for the MTBE oxidation was not elucidated; Rhodococcus sp. strain EH831 was able to degrade MTBE (27).
There are few data in the literature regarding the biodegradability of isooctane; Mycobacterium austroafricanum IFP 2173 was the only strain described for its ability to use isooctane as the sole carbon and energy source (42), and more recently, Cho et al. (9) demonstrated the biodegradability of isooctane using previously acclimated biomass. Regarding the biodegradability of 2-EHN, only M. austroafricanum IFP 2173 was recently reported to degrade 2-EHN to 4-ethyldihydrofuran-2(3H)-one (36).
The biodegradation of complex mixtures of hydrocarbons has generally been studied only under the highest-performing conditions using different processes (e.g., biofilters) in which the microorganisms and the role played by each of them have not necessarily been elucidated. Individual microorganisms have generally been characterized for their ability to degrade individual petroleum compounds or classes of compounds, i.e., monoaromatics. There is much less work addressing the issue of the biodegradation by individual, characterized microorganisms of complex mixtures generally found in sites polluted by hydrocarbons, even though some bacterial genera (Pseudomonas and Rhodococcus, for example) are known to degrade a wide range of xenobiotics (17, 25, 41). Some authors have investigated the range of biodegradation capacities of given individual strains. Solano-Serena et al. (42) previously isolated M. austroafricanum IFP 2173 from gasoline-contaminated groundwater, and this strain, tested on a mixture of petroleum compounds, showed extended biodegradation capacities toward various hydrocarbons. More recently, Rhodococcus sp. strain EC1 was shown to degrade BTEXs, short-chain alkanes, pyrene, and MTBE (26).
The selection and the study of strains with capacities to use a broad spectrum of various hydrocarbons is of great interest because it could facilitate the study of the effect of selective pressure in terms of gene acquisition. From a bacterial consortium, including bacteria from soil at a gas station polluted by leaking tanks and enriched on a mixture of various hydrocarbons, we isolated two strains of Rhodococcus wratislaviensis and Rhodococcus aetherivorans and studied their biodegradation capacities toward hydrocarbons or additives added individually or in mixtures.
MATERIALS AND METHODS
Media composition and culture conditions.
The mineral medium (MM) that was used contained KH2PO4, 1.40 g liter−1; K2HPO4, 1.70 g liter−1; MgSO4·7 H2O, 0.5 g liter−1; NH4NO3, 1.5 g liter−1; CaCl2·2 H2O, 0.04 g liter−1; FeSO4·7 H2O, 1 mg liter−1. A vitamin solution and an oligo-element solution were added as previously described (37). After inoculation, the adequate carbon source was added, and the cultures were incubated at 30°C with constant agitation (120 rpm).
The pure strains were also cultivated on 150 ml tripticase-soy (TS) in 500-ml cotton-plugged Erlenmeyer flasks incubated at 30°C with constant agitation (120 rpm).
The solid medium used in petri dishes was TS agar (Bacto). The plates were incubated statically at 30°C.
Isolation procedure.
A bacterial consortium (Mix3) constituted of microorganisms originating from different environments—a sample from a wastewater treatment plant (Achères, France), a sample of pristine forest soil (Ile-de-France, France), and a sample of soil from a gas station polluted by hydrocarbons (Bourgogne, France)—was obtained after three successive transfers on MM in the presence of a mixture of 16 substrates (see below). The samples from these environments were combined to ensure a deep microbial diversity in the starting mix and a high likelihood of containing degrader strains. Each transfer step (Mix1, Mix2, and then Mix3) was carried out in triplicate, and at each transfer step, two of the flasks were used for analyzing residual substrates after different incubation times in order to determine the extent of degradation and to optimize the time for the following transfer.
Samples of Mix3 were serially diluted and plated onto TS agar plates. The plates were incubated at 30°C. Individual colonies were streaked onto TS agar for isolation. Each isolate was checked for purity by restreaking on TS agar.
Strains and preservation.
Stock cultures of Rhodococcus wratislaviensis IFP 2016 and Rhodococcus aetherivorans IFP 2017 were kept frozen at −80°C in MM containing 20% (vol/vol) glycerol, and both strains were deposited at the CNCM (Pasteur Institute, Paris, France) under numbers I-4088 and I-4089, respectively.
Degradation capacities of the mixture of 16 compounds.
All the experiments carried out with the mixture of 16 compounds (i.e., the initial enrichment cultures Mix1, Mix2, and Mix3 or the determination of pure strains' degradation capacities) were carried out under similar conditions on 150 ml MM in 500-ml Schott closed flasks equipped with a side-arm sealed with a rubber stopper allowing sampling with a syringe and injection of the solvent for total extraction (see below). The volume of headspace was sufficient to prevent any limitation of O2 during the experiment.
The inoculation procedures were as follows: during the initial enrichment procedure, at each step the new flasks were inoculated from the previous ones at 15% (vol/vol). When pure strains were tested under these conditions, R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were previously precultivated on TS at 30°C under agitation for 72 h. Cells were washed, resuspended in MM, and inoculated individually or in coculture in the 500-ml Schott flasks equipped with a side-arm and containing 150 ml MM. The final optical density at 600 nm (OD600) in these experiments was 0.5, whether the strain was added individually or not.
After inoculation, the carbon source constituted by a model mixture of 16 hydrocarbons and additives of gasoline and gazole was added (23 μl). The carbon source mixture was composed of nearly equal concentrations of each compound as follows: benzene, 7.2 mg liter−1; toluene, 7.2 mg liter−1; ethylbenzene, 7.1 mg liter−1; m-xylene, 7.0 mg liter−1; p-xylene, 7.1 mg liter−1, o-xylene, 7.1 mg liter−1; octane, 7.9 mg liter−1; hexadecane, 7.4 mg liter−1; isooctane, 7.9 mg liter−1; cyclohexane, 7.7 mg liter−1; cyclohexanol, 6.8 mg liter−1; naphthalene, 8.3 mg liter−1; MTBE, 7.9 mg liter−1; ETBE, 8.0 mg liter−1; TBA, 7.3 mg liter−1; and 2-EHN, 8.2 mg liter−1. The flasks were then incubated at 30°C with constant agitation (120 rpm).
The controls for these experiments were similarly uninoculated flasks containing the mixture of substrates and incubated under the same conditions.
At the end of this type of experiment, the cultures were sampled for the measurement of residual MTBE, ETBE, TBA, and cyclohexanol by gas chromatography (GC)/flame ionization detection (FID). Then, pentane containing 1,1,2-trichloroethane (1,1,2-TCA) was injected through the septum to extract the 12 other substrates. The substrates extracted by pentane were analyzed by GC/FID. The calculation of degradation for each compound was [(residual substrate in control − residual substrate in assay)/residual substrate in control] × 100.
Mineralization measurements of individual compounds.
Cells of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 grown on TS were harvested by centrifugation (23,000 × g for 15 min), washed twice, and suspended in MM at a final OD600 of about 0.5. Serum bottles (160 ml) containing 20 ml MM were seeded at an initial OD600 of 0.2. Each compound was added at 5 μl/flask. In some cases, the use of 2,2,4,4,6,8,8-heptamethylnonane (HMN) as a cosolvent was required, so 5 μl of the substrate was dissolved in 0.5 ml of HMN before introduction into the flask. The bottles were closed with a butyl rubber stopper and sealed. The volume of the headspace was sufficient to prevent any O2 limitation during the experiment. Flasks were incubated under agitation at 30°C. After incubation (42 days), 2 ml of HNO3 (68% [vol/vol]) were added to each serum bottle through the stopper to strip the CO2 dissolved in the aqueous phase to the headspace, and the headspace was then sampled with a gas-tight syringe for analysis by GC/thermal conductivity detection (TCD) to determine the total amount of CO2 produced in each bottle. The endogenous respiration (controls) was estimated with similar flasks without addition of any substrate, and the corresponding CO2 value was subtracted for further calculations. The experiment was carried out in duplicate for each substrate. We calculated the mineralization yield by determining the molar ratios of the difference between the amount of carbon in the total CO2 produced in a test flask (mole 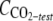 ) and the amount of carbon in the CO2 produced by endogenous respiration (mole
) and the amount of carbon in the CO2 produced by endogenous respiration (mole 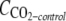 ) to the amount of carbon in the substrate consumed (mole Csubstrate consumed) as follows: [(mole CCO2-test − mole CCO2-control)/mole Csubstrate consumed] × 100.
) to the amount of carbon in the substrate consumed (mole Csubstrate consumed) as follows: [(mole CCO2-test − mole CCO2-control)/mole Csubstrate consumed] × 100.
Degradation capacities of individual compounds.
R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were prepared and tested under the conditions described in the previous paragraph. In this experiment, the controls consisted of noninoculated flasks containing the substrate and incubated under similar conditions. As described above, in some cases, HMN was used to dissolve the substrate. At the end of the experiment, the cultures incubated in the presence of MTBE, ETBE, TBA, or cyclohexanol were sampled directly for determination of residual substrate concentration by GC/FID. In the flasks containing one of the 12 other substrates, pentane containing 1,1,2-TCA as the internal standard was injected to extract any residual substrate. The substrates extracted by pentane were analyzed by GC/FID. The experiment was carried out in duplicate for each substrate. The calculation of degradation for each compound was as follows: [(residual substrate in control − residual substrate in assay)/residual substrate in control] × 100.
Monitoring of CO2 production.
After preculturing on TS, R. wratislaviensis IFP 2016 cells were harvested by centrifugation (23,000 × g for 15 min), washed twice, and suspended in MM to a final OD600 of about 0.2. The culture was incubated in 500-ml Schott flasks containing 200 ml MM in the presence of the desired concentration of substrate(s). The flasks were closed with a stopper equipped with a septum. The CO2 produced in the gaseous phase was sampled regularly through the septum using a gas-tight syringe and measured by GC/TCD.
Degradation assay using resting cells.
Cells of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 grown on TS were harvested by centrifugation (23,000 × g for 15 min), washed twice, and suspended in 100 ml phosphate buffer (20 mM, pH 7) containing the test substrate (ETBE, MTBE, a mixture of ETBE and BTEXs, a mixture of MTBE and BTEXs, or MTBE-ETBE) in 240-ml sealed serum bottles (initial OD600 of ca. 0.5). Five μl of substrate was added to each flask. For the BTEXs mixture, an equimolar mixture of BTEXs was prepared at a concentration of 876 mg ml−1, and 5 μl (ca. 4.4 mg) was added per flask. The volume of headspace was sufficient to prevent any limitation in O2 along the experiment. After inoculation, the flasks were incubated at 30°C on an orbital shaker. All experiments were performed in triplicate. Filtered samples were analyzed by GC. Substrate concentration was measured at 0, 4, 6, 24, 48, and 52 h. The dry weight of the cells was measured by filtration through a 0.22-μm filter at the end of the experiment. The filters were dried at 105°C and weighed to calculate the biomass (dry weight) concentration. The rate of ether degradation was expressed in μmoles of ether degraded g−1 biomass (dry weight) h−1.
Analytical procedures.
MTBE, ETBE, TBA, and cyclohexanol were quantified by flame ionization detection on a Varian 3300 gas chromatograph (Varian, France) as previously described (18). Samples filtered through 0.22-μm filters (Prolabo, Fontenay-sous-Bois, France) were injected without further treatment.
After extraction by pentane containing 1,1,2-TCA as the internal standard, the 12 other compounds were quantified by FID on a Varian gas chromatograph (Varian, France) equipped with a 0.2-mm by 50-m PONA capillary column with a 0.5-μm stationary phase composed of methylpolysiloxane (J&W Scientific, Chromoptic, Auxerre, France), using a two-step temperature gradient ranging from 35°C to 114°C at 1.1°C min−1 and then from 114°C to 310°C at 1.7°C min−1. Helium (0.8 ml min−1) was used as the carrier gas.
Carbon dioxide was quantified by GC/TCD on a Varian 3800 gas chromatograph fitted with a Porapak Q column (1,830 mm by 2 mm). Oven and detector temperatures were 100 and 130°C, respectively. The carrier gas was helium (30 ml min−1), and the column was maintained at 50°C.
Total DNA extraction.
For the initial identification of the 16S rRNA gene from different isolates, DNA from isolated colonies was prepared by boiling lysis; a loopful of isolated colonies was resuspended in 50 μl of sterilized water in Eppendorf tubes and placed in a boiling-water bath for 10 min. After centrifugation (10,000 × g, 10 min, 4°C), supernatants were transferred to new Eppendorf tubes to be used for PCR amplification (5 μl/reaction).
The extraction of DNA from the pure cultures, R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017, was carried out according to the protocol of Pospiech and Neumann (38). The DNA was used to test different catabolic primers by PCR to identify which genes were present in the pure strains.
Primers used for PCR and DNA sequencing.
The PCR amplification and the sequencing of the 16S rRNA gene were performed using the forward primer 5′-GAGAGTTTGATCCTGGCTCAG-3′ (primer 8F) and the reverse primer 5′-TGCACACAGGCCACAACCCA-3′ (primer 1492R).
Other primer pairs used for amplifying and sequencing catabolic genes specific to each chemical compound tested are described in Table 1.
TABLE 1.
PCR detection and DNA sequencing of degradation genes in R. wratislaviensis and R. aetherivorans
| Primer set | Target gene (protein encoded) | Reference | Primers used | Length (bp) of PCR fragment sequenced (expected amplicon size) | GenBank accession no. |
|---|---|---|---|---|---|
| R. wratislaviensis | |||||
| alk-H1F, alk-H3R | alkB (alkane hydroxylase) | 8 | alk-H1F, 5′-CIG IIC ACG AII TIG GIC ACA AGA AGG-3′; alk-H3R, 5′-IGC ITG ITG ATC III GTG ICG CTG IAG-3′ | 429 (549) | FJ590423 |
| nidA-F, -R | nidA | 29 | nidA-F, 5′-GGACTACCTCGGCGATATGA-3′; nidA-R, 5′-TGTGGACGTGCTCTCCATAG-3′ | 310a (233) | FJ590424 |
| PAH-RHD-GP F, R | (Naphthalene inducible dioxygenase) | 5 | PAH-RHD-GP F, 5′-CGGCGCCGACAAYTTYGTNGG-3′; PAH-RHD-GP R, 5′-GGGAACACGGTGCCRTGDATRAA-3′ | 292 | |
| R. aetherivorans | |||||
| alk-H1F, alk-H3R | alkB (alkane hydroxylase) | 8 | alk-H1F, 5′-CIG IIC ACG AII TIG GIC ACA AGA AGG-3′; alk-H3R, 5′-IGC ITG ITG ATC III GTG ICG CTG IAG-3′ | 309 (549) | FJ590422 |
| ethB-F2, -R2 | ethB (cytochrome P450) | 6 | ethB-F2, 5′-CACGCGCTCGGCGACTGGCAGACGTTCAGT-3′; ethB-R2, 5′-TCCGACGCACATGTGCGGGCCGTACCCGAA-3′ | 698 (881) | FJ607040 |
Complete DNA sequence determined and assembled from the nidA and PAH overlapping PCR fragments.
PCR mixes contained 100 ng of total DNA, 0.4 μM of each oligonucleotide primer, 200 μM of each dNTP, and 2.5 U of Taq DNA polymerase in the provided 1× standard buffer (New England Biolabs, Pickering, Canada) supplemented with 1.25 mM of MgCl2. After an initial incubation at 95°C for 5 min, 35 cycles of the following conditions were used, with each cycle consisting of 94°C for 30 s; 30 s at an annealing temperature of 55°C (alk), 57°C (nidA), 54°C (PAH-RHD GP), or 68°C (ethB); and 72°C for 1 min followed by a final extension at 72°C for 7 min. The PCR products were purified on a MinElute column (MinElute PCR purification kit; Qiagen, Mississauga, Canada), and 20 ng was submitted to sequencing on both strands.
Sequences were determined by Cogenics (Grenoble, France) or the Plate-forme d'analyses biomoléculaires at Université Laval (Québec, Canada). The sequences obtained were compared to sequences deposited in GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and identities were evaluated (1).
Chemicals.
All chemicals used in the study were of the highest purity available from the manufacturers.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were deposited in GenBank under accession no. FJ590420 and FJ590421, respectively. The accession numbers corresponding to the partial sequences of the amplified degradation genes in the two strains are indicated in Table 1.
RESULTS
Isolation of pure strains from the Mix3 consortium.
The Mix3 consortium was an enrichment derived from an initial inoculum composed of three samples (wastewater treatment plant residue, petroleum-contaminated soil from a gas station, and a pristine forest soil). The enrichment was cultivated in MM in the presence of the mixture of 16 compounds (final concentrations of each substrate of 7 to 8 mg liter−1) and transferred twice to fresh medium after degradation was confirmed by GC/FID. The Mix3 consortium was able to degrade more than 90% of 12 of the compounds; isooctane and MTBE were degraded to a lesser extent (40 and 35%, respectively). TBA was not degraded, and more was produced through the degradation of MTBE and ETBE (results not shown).
Isolation of pure strains was carried out by plating on TS-diluted samples of the Mix3 culture. After growth, 63 individual colonies were isolated and purified. DNA of the 63 isolates was extracted by boiling lysis, and the 16S rRNA genes were amplified and sequenced using the 8F/1452R primer pair. Comparison to sequences deposited in GenBank allowed us to determine that the isolates were closely related to the following 10 different strains (percentage of identity in parentheses): Sphingopyxis granuli (99%), Rhodopseudomonas sp. (97%), Mesorhizobium sp. (98%), Acidovorax sp. (98%), Rhodanobacter lindaniclasticus (96%), an uncultured Betaproteobacteria sp. (97%), Pigmentiphaga kullae (97%), Pseudomonas sp. (97%), Rhodococcus aetherivorans (98%), and Rhodococcus wratislaviensis (97%). All strains were preliminarily tested for their degradation capacities (results not shown), and the two strains with the most interesting potential, R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017, were chosen for an extensive study of their capacities. R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were 97% identical to strain CCM4930 (accession no. AJ786666) and 98% identical to isolate AK44 (accession no. AY785745), respectively.
Degradation capacities of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017.
R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were tested for their capacities to degrade a mixture of the following 16 compounds, each provided at an average concentration of 7.5 mg liter−1: benzene, toluene, ethylbenzene, m-xylene, p-xylene, o-xylene, octane, hexadecane, isooctane, cyclohexane, cyclohexanol, naphthalene, MTBE, ETBE, TBA, and 2-EHN. The two strains were tested individually and in coculture (the final OD600 was 0.5 in both cases). After 8 weeks of incubation, the residual substrates were measured, and the results are presented in Table 2. R. wratislaviensis IFP 2016 showed a remarkable potential, as it was able to completely degrade 11 compounds in the mixture (BTEXs, octane and hexadecane, cyclohexane and cyclohexanol, and naphthalene), two compounds were ∼70% degraded (MTBE and 2-EHN), and ETBE and isooctane were degraded to a lesser extent (∼ 50 and 25%, respectively). TBA was the only compound not degraded; it actually accumulated from the degradation of both MTBE and ETBE. R. aetherivorans IFP 2017 showed a more limited capacity, completely degrading only hexadecane, cyclohexanol, and ETBE, whereas 2-EHN, MTBE, and ethylbenzene were degraded to a lesser extent.
TABLE 2.
Degradation of the mixture of 16 compounds by R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 tested alone or in coculture
| Compounds in the mixture of substrates | Degradation capacities (%) ofa: |
||
|---|---|---|---|
| R. wratislaviensis IFP 2016 | R. aetherivorans IFP 2017 | Coculture of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 | |
| Benzene | 100 | 5.7 ± 0.02 | 100 |
| Ethylbenzene | 100 | 24.0 ± 2.2 | 97.5 ± 0.3 |
| Toluene | 100 | 2.0 ± 1.0 | 100 |
| m-Xylene | 100 | 2.4 ± 4.3 | 95.9 ± 0.4 |
| p-Xylene | 100 | 0 | 96.0 ± 0.3 |
| o-Xylene | 100 | 1.7 ± 2,4 | 96.0 ± 0.3 |
| Cyclohexane | 100 | 5.1 ± 1.0 | 100 |
| Octane | 94.3 ± 1.4 | 9.0 ± 1.2 | 91.6 ± 0.3 |
| Hexadecane | 100 | 96.5 ± 4.4 | 98.1 ± 2.7 |
| Isooctane | 26.3 ± 10.7 | 2.6 ± 1.2 | 29.8 ± 1.6 |
| Cyclohexanol | 100 | 100 | 100 |
| MTBE | 78.2 ± 1.3 | 32.4 ± 0.1 | 100 |
| ETBE | 50.8 ± 1.4 | 100 | 100 |
| TBA | No degradationb | No degradationb | No degradationb |
| 2-EHN | 67.9 ± 13.3 | 37.8 ± 4.7 | 72.9 ± 1.7 |
| Naphthalene | 100 | 5.5 ± 7.3 | 97.8 ± 0.1 |
The values are means based on the values obtained for three test flasks. Extent of degradation was calculated as described in Materials and Methods.
Production of TBA from MTBE and ETBE degradation.
R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were also tested for their capacity to degrade and to mineralize the 16 compounds individually at a final concentration of ∼200 mg liter−1 (Table 3). In some cases, no degradation or mineralization was observed when the compounds were tested individually, although the compounds were degraded when provided in the mixture (Table 2). To explore this further, we provided the substrates in HMN as a cosolvent. This was performed for R. wratislaviensis IFP 2016 with m-xylene, p-xylene, o-xylene, and cyclohexane and for both strains with 2-EHN (Table 3). When tested individually and at a higher concentration (200 mg liter−1), the degradation capacity of R. wratislaviensis IFP 2016 was determined more accurately; (i) some compounds were degraded and mineralized easily when added directly in the culture medium (benzene, toluene, ethylbenzene, naphthalene, octane, hexadecane, and cyclohexanol); (ii) other compounds were degraded and mineralized at a higher concentration only when added in a cosolvent like HMN (m-xylene, o-xylene, and cyclohexane); (iii) a compound was transformed at high concentration in the presence of HMN but was not mineralized (p-xylene), suggesting a metabolism leading to a dead-end compound; and (iv) some compounds were not degraded when provided alone and/or at a high concentration (isooctane, MTBE, ETBE, and 2-EHN), suggesting cometabolic degradation when provided in the mixture.
TABLE 3.
Degradation and mineralization of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 toward the 16 compounds tested individuallye
| Compound tested |
R. wratislaviensis IFP 2016 |
R. aetherivorans IFP 2017 |
||
|---|---|---|---|---|
| Degradation (%)c | Mineralization yield (%)d | Degradation (%)c | Mineralization yield (%)d | |
| Benzene | 64.9a | 24.9a | 4.1a | 0a |
| Ethylbenzene | 95.2a | 46.6a | 0a | ND |
| Toluene | 99.7a | 65.8a | 0a | ND |
| m-Xylene | 0a | ND | 0a | ND |
| 63.3b | 100b | |||
| p-Xylene | 0a | ND | 0a | ND |
| 52.1b | 0b | |||
| o-Xylene | 13.5a | 0a | 0a | ND |
| 48.1b | 42.1b | |||
| Cyclohexane | 0a | ND | 0a | ND |
| 100b | 50.4b | |||
| Octane | 97.5a | 44.9a | 21.4a | 61.9a |
| Hexadecane | 32.1a | 100a | 96.3a | 20.1a |
| Isooctane | 16.8a | 0a | 17.3a | 5.3a |
| Cyclohexanol | 100a | 43.6a | 100a | 46.7a |
| MTBE | 3.2a,f | 0a | 22.3a,f | 0a |
| ETBE | 0a,f | ND | 100a,f | 18.2a |
| TBA | 0a | ND | 0a | ND |
| 2-EHN | 0b | ND | 26.2b | 12b |
| Naphthalene | 100a | 36.4a | NT | NT |
The substrate was added directly to the culture medium (5 μl) at a final concentration of ∼200 mg liter−1.
The substrate was added after dissolution into HMN (5 μl/0.5 ml of HMN).
The values are means based on the values obtained for two test flasks; the extent of degradation was calculated as described in Materials and Methods.
The values are means based on the values obtained for two test flasks; the extent of mineralization was calculated as described in Materials and Methods and with respect to the substrate degraded.
The results are the average value of two independent measurements. ND, not determined; NT, not tested.
TBA was produced stoichiometrically, confirming the ether degradation measurement.
The degradation results obtained with R. aetherivorans IFP 2017 in the mixture of compounds showed a more limited degradation profile, with complete degradation restricted to hexadecane, cyclohexanol, and ETBE.
Interactions between substrates.
The interaction of some of the compounds on the biodegradation or mineralization capacities of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were investigated in two-substrate systems.
The effect of the presence of a mixture of BTEXs on ETBE or MTBE degradation and the effect of the presence of one of the ether fuels (MTBE or ETBE) on the degradation capacities of the other ether fuel (ETBE or MTBE) was examined. Resting cells of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 were used for this assay, and the results are presented in Table 4. R. aetherivorans IFP 2017 had the higher degradation capacity toward ether fuels. ETBE was the preferred substrate, with the degradation rate being eightfold higher than that for MTBE. The presence of MTBE had no significant effect on the ETBE degradation rate, whereas no MTBE degradation was observed in the presence of ETBE. The presence of a mixture of BTEXs inhibited ETBE degradation (1.5-fold lower in the presence of the mixture of BTEXs), and MTBE degradation activity was even more affected by the presence of BTEXs (24-fold-lower degradation rate in the presence of BTEXs). In the case of R. wratislaviensis IFP 2016, MTBE degradation was largely inferior to that of R. aetherivorans IFP 2017 (∼150-fold lower) and the activity toward ETBE was not significant. The presence of BTEXs was also shown to inhibit the degradation of MTBE.
TABLE 4.
Degradation rates of ETBE and MTBE by R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017
| Strain | Sp act (μmol ether degraded h−1 g−1 dry wt)c |
||||
|---|---|---|---|---|---|
| MTBE | ETBE | MTBE/ETBE | MTBE/BTEXs | ETBE/BTEXs | |
| R. aetherivorans IFP 2017 | 14.3 ± 1.4a | 116.1 ± 8.0a | 111.7 ± 10.8a,d | 0.59 ± 0.77a | 75.5 ± 12.6a |
| R. wratislaviensis IFP 2016 | 0.09 ± 0.01b | 0.005 ± 0.001b | ND | 0.020 ± 0.005b | ND |
Rate of degradation was calculated between 4 and 6 h of incubation.
Rate of degradation was calculated between 48and 52 h of incubation.
The experiment was carried out as described in Materials and Methods. Initial BTEX mixture concentration, 44 mg liter−1; initial MTBE concentration, 37 mg liter−1; initial ETBE concentration, 37 mg liter−1. The values are means based on the values obtained for three test flasks.
Only the ETBE degradation rate was calculated; no degradation of MTBE was observed during this interval of time.
The effect of the addition of octane on MTBE biodegradation by R. wratislaviensis IFP 2016 was also investigated. The strain was incubated under growing conditions in the presence of MTBE (initial concentration: 10.1 ± 1.7 mg liter−1) and octane (initial concentration: 13.5 ± 1.7 mg liter−1) or in the presence of MTBE alone at the same concentration. After 5 weeks of incubation, the residual concentrations of substrates were measured. The extent of MTBE degradation, when provided alone, was 14.8 ± 5.1%, whereas it was 71.8 ± 1.8% when R. wratislaviensis IFP 2016 was incubated in the presence of both MTBE and octane. No positive effect was observed in the presence of hexadecane (results not shown).
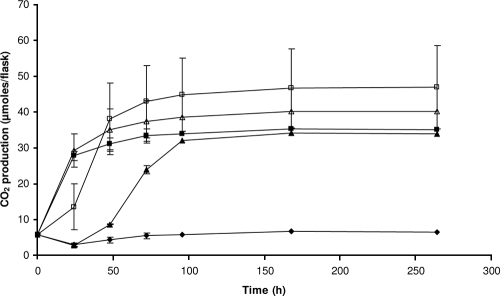
The production of CO2 by R. wratislaviensis IFP 2016 on isooctane was measured in the presence of octane or hexadecane (Fig. 1). In both octane and isooctane flasks, CO2 production was 16% higher than the sum of CO2 produced on octane or isooctane, when tested separately. At the same time, the CO2 production levels in both hexadecane and isooctane flasks were in ranges very similar to that of the sum of CO2 produced on hexadecane or isooctane, when tested separately. The effect of octane addition on isooctane biodegradation by R. wratislaviensis IFP 2016 was investigated in more detail. The strain was incubated under growing conditions in the presence of isooctane (initial concentration of 12.4 ± 1.5 mg liter−1) and octane (initial concentration of 14.7 ± 3.5 mg liter−1) or in the presence of isooctane alone at the same concentration. After 5 weeks of incubation, the residual concentration of substrates was measured. No degradation of isooctane was observed when provided alone, whereas it was 24.0 ± 10.4% degraded when both isooctane and octane were provided.
FIG. 1.
Production of CO2 during growth of R. wratislaviensis IFP 2016 on isooctane, octane, and hexadecane. R. wratislaviensis IFP 2016 was cultivated in 500-ml Schott flasks on 200 ml MM in the presence of isooctane, octane, or hexadecane (4 μl) provided alone or in mixture. CO2 produced was measured with the gaseous phase. ⧫-⧫, isooctane; ▴-▴, octane; ▪-▪, hexadecane; □-□, isooctane and hexadecane; Δ-Δ, isooctane and octane.
Similarly, we investigated the effect of the presence of BTEXs (benzene, toluene, ethylbenzene, and m-xylene), which were readily biodegraded by R. wratislaviensis IFP 2016 on the mineralization of o-xylene and p-xylene (Table 5). CO2 production in the presence of a mixture of two BTEX components was compared to the sum of CO2 produced when the same BTEX components were tested individually at the same concentration. The results showed that p-xylene, or one of its metabolites, had a detrimental effect on the assimilation of o-xylene and m-xylene (2.8- and 1.9-fold-lower CO2 production in the presence of p-xylene), whereas this effect was not observed in the presence of benzene, toluene, or ethylbenzene. A beneficial effect was observed when o-xylene was added to benzene, toluene, or ethylbenzene, with CO2 production 14%, 17%, or 18% higher, respectively, than when these substrates were provided alone.
TABLE 5.
Cross-effects of BTEXs on their own mineralization by R. wratislaviensis IFP 2016
| BTEX(s) (alone or in a mixture)a | CO2 produced (μmoles per flask)b |
|---|---|
| Benzene | 59.8 |
| Toluene | 44.1 |
| Ethylbenzene | 59.6 |
| o-Xylene | 63.4 |
| m-Xylene | 48.6 |
| p-Xylene | 18.8 |
| Benzene and p-xylene | 87.5 (78.8) |
| Toluene and p-xylene | 70.6 (62.9) |
| Ethylbenzene and p-xylene | 91.3 (78.4) |
| m-Xylene and p-xylene | 35.2 (67.4) |
| o-Xylene and p-xylene | 29.2 (82.2) |
| Benzene and o-xylene | 140.4 (123.2) |
| Toluene and o-xylene | 126.3 (107.5) |
| Ethylbenzene and o-xylene | 145.1 (123) |
| m-Xylene and o-xylene | 112.3 (112) |
Four μl of each BTEX component was added to each flask containing 200 ml MM.
Values given in parentheses indicate the sum of CO2 produced on the corresponding compounds tested individually.
PCR detection of degradation genes in R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017.
Total DNA from R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 was analyzed for the presence of different genes known to be involved in the degradation of some of the compounds tested in our study (Table 1).
PCR products of the expected size were obtained from both strains using the alkB consensus primers alk-H1F and -H3R. Sequencing of the R. wrastislaviensis IFP 2016 and R. aetherivorans IFP 2017 amplicons and BLAST analyses revealed 99% identity with alkB genes coding for putative alkane hydroxylases in Rhodococcus sp. 1BN (2) and in Rhodococcus ruber strain IV11 (S. Heiss et al., unpublished results). Polyaromatic hydrocarbon-ring-hydroxylating dioxygenase genes were detected only in R. wratislaviensis IFP 2016 DNA amplified with the nidA-F and -R and PAH-RHD GP F and R primer sets. When sequenced, these two PCR fragments gave overlapping DNA sequences, which were assembled to produce a final sequence that was 99% identical with the nidA gene coding for the large subunit of a dioxygenase iron-sulfur protein present in Rhodococcus opacus TKN14 (32).
A gene highly similar (99% identity) to ethB from R. ruber IFP 2001 was amplified from the DNA of R. aetherivorans IFP 2017. The ethB gene encodes a cytochrome P450 responsible for ETBE and MTBE degradation in R. ruber IFP 2001 (6). No such gene was detected with R. wratislaviensis IFP 2016 using the same primers (result not shown).
DISCUSSION
Two strains belonging to the genus Rhodococcus were isolated during this study: R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017. Recently, R. wratislaviensis DLC-cam was shown to hydroxylate 2-methylisoborneol, a bacterial product structurally similar to camphor (15), and R. aetherivorans Bc663 was described as a novel species related to the subgroup containing R. rhodochrous, which was identified for its ability to degrade MTBE (20).
Degradation capacity of R. wratislaviensis IFP 2016.
R. wratislaviensis IFP 2016 had a very broad substrate degradation capacity. Various degradation mechanisms were identified, including the use of the substrate as a carbon and energy source (benzene, toluene, ethylbenzene, m-xylene and o-xylene, octane, hexadecane, cyclohexane, cyclohexanol, and naphthalene) and cometabolic processes, as shown in Table 3, by the partial biodegradation of the compounds without mineralization when tested alone at a high concentration (p-xylene, isooctane, and MTBE) or by partial biodegradation when tested at a low concentration in the mixture of compounds (isooctane, MTBE, ETBE, and 2-EHN). In some cases, the use of HMN to dissolve the substrate before addition to the culture medium was shown to be beneficial (m-, p-, and o-xylenes and cyclohexane), suggesting that HMN improved substrate transfer to the microorganism and/or reduced substrate toxicity/inhibition (33).
(i) Aromatic compounds.
The ability of Rhodococcus spp. to degrade a large variety of organic compounds, including monoaromatics, has been documented (26).
Cometabolic biodegradation of xylenes by a Rhodococcus rhodochrous strain isolated from a consortium able to utilize benzene, toluene, and ethylbenzene has been described (12), and although the initial degradation rates were studied, the ultimate fate of the compounds was not determined. By evaluating the mineralization capacity in parallel with the degradation capacity, it was shown that R. wratislaviensis IFP 2016 transformed p-xylene to a potential dead-end product, as there was no mineralization measured when p-xylene was provided as the only carbon and energy source. The transformation of p-xylene to the dead-end metabolite 2,5-dimethylhydroquinone was reported with Rhodococcus sp. DK17 (24). Although we did not identify the transformation product of p-xylene in R. wratislaviensis IFP 2016, we did show that p-xylene itself or its transformation product had a strong inhibitory effect on the degradation of m- and o-xylene, but no effect on benzene, toluene, or ethylbenzene degradation.
In contrast to its interaction with p-xylene, R. wratislaviensis IFP 2016 degraded and mineralized both m- and o-xylene. The ability to degrade o-xylene is important since it is considered a difficult-to-degrade hydrocarbon and only a few Rhodococcus strains with this ability have been isolated (14, 23, 24, 29, 45). A nidA gene in R. wratislaviensis IFP 2016 was identified with 99% identity to a gene from Rhodococcus opacus TKN14 (29) that encodes the large subunit of a dioxygenase iron-sulfur protein. This gene is part of a group of genes (nidABEF) induced in R. opacus TKN14 by o-xylene. It is possible that this gene was responsible for the conversion of xylenes and possibly naphthalene in R. wratislaviensis IFP 2016, as was shown with R. opacus TKN14.
(ii) Alkanes.
The ability to degrade n-alkanes of various chain lengths and the presence of alkB genes, often multiple copies, have frequently been reported for Rhodococcus strains (25). R. wratislaviensis IFP 2016 was able to use short- and long-chain alkanes (C8 and C16) as the sole carbon and energy source, and the presence of an alkB gene with 99% identity to the alkB gene from Rhodococcus sp. 1BN (2) was identified.
(iii) Other recalcitrant compounds.
R. wratislaviensis IFP 2016 was able to degrade other known recalcitrant compounds, such as cyclohexane, isooctane, MTBE, ETBE, and 2-EHN, when provided in a mixture of hydrocarbons. Among these compounds, only cyclohexane was used as a source of carbon and energy, as shown by both degradation and mineralization analyses. Of interest, the presence of HMN as a cosolvent was beneficial for these degradations. The cytotoxicity of cyclohexane was explained by its inhibitory effect (26) when used at high substrate concentration (9.2 mM), higher than that used in our study (about 2.5 mM). Cyclohexane is considered a recalcitrant compound, and when degraded, it frequently requires a syntrophic association of at least two different bacteria, one performing the oxidation of cyclohexane to cyclohexanone and the other using cyclohexanone as a carbon and energy source (26, 42). In our study, cyclohexane and cyclohexanol degradation were both accomplished by R. wratislaviensis IFP 2016 alone.
Isooctane, another well known recalcitrant compound, was degraded to about 26% by R. wratislaviensis IFP 2016, when provided in the mixture of compounds, indicating that there are enzymes in this bacterium that are able to metabolize this compound. The presence of octane, but not of hexadecane, in the mixture was shown to improve isooctane degradation and mineralization. This suggested the possible role of an alkane hydroxylase that would be able, once induced, to attack branched alkanes. It also suggested that in R. wratislaviensis IFP 2016 there may be at least two different alkane hydroxylases: one attacking long-chain alkanes (like hexadecane) and having no capacity toward isooctane and the other attacking short-chain alkanes (like octane) and also isooctane. The biodegradability of isooctane is poorly documented; Mycobacterium austroafricanum IFP 2173 is the only strain for which the capacity to grow on isooctane as a sole carbon and energy source has been characterized, and it was shown by reverse transcription-PCR that this probably involved the expression of an alkB gene (43). It is also interesting that isooctane utilization was reduced considerably by the presence of BTEXs (9). This lack of information concerning isooctane biodegradation is an issue when considering the possibility of using it as a fuel additive (45).
The biodegradation of 2-EHN, a recently used additive of diesel fuel, by M. austroafricanum IFP 2173 was recently reported, but metabolites accumulated in the medium during the degradation (36). R. wratislaviensis IFP 2016 partially degraded 2-EHN when provided in a mixture of compounds, but no degradation or mineralization was observed when 2-EHN was the only carbon and energy source.
MTBE was degraded at a low rate by R. wratislaviensis IFP 2016, as has been reported for other Rhodococcus strains able to degrade ether fuels (16, 21; Malandain et al., submitted). TBA was produced stoichiometrically during MTBE degradation and was not further degraded. The presence of the eth genes in the Rhodococcus strains with fuel ether degradation capacities that are cited above was not detected with R. wratislaviensis IFP 2016. The simultaneous presences of BTEXs and MTBE had a negative impact on the MTBE degradation rate (nearly fivefold lower) as previously reported (12, 27, 40, 47), whereas no negative effect was shown on strain UC1, a strain closely related to the well-known Methylibium petroleiphilum PM1 (39). The negative effect of the presence of BTEXs on MTBE biodegradation has consequences for the environmental fate of MTBE in contaminated aquifers. The presence of octane had a positive effect on MTBE degradation (72% versus 15% degradation), suggesting the possibility of related metabolic processes, and the positive effect of short-chain alkanes on MTBE degradation has also been reported for Pseudomonas (32). The possible role of an alkane hydroxylase in the degradation of MTBE by R. wratislaviensis IFP 2016 needs to be determined. ETBE was partially degraded when provided in a mixture of compounds, and as in the case of MTBE, its degradation led to the production of TBA. The ETBE degradation rate was 18-fold lower than the MTBE degradation rate. TBA was the only compound tested that was not degraded to any extent by R. wratislaviensis IFP 2016.
Degradation capacities of R. aetherivorans IFP 2017.
By comparison, R. aetherivorans IFP 2017 showed a more- restricted degradation profile, able to totally degrade hexadecane and cyclohexanol. ETBE was completely transformed to TBA, and the mineralization yield corresponded to the utilization of the C2 compound produced after cleavage of the ether bond, whereas TBA was not assimilated. Several of the other tested compounds were degraded to a much lower extent (2-EHN, 26% degraded; MTBE, 22% degraded; isooctane, 17% degraded) and were not, or very poorly, mineralized, suggesting cometabolic biodegradation. No BTEXs were degraded by this strain. The ETBE degradation rate of R. aetherivorans IFP 2017 measured after preculture on TS was in the same range as that determined with R. ruber IFP 2001 and R. zopfii IFP 2005 under similar conditions (Malandain et al., submitted), i.e., 116, 210, and 268 μmol of ETBE degraded h−1 g−1 dry weight. The degradation rate measured for MTBE was 14.3 μmol of MTBE degraded h−1 g−1 dry weight. With R. ruber IFP 2001 and R. zopfii IFP 2005, the biodegradation of ETBE and MTBE was associated with a cytochrome P450 system encoded by the eth genes (4, 6; Malandain et al., submitted). A cytochrome P450 is most likely responsible for ETBE and MTBE degradation in R. aetherivorans IFP 2017, since an ethB gene was detected that was 99% identical to ethB from R. ruber IFP 2001. This is the first report for R. aetherivorans, a new species that was characterized only recently (20), of the presence of an ethB gene known to encode a cytochrome P450 and previously shown with other Rhodococcus strains to be responsible for ether fuel degradation. As previously shown with R. ruber IFP 2001 and R. zopfii IFP 2005, ETBE and MTBE were degraded stoichiometrically to TBA, which accumulated in the medium and was not used as a carbon source by the strain. Growth on ETBE, confirmed by the mineralization yield, was at the expense of the C2 moiety, liberated by the cleavage of the ether bond. Similarly, TBA accumulated during ETBE or MTBE degradation by R. aetherivorans IFP 2017.
The presence of MTBE did not have an impact on the ETBE degradation rate, but no MTBE was degraded in the presence of ETBE. This was likely due to a difference of affinities of the cytochrome for these compounds, the preferred substrate being ETBE (Malandain et al., submitted). As mentioned above for R. wratislaviensis IFP 2016, the presence of a mixture of BTEXs had a strong negative impact on MTBE degradation and decreased the ETBE degradation rate by 1.5-fold.
These data on the effect of the simultaneous presences of MTBE and ETBE on their respective degradation rates are of importance because in countries that shifted from MTBE to ETBE as a gasoline additive, both compounds can occur in contaminated sites (F. Fayolle-Guichard, unpublished results). It is therefore important to obtain information on the behavior of microorganisms under these conditions.
Bioremediation of polluted sites.
A coculture of R. wratislaviensis IFP 2016 and R. aetherivorans IFP 2017 showed results similar to or even better than those obtained with Mix3, as MTBE was completely degraded by the coculture and only partially by Mix3. This result emphasizes the advantage of working with pure strains rather than undefined enrichment cultures in the bioremediation process. The renewed interest in bioaugmentation processes, which were previously often rejected due to inefficiency, have been reexamined over the last decade as conditions for their use have been better determined (22).
The degradation of some toxic organic compounds at high concentrations in the environment poses challenges to conventional biological treatment. An ex situ treatment applying a system in a two-phase partitioning bioreactor with these pure strains may be an effective alternative (11, 33).
Another advantage of using mixed cultures of defined microorganisms is the ability to create specific probes to specifically monitor the fate of the microorganisms released into the environment by quantitative PCR (34) or fluorescent in situ hybridization (19).
In conclusion, the capacity of two Rhodococcus strains with the ability to degrade a variety of hydrocarbon and fuel additive compounds was determined. To the best of our knowledge, R. wratislaviensis IFP 2016 is the first pure strain with such broad degradation capacity that has been characterized in detail. This study also emphasized (i) the positive role of cometabolism in the biodegradation of a complex mixture of chemicals with different structures in pure strains and (ii) the problems due to inhibitory effects of some compounds on the degradation of other compounds when present in complex mixtures.
Acknowledgments
Marc Auffret was partly supported by a CIFRE (Convention Industrielle de Formation par la Recherche) fellowship provided by ANRT (Association Nationale de la Recherche Technique) and by IFP.
We thank Isabelle Durand and Françoise Le Roux for hydrocarbon analyses. We thank Nicolas Lopes Ferreira for help in PCR amplification.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, J. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoni, V., S. Bernasconi, M. Colombo, J. B. van Beilen, and L. Cavalca. 2000. Detection of genes for alkane and naphthalene catabolism in Rhodococcus sp. strain 1BN. Environ. Microbiol. 2:572-577. [DOI] [PubMed] [Google Scholar]
- 3.Babé, A., D. Labbé, F. Monot, C. W. Greer, and F. Fayolle-Guichard. 2007. Biodegradability of oxygenates by microflora from MTBE-contaminated sites: new molecular tools. Handb. Environ. Chem. 5(Part R):75-98. [Google Scholar]
- 4.Beguin, P., S. Chauvaux, I. Miras, A. Francois, F. Fayolle, and F. Monot. 2003. Genes involved in the degradation of ether fuels by bacteria of the Mycobacterium/Rhodococcus group. Oil Gas Sci. Technol. 58:489-495. [Google Scholar]
- 5.Cébron, A., M. P. Norini, T. Beguiristain, and C. Leyval. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148-159. [DOI] [PubMed] [Google Scholar]
- 6.Chauvaux, S., F. Chevalier, C. Le Dantec, F. Fayolle, I. Miras, F. Kunst, and P. Beguin. 2001. Cloning of a genetically unstable cytochrome P-450 gene cluster involved in degradation of the pollutant ethyl tert-butyl ether by Rhodococcus ruber. J. Bacteriol. 183:6551-6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, K. F., C. M. Kao, J. Y. Wang, T. Y. Chen, and C. C. Chien. 2005. Natural attenuation of MTBE at two petroleum-hydrocarbon spill sites. J. Hazard. Mater. 125:10-16. [DOI] [PubMed] [Google Scholar]
- 8.Chénier, M. R., D. Beaumier, R. Roy, B. T. Driscoll, J. R. Lawrence, and C. W. Greer. 2003. Impact of seasonal variations and nutrient inputs on nitrogen cycling and degradation of hexadecane by replicated river biofilms. Appl. Environ. Microbiol. 69:5170-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, J., M. M. Zein, M. T. Suidan, and A. D. Venosa. 2007. Biodegradability of alkylates as a sole carbon source in the presence of ethanol and BTEX. Chemosphere 68:266-273. [DOI] [PubMed] [Google Scholar]
- 10.Dakhel, N., G. Pasteris, D. Werner, and P. Höhener. 2003. Small-volume releases of gasoline in the vadose zone: impact of the additives MTBE and ethanol on groundwater quality. Environ. Sci. Technol. 37:2127-2133. [DOI] [PubMed] [Google Scholar]
- 11.Daugulis, A. J. 2001. Two-phase partitioning bioreactors: a new technology platform for destroying xenobiotics. Trends Biotechnol. 19:457-462. [DOI] [PubMed] [Google Scholar]
- 12.Deeb, R. A., and L. Alvarez-Cohen. 1999. Temperature effects and substrate interactions during the aerobic biotransformation of BTEX mixtures by toluene-enriched consortia and Rhodococcus rhodochrous. Biotechnol. Bioeng. 62:526-536. [PubMed] [Google Scholar]
- 13.Deeb, R. A., H. Hu, J. R. Hanson, K. M. Scow, and L. Alvarez-Cohen. 2001. Substrate interactions in BTEX and MTBE mixtures by an MTBE-degrading isolate. Environ. Sci. Technol. 35:312-317. [DOI] [PubMed] [Google Scholar]
- 14.Di Gennaro, P., E. Rescalli, E. Galli, G. Sello, and G. Bestetti. 2001. Characterization of Rhodococcus opacus R7, a strain able to degrade naphthalene and o-xylene isolated from a polycyclic aromatic hydrocarbon-contaminated soil. Res. Microbiol. 152:641-651. [DOI] [PubMed] [Google Scholar]
- 15.Eaton, R. W., and P. Sandusky. 2009. Biotransformations of 2-methylisoborneol by camphor-degrading bacteria. Appl. Environ. Microbiol. 75:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fayolle, F., G. Hernandez, F. Le Roux, and J.-P. Vandecasteele. 1998. Isolation of two aerobic bacterial strains that degrade efficiently ethyl t-butyl ether (ETBE). Biotechnol. Lett. 20:283-286. [Google Scholar]
- 17.Finnerty, W. R. 1992. The biology and genetics of the genus Rhodococcus. Annu. Rev. Microbiol. 46:193-218. [DOI] [PubMed] [Google Scholar]
- 18.François, A., H. Mathis, D. Godefroy, P. Piveteau, F. Fayolle, and F. Monot. 2002. Biodegradation of methyl tert-butyl ether and other fuel oxygenates by a new strain, Mycobacterium austroafricanum IFP 2012. Appl. Environ. Microbiol. 68:2754-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glökner, F. O., R. Amann, A. Alfleider, L. Pernthaler, R. Psenner, K. Trebesius, and K. G. Field. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 20.Goodfellow, M., A. L. Jones, L. A. Maldonado, and J. Salanitro. 2004. Rhodococcus aetherivorans sp. nov., a new species that contains methyl t-butyl ether-degrading actinomycetes. Syst. Appl. Microbiol. 27:61-65. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez-Perez, G., F. Fayolle, and J. P. Vandecasteele. 2001. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl. Microbiol. Biotechnol. 55:117-121. [DOI] [PubMed] [Google Scholar]
- 22.Iwamoto, T., and M. Nasu. 2001. Current bioremediation practice and perspective. J. Biosci. Bioeng. 92:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Jang, J. Y., D. Kim, H. W. Bae, K. Y. Choi, J.-C. Chae, G. J. Zylstra, Y. M. Kim, and E. Kim. 2005. Isolation and characterization of a Rhodococcus species strain able to grow on ortho- and para-xylene. J. Microbiol. 43:325-330. [PubMed] [Google Scholar]
- 24.Kim, D., J.-C. Chae, G. J. Zylstra, Y.-S. Kim, S.-K. Kim, M. H. Nam, Y. M. Kim, and E. Kim. 2004. Identification of a novel dioxygenase involved in metabolism of o-xylene, toluene, and ethylbenzene by Rhodococcus sp. strain DK17. Appl. Environ. Microbiol. 70:7086-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin, M. J., L. A. Kulakov, and C. C. Allen. 2005. Biodegradation and Rhodococcus—masters of catabolic versatility. Curr. Opin. Biotechnol. 16:282-290. [DOI] [PubMed] [Google Scholar]
- 26.Lee, E.-H., and K.-S. Cho. 2008. Characterization of cyclohexane and hexane degradation by Rhodococcus sp. EC1. Chemosphere 71:1738-1744. [DOI] [PubMed] [Google Scholar]
- 27.Lee, E.-H., and K.-S. Cho. 2009. Effect of substrate interaction on the degradation of methyl tert-butyl ether, benzene, toluene, ethylbenzene, and xylene by Rhodococcus sp. J. Hazard. Mater. 167:669-674. [DOI] [PubMed] [Google Scholar]
- 28.Lopes Ferreira, N., C. Malandain, and F. Fayolle-Guichard. 2006. Enzymes and genes involved in the aerobic biodegradation of methyl tert-butyl ether (MTBE). Appl. Microbiol. Biotechnol. 72:252-262. [DOI] [PubMed] [Google Scholar]
- 29.Maruyama, T., M. Ishikura, H. Taki, K. Shindo, H. Kasai, M. Haga, Y. Inomata, and N. Misawa. 2005. Isolation and characterization of o-xylene oxygenase genes from Rhodococcus opacus TKN14. Appl. Environ. Microbiol. 71:7705-7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mihelcic, J. R. 2007. Modeling the potential effect of additives on enhancing the solubility of aromatic solutes contained in gasoline. Ground Water Monit. Remediation 10:132-137. [Google Scholar]
- 31.Mo, K., C. O. Lor, A. E. Wanken, and J. Javanmardian. 1997. Biodegradation of methyl tert-butyl ether by pure bacterial cultures. Appl. Microbiol. Biotechnol. 47:68-72. [DOI] [PubMed] [Google Scholar]
- 32.Morales, M., V. Nava, E. Velasquez, E. Razo-Flores, and S. Revah. 2009. Mineralization of methyl tert-butyl ether and other gasoline oxygenates by Pseudomonads using short n-alkanes as growth source. Biodegradation 20:271-280. [DOI] [PubMed] [Google Scholar]
- 33.Munoz, R., S. Villaverde, B. Guieysse, and S. Revah. 2007. Two-phase partitioning bioreactors for treatment of volatile organic compounds. Biotechnol. Adv. 25:410-422. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, K., H. Ishida, T. Iizumi, K. Shibuya, and K. Okamura. 2000. Quantitative PCR detection of a phenol-utilizing bacterium, Ralstonia eutropha KT-1, injected to a trichlorethylene-contaminated site. Environ. Eng. Res. 37:267-278. [Google Scholar]
- 35.NESCAUM. 2001. Health, environmental, and economic impacts of adding ethanol to gasoline in the Northeast states. Vol. 2: air quality, health and economic impacts. http://www.nescaum.org/documents/health-environmental-and-economic-impacts-of-adding-ethanol-to-gasoline-in-the-northeast-states/.
- 36.Nicolau, E., L. Kerhoas, M. Lettere, Y. Jouanneau, and R. Marchal. 2008. Biodegradation of 2-ethylhexyl nitrate by Mycobacterium austroafricanum IFP 2173. Appl. Environ. Microbiol. 74:6187-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piveteau, P., F. Fayolle, J.-P. Vandecasteele, and F. Monot. 2001. Biodegradation of tert-butyl alcohol and related xenobiotics by a methylotrophic bacterial isolate. Appl. Microbiol. Biotechnol. 55:369-373. [DOI] [PubMed] [Google Scholar]
- 38.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 6:217-218. [DOI] [PubMed] [Google Scholar]
- 39.Pruden, A., and M. Suidan. 2004. Effect of benzene, toluene, ethylbenzene, and p-xylene (BTEX) mixture on biodegradation of methyl tert-butyl ether (MTBE) and tert-butyl alcohol (TBA) by pure culture UC1. Biodegradation 15:213-227. [DOI] [PubMed] [Google Scholar]
- 40.Raynal, M., and A. Pruden. 2008. Aerobic MTBE biodegradation in the presence of BTEX by two consortia under batch and semi-batch conditions. Biodegradation 19:269-282. [DOI] [PubMed] [Google Scholar]
- 41.Sayler, G. S., S. W. Hooper, A. C. Layton, and J. M. Henry King. 1990. Catabolic plasmids of environmental and ecological significance. Microbiol. Ecol. 19:1-20. [DOI] [PubMed] [Google Scholar]
- 42.Solano-Serena, F., R. Marchal, S. Casaregola, C. Vasnier, J. M. Lebeault, and J.-P. Vandecasteele. 2000. A Mycobacterium strain with extended capacities for degradation of gasoline hydrocarbons. Appl. Environ. Microbiol. 66:2392-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solano-Serena, F., R. Marchal, S. Heiss, and J.-P. Vandecasteele. 2004. Degradation of isooctane by Mycobacterium austroafricanum IFP 2173: growth and catabolic pathway. J. Appl. Microbiol. 97:629-639. [DOI] [PubMed] [Google Scholar]
- 44.Solano-Serena, F., R. Marchal, and J.-P. Vandecasteele. 2008. Biodegradation of aliphatic and alicyclic hydrocarbons, p. 170-240. In J.-P. Vandecasteele (ed.), Petroleum microbiology: concepts, environmental implications, industrial applications, vol. 1. Editions Technip, Paris, France. [Google Scholar]
- 45.Taki, H., K. Syutsubo, R. G. Mattison, and S. Haramaya. 2007. Identification and characterization of o-xylene-degrading Rhodococcus spp. which were dominant species in the remediation of o-xylene-contaminated soils. Biodegradation 18:17-26. [DOI] [PubMed] [Google Scholar]
- 46.Vandecasteele, J.-P., F. Monot. 2008. Biodegradation of monoaromatic and chloroaromatic hydrocarbons, p. 240-339. In J.-P. Vandecasteele (ed.), Petroleum microbiology: concepts, environmental implications, industrial applications, vol. 1. Editions Technip, Paris, France. [Google Scholar]
- 47.Wang, X., and M. A. Deshusses. 2007. Biotreatment of groundwater contaminated with MTBE: interaction of common environmental co-contaminants. Biodegradation 18:37-50. [DOI] [PubMed] [Google Scholar]