Abstract
Biofuels derived from lignocellulosic biomass hold promises for a sustainable fuel economy, but several problems hamper their economical feasibility. One important problem is the presence of toxic compounds in processed lignocellulosic hydrolysates, with furfural as a key toxin. While Saccharomyces cerevisiae has some intrinsic ability to reduce furfural to the less-toxic furfuryl alcohol, higher resistance is necessary for process conditions. By comparing an evolved, furfural-resistant strain and its parent in microaerobic, glucose-limited chemostats at increasing furfural challenge, we elucidate key mechanism and the molecular basis of both natural and high-level furfural resistance. At lower concentrations of furfural, NADH-dependent oxireductases are the main defense mechanism. At furfural concentrations above 15 mM, however, 13C-flux and global array-based transcript analysis demonstrated that the NADPH-generating flux through the pentose phosphate pathway increases and that NADPH-dependent oxireductases become the major resistance mechanism. The transcript analysis further revealed that iron transmembrane transport is upregulated in response to furfural. While these responses occur in both strains, high-level resistance in the evolved strain was based on strong induction of ADH7, the uncharacterized open reading frame (ORF) YKL071W, and four further, likely NADPH-dependent, oxireductases. By overexpressing the ADH7 gene and the ORF YKL071W, we inversely engineered significantly increased furfural resistance in the parent strain, thereby demonstrating that these two enzymes are key elements of the resistance phenotype.
A whole range of biofuels are currently being investigated as potential candidates to contribute to future energy and environmental goals (8, 11, 16, 49). The basis for such processes is mainly plant-based carbohydrates. Despite the advantage of their high abundance and low cost, biofuel production remains an economic challenge for several reasons that include expensive and inefficient hydrolysis prior to fermentation, suboptimal yields and production rates, and process scale-up problems (31).
One problem that influences yields and production rates is the primary occurrence of sugars in polymeric forms that are tightly packed and closely associated with other cellular components and hence inaccessible for good-fermenting microorganisms. Therefore, plant biomass is pretreated under harsh physicochemical conditions to break down the pentose-containing hemicelluloses and to partially fragment the almost crystalline cellulose fibers. An inevitable consequence of this pretreatment is undesired side reactions such as lignin breakdown and water elimination from monomeric sugars, which generate toxic compounds that inhibit Saccharomyces cerevisiae and other microorganisms (10, 23, 24, 51). A key compound is the aldehyde furfural, an elimination product from 5-carbon sugars that occurs under the typical high-temperature and low-pH conditions during biomass pretreatment (46). It determines the toxicity of lignocellulosic hydrolysates to a large extent (18).
Owing to its relevance to biofuels, several studies assessed furfural toxicity and resistance mechanisms. Since it is a quite reactive aldehyde, the primary resistance mechanism is its conversion to less-toxic compounds. The key conversion is the reduction to furfuryl alcohol (18, 29, 32, 34), but the oxidation to furoic acid has also been observed, especially under respirative conditions (45). It is known that overexpression of several endogenous genes can increase yeasts' furfural reduction capacity, at least in vitro (28, 36). The reduction is generally believed to depend on NADH (34, 35), although there are strong indications that NADPH-dependent processes are also involved (28, 36).
In more global discovery approaches, genes that are vital for furfural resistance have been identified. Screening of a gene disruption library revealed that several dehydrogenases and some pentose phosphate pathway genes are important resistance elements (17). Recently, untargeted comparative proteomics demonstrated the importance of dehydrogenases and the tricarboxylic acid cycle when growth was stopped instantaneously by the addition of furfural (27). Moreover, yeasts have been adapted in long-term evolution experiments to higher concentrations of furfural, yielding strains with increased reduction activities (29) or increased viability (18). Despite these studies, we lack a clear understanding of resistance, with open key questions that include the in vivo source of reduction equivalents, the molecular identification of enzymes that catalyze the reduction, and whether other cellular processes are also relevant. Notably, the in vivo relevance of the above-identified genes to actually improve furfural tolerance has not yet been demonstrated, with the exception of the overexpression of ZWF1 in the study by Gorsich and coworkers (17).
Here, we elucidate the functional basis of furfural resistance by comparing a semi-industrial, furfural-adapted Saccharomyces cerevisiae strain with increased viability under high concentrations of furfural to its parent by means of a metabolic flux (41) and global transcript analysis. Pivotal for this study is the use of chemostat cultures that allows us to study the furfural impact on steady-state growth, specifically avoiding the otherwise inevitable indirect effects of reduced or stopped growth (1, 9, 13, 19, 27). We reveal physiological and molecular features crucial for resistance and identify several genes involved in resistance. By overexpression of two identified key genes, we demonstrate the improved resistance of S. cerevisiae against furfural.
MATERIALS AND METHODS
Strains and cultivation.
Throughout, we used the two S. cerevisiae strains TMB3400 (48) and TMB3400-FT30-3 (18). For chemostat and batch experiments, minimal media containing main and tracer salts, as described in reference 47, with 5 g liter−1 of glucose buffered with 50 mM potassium hydrogen phthalate at pH 5, were used. Furfural was added at the desired concentrations, and the media were sterilized by filtration.
We performed chemostat fermentations in 300-ml bioreactors filled with 200 ml media at 30°C and stirred at 250 rpm, and the dilution rate was set to 0.1 h−1. To ensure microaerobic conditions, the reactors were flushed with N2 (O2 < 2 ppm) at a rate of 100 ml min−1. This leads to a maximal oxygen feed of 1.3 μmol h−1 that suffices to ensure the S. cerevisiae minimal requirement for oxygen to synthesize unsaturated fatty acids and sterols of 70 pmol g of cells (dry weight)−1 (gcdw−1) h−1 (adapted from reference 38). After the batch phase, a furfural-free feed was started, and the cells were allowed to attain steady state for at least 4 volume changes. Then, the feed was exchanged by with furfural-containing feed.
Batch cultures were done in an aerobic shake flask with the identical medium that was used for the chemostat experiments, with the exception of 11 mM furfural. Precultures were grown to mid-exponential phase in cultures supplemented with G418 in the case of the plasmid-based overexpression strains and then used to inoculate the culture.
Metabolic flux analysis.
Generally, metabolic fluxes were determined as described previously (12, 52, 53). The furfural-containing feed was replaced after at least 5 volume changes by an identical medium containing [1-13C]glucose instead of naturally labeled glucose. Samples for metabolic flux analysis were withdrawn after at least 0.7 volume changes with the labeled feed. The exact time span between feed change and sample withdrawal was recorded for later calculation of the percentage of unlabeled biomass at the sampling time point. The cells were washed twice with ultrapure water and stored at −20°C for later analysis. Hydrolysis of cellular proteins was performed in 6 M HCl overnight at 105°C, and the samples were dried under airflow. The amino acids were then derivatized with N-tert-butyl-N-dimethylsilylacetamide in dimethylformamide at 95°C for 1 h. These samples were injected into a gas chromatograph coupled to a mass spectrometer to measure the fractional labeling of amino acids. Data analysis and determination of the fraction of serine coming from glucose metabolized through glycolysis were performed with the “ratio” module from the FiatFlux software package (53). The fraction of unlabeled biomass was estimated with e−Dt, where D is the dilution rate and t the time span between the switch to the labeled feed and sampling. The relative pathway usage between the pentose phosphate pathway and glycolysis was estimated with the “netto” module of the FiatFlux software. A detailed experimental guideline is given elsewhere (52). For flux estimations used to calculate the regeneration and consumption of redox equivalents and carbon balances, the following simplified stoichiometric model was used for the flux analysis (adapted from reference 4): glucose → glucose-6-phosphate, glucose-6-phosphate + 2 NADP+→ pentose-5-phosphate + 2 NADPH, 2 pentose-5-phosphate → seduheptulose-7-phosphate + glyceraldehyde-3-phosphate, seduheptulose-7-phosphate + glyceraldehyde-3-phosphate → erythrose-4-phosphate + fructose-6-phosphate, pentose-5-phosphate + erythrose-4-phosphate → fructose-6-phosphate + glyceraldehyde-3-phosphate, glyceraldehyde-3-phosphate + NAD+ → serine + NADH, serine + NADH → glycine + NAD+, glucose-6-phosphate → fructose-6-phosphate, fructose-6-phosphate → glyceraldehyde-3-phosphate, glyceraldehyde-3-phosphate + NAD+ → phospho-enol-pyruvate + NADH, glyceraldehyde-3-phosphate + NADH → glycerol + NAD+, phospho-enol-pyruvate → pyruvate, pyruvate + NADP+→ acetate + NADPH, pyruvate + NADH → ethanol + NAD+, pyruvate → oxoglutarate, oxaloacetate, acetyl-coenzyme A, furfural + 1/2 NADP+ + 1/2 NAD+ → 2-furoic acid + 1/2 NADPH + 1/2 NADH.
The cofactor preference for the last reaction is not known. For this reason and because 2-furoic acid production was less than 0.5% of total cofactor turnover, we evenly partitioned it between the two cofactors. Biomass was assumed to be composed as follows (values in mmol gcdw−1) (adapted from reference 25): glucose-6-phosphate, 1.911; pentose-5-phosphate, 0.351; gylceraldehyde-3-phosphate, 0.363; erythrose-4-phosphate, 0.289; phospho-enol-pyruvate, 0.579; oxaloacetate, 1.332; serine, 0.203; glycine, 0.318; acetyl-coenzyme A, 1.552; oxaloglutarate, 0.997; pyruvate, 2.386; NADPH, 11.249; and NADH, 0.118.
Transcript analysis.
Samples for analysis were withdrawn after at least 3 volume changes after the switch from furfural-free feed to furfural-containing feed. Three biological replicates were taken from separate chemostat runs for each condition and strain. After centrifugation for 3 min at room temperature, the samples were washed with RNase-free buffer containing 10 mM Tris-HCl and 10 mM EDTA at pH 5. Cell pellets were stored at −80°C for later processing. The cells were disrupted using 500-μm glass beads on a Vortex-Genie II TurboMix tissue lyser. Subsequently, RNA was extracted with a Qiagen RNA extraction kit according to the manufacturer's protocol and stored at −80°C. The obtained RNA was hybridized to Affymetrix GeneChip yeast genome 2.0 arrays that were scanned with a GeneChip scanner 3000.
Transcript data analysis was conducted with the R Bioconductor package (release 2.2) (http://www.bioconductor.org). Analysis was performed as described previously (14). In brief, the raw intensity data were normalized on quantiles, and background and perfect match/mismatch were corrected with the MAS algorithm and summarized according to the Li-Wong method to yield probe set intensities. Quality control of obtained data was performed as described previously (see chapter 3 of reference 14). To find differentially expressed genes, a moderated t statistic was used (42). For further analyses such as gene ontology searches, only open reading frames (ORFs) with a P value of less than 0.01 were considered. Clustering of probe set intensities was performed with the R cluster package. Gene ontology search was performed with the GO term finder of the Saccharomyces Genome Database (http://www.yeastgenome.org) (5), the P value was set to <0.01, and the background set consisted of all features that have GO annotations in the database. At the time of query, this set counted 7,163 features.
Transcript data can be downloaded from the GEO database under the accession number GSE18314 (http://www.ncbi.nlm.nih.gov/projects/geo/).
Sporulation and analysis of segregants.
To force the diploid strains to undergo meiosis, the yeasts were spread on agar slants containing 6 g liter−1 potassium acetate and 0.5 g liter−1 glucose at pH 7 and incubated for 3 to 5 days at 25°C. Tetrads were digested with 500 U ml−1 zymolyase and dissected with a dissection scope on agar slants containing 1% yeast extract, 2% peptone, and 2% glucose. The segregants were allowed to grow at room temperature for 3 to 4 days. From a mid-exponential-phase preculture in minimal medium, an aerobic shake flask with minimal medium containing 17 mM furfural was inoculated at 0.025 gcdw liter−1. Lag phases are determined by the x-axis intercept of the intersection from the linearized exponential-growth curve, with the baseline showing the absence of growth.
Genetic constructs and sequencing.
Standard molecular biology techniques were used for all cloning steps (39). Yeast plasmid transformations were carried out according to Gietz and Schiestl (15). The Phusion high-fidelity DNA polymerase (Finnzymes) was used for all PCRs. A plasmid carrying a G418 resistance marker and the GPD promoter was cloned by ligating the KpnI/NotI fragment of pRS420 (Addgene) with the KpnI/NotI-digested PCR product amplified from p426GPD with the primers 5′-TCTAGGCGGCCGCGAGCTCAGTTTATCATTATC-3′ and 5′-GTAAAACGACGGCCAG-3′, yielding the plasmid pDH20. ADH7 was amplified from a TMB3400-FT30-3 segregant with the primers 5′-GTACACTAGTCCTGCTTTAGATGTTGCTTGA-3′ and 5′-TCTTAGTACTCGAGTGGGTCATGATAAATTCCG-3′, digested with SpeI/XhoI, and ligated into the SpeI/XhoI vector fragment of pDH20. YKL071W was amplified with the primers 5′-TAGGGATCCGCCTTTACCAGTGGCATAAATC-3′ and 5′-TCTTAGTACTCGAGTTGCACTTATCTAAAAGACGC-3′, digested with BamHI/XhoI, and ligated into the BamHI/XhoI vector fragment of pDH20.
To sequence the DNA upstream of ADH7, genomic DNA of four segregants for each strain was PCR amplified with the primers 5′-TCC ACT AGA TTG AGA CCA GCC-3′ and 5′-TTG CGT TGG AAA TAC CGA TG-3′. The same primers were used for sequencing with the Sanger method. To sequence the coding region of ADH7, the same primers used for cloning the ADH7 genes were used to amplify and to perform the sequencing.
RESULTS
Metabolic characterization of parent and evolved strains.
Since yeasts' main resistance strategy of furfural reduction (18, 29, 32, 34) requires a constant supply of reduction equivalents, we used metabolic flux analysis to elucidate which redox cofactor is used in vivo. An increased flux through glycolysis or the tricarboxylic acid cycle, accompanied by a decreased production of glycerol and ethanol, would be indicative for a larger supply of NADH. An increased flux through the pentose phosphate pathway, through the malic enzyme, or to acetate would indicate a larger supply of NADPH.
To address this question, we chose an environment with a constant growth rate and steady furfural stress. Specifically, we cultivated the previously evolved furfural-resistant strain TMB3400-FT30-3 (18), with increased viability at high concentrations of furfural, and its semi-industrial parent TMB3400 (48) in glucose-limited chemostats at a dilution rate of 0.1 h−1. To mimic industrial process conditions, the cultivation was done under microaerobic conditions by flushing with nitrogen containing less than 2 ppm oxygen at a rate that suffices to support anaerobic yeast growth without the addition of ergosterol or Tween.
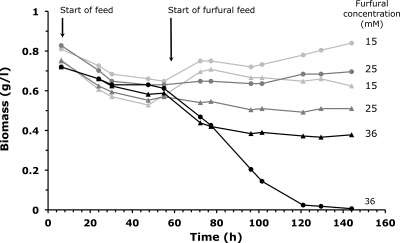
Upon achieving a physiological steady state in the unchallenged culture, we added furfural to the feed and observed the dynamics of attaining a new steady state. We probed concentrations ranging from 6 to 42 mM. There were no significant physiological differences between the strains at the lower concentrations and those at 42 mM, and both parent and evolved strain washed out. We then chose three interesting concentrations for further analysis. At 15 mM furfural, we observed only minor differences between the strains, at 25 mM, the physiological differences became apparent, and at 36 mM, the parent strain washed out but the evolved strain could cope with the high and constant furfural challenge and attained a steady state (Fig. 1). As expected, furfural reduction rates (measured as furfuryl alcohol production rates) increased with increasing concentrations of furfural in the feed for both strains (Table 1).
FIG. 1.
Impact of increasing concentrations of furfural on steady-state growth in glucose-limited chemostat culture of S. cerevisiae TMB3400 (circles) and TMB3400-FT30-3 (triangles). Upon steady state in the unchallenged culture at around 60 h, the feed medium was switched to one containing 15, 25, or 36 mM furfural.
TABLE 1.
Selected physiological parameters of chemostat cultivationsa
| Strain | Furfural feed concn (mM) | Specific production rates (mmol gcdw−1 h−1) |
Biomass yield (gcdw mol−1) | Glucose uptake rate (mmol gcdw−1 h−1) | Residual furfural concn (mM) | Ratio of serine through PP pathway | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ethanol | Acetate | Glycerol | Furoic acid | Furfuryl alcohol | ||||||
| TMB3400 | 15 | 5.5 ± 0.7 | 0.13 ± 0.02 | 0.048 ± 0.007 | 0.051 ± 0.007 | 1.8 ± 0.2 | 0.027 ± 0.003 | 3.7 ± 0.4 | 0.8 ± 0.3 | ND |
| 25 | 6.5 ± 1.2 | 0.20 ± 0.04 | 0.034 ± 0.006 | 0.08 ± 0.01 | 3.1 ± 0.6 | 0.024 ± 0.003 | 4.2 ± 0.5 | 3.0 ± 0.8 | 0.18 ± 0.02 | |
| 36 | — | — | — | — | — | — | — | — | — | |
| TMB3400-FT30-3 | 15 | 7.0 ± 0.46 | 0.18 ± 0.01 | 0.057 ± 0.004 | 0.052 ± 0.003 | 2.0 ± 0.1 | 0.023 ± 0.001 | 4.3 ± 0.2 | 1.4 ± 0.3 | 0.08 ± 0.01 |
| 25 | 7.4 ± 0.7 | 0.30 ± 0.03 | 0.071 ± 0.006 | 0.090 ± 0.008 | 3.7 ± 0.3 | 0.019 ± 0.001 | 5.2 ± 0.2 | 4.0 ± 0.6 | 0.15 ± 0.02 | |
| 36 | 8.7 ± 1.8 | 0.44 ± 0.09 | 0.12 ± 0.01 | 0.12 ± 0.02 | 6.0 ± 1.2 | 0.015 ± 0.001 | 6.5 ± 0.8 | 8.8 ± 0.3 | 0.33 ± 0.04 | |
PP, pentose phosphate; ND, not determined; —, washout occurred.
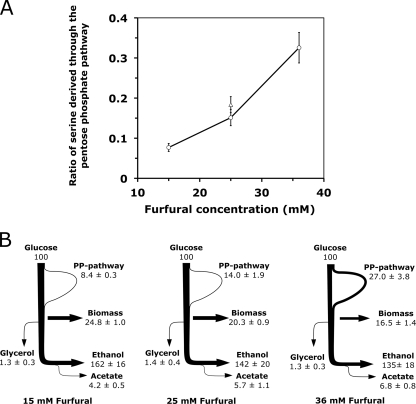
For each attained, furfural-challenged steady state, we then determined metabolic fluxes with a labeling experiment using 1-13C-labeled glucose (12, 52). The carbon at the C-1 position is lost from CO2 when glucose is metabolized through the NADPH-generating pentose phosphate pathway but retained when glucose takes the path through glycolysis. We then measured the label content in protein-bound serine, an amino acid that originates from the merge point of glycolysis and the pentose phosphate pathway, which gives information about the relative pathway usage (12). From this label content, we calculated the ratio of serine derived through the pentose phosphate pathway, as described earlier (12), with the Matlab-based FiatFlux software (53). This ratio showed a marked dependence on the furfural concentration for both strains (Fig. 2A). In the evolved strain, it rises from 0.08 at 15 mM furfural to 0.33 at 36 mM furfural. The ratio in the parent strain was 0.18 at 25 mM furfural and indistinguishable from the evolved strain (Table 1 and Fig. 2A).
FIG. 2.
Effect of furfural on metabolic fluxes. (A) Ratio of serine derived through glycolysis in response to furfural for strains TMB3400 (triangle) and TMB3400-FT30-3 (circles). (B) Estimated flux distributions based on [1-13C]glucose experiments and culture physiology. The thickness of the lines corresponds to the flux magnitude. The values are expressed as percent mole products per moles of consumed glucose. PP, pentose phosphate.
We then determined absolute metabolic fluxes using the ratio of serine derived through the pentose phosphate pathway and the measured extracellular uptake and secretion rates as constraints for a simplified stoichiometric network (Fig. 2B). In addition to the increased pentose phosphate pathway flux, furfural caused a slightly increased flux to acetate but did not affect the secretion of glycerol. The tricarboxylic acid cycle was omitted from the network because its redox reactions require molecular oxygen in stoichiometric amounts for them to occur. Our microaerobic conditions provide oxygen that allow a maximal tricarboxylic acid cycle flux of around 4 μmol gcdw−1 h−1 (calculated for the 36 mM furfural condition), which is negligible (also see reference 22). Since our carbon balances closed to at least 96%, we have sufficient evidence for only marginal tricarboxylic acid cycle flux. Based on these data, we conclude that the increased demand for reduction equivalents for furfural reduction is supplied mainly by the in vivo increase of pentose phosphate pathway activity.
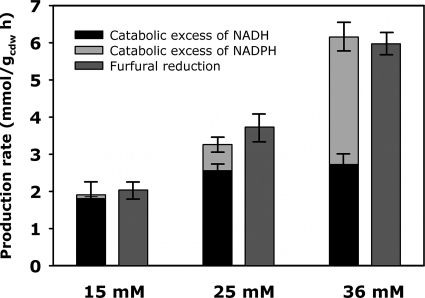
To quantify the in vivo contribution of each redox factor to furfural reduction, we calculated the rates of NADH and NADPH regeneration and consumption based on the above-determined fluxes. The regeneration exceeded the consumption rate under all conditions, and this excess matched the observed furfural reduction rate quantitatively (Fig. 3). At 15 mM furfural, the reduction can be covered entirely by NADH-dependent reduction mechanisms, whereas NADH is not sufficient anymore at 25 mM furfural. At the high concentration of 36 mM furfural, more than half of the furfural reduction depends on NADPH. We therefore conclude that at low concentrations of furfural, NADH-dependent reduction prevails. As the furfural concentration rises above a critical limit that is above 15 mM, S. cerevisiae increases its pentose phosphate pathway flux to provide NADPH that is used for the reduction reaction. Both strains increase their relative flux through the pentose phosphate pathway upon furfural concentration increase. We therefore conclude that the observed increases are not specific to our evolved strain but are a general response of yeast to furfural.
FIG. 3.
Redox cofactor usage for furfural reduction in TMB3400-FT30-3 at 15, 25, and 36 mM furfural. The excess is calculated as the difference between regeneration and the consumption rate of reduction equivalents, which were calculated based on the estimated metabolic flux distributions.
Transcriptional response to furfural stress.
Which enzyme does S. cerevisiae use to catalyze the reduction of furfural? The usage of both NADH and NADPH as cofactors for reduction and the many oxireductases present in yeast render a large number of known and unknown enzymes possible. To identify the key genes, we performed a global microarray-based transcript analysis with RNA samples from all conditions and both strains (9).
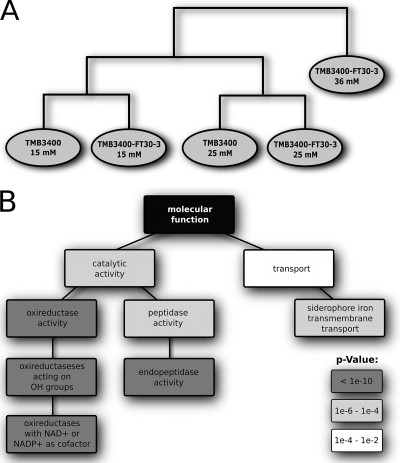
For a first overview and characterization of the transcriptional response to furfural stress, we clustered the probe set intensities for all strains and conditions with a hierarchical clustering algorithm (Fig. 4A). This reveals that the condition (furfural concentration) rather than the different strains is the discriminating feature. To identify genes that are causally involved in resistance, we applied differential expression analysis. A remarkably small number of 14 and 21 genes changed their expression more than twofold upon furfural increase from 15 to 25 mM in the parent and the evolved strain, respectively. This low number indicates that only a few changes are responsible for the observed physiological changes. It is also an indication that the chemostat setup indeed ensured a constant growth environment that minimizes unspecific growth rate-related responses. Among the most upregulated genes in both strains upon furfural increase from 15 to 25 mM were the oxireductases ADH7, GRE2, OYE3, AAD4, YML131W, YDL124W, OYE2, and GCY1 (see Table S1 in the supplemental material). Six of them prefer NADPH as the cofactor (6, 7, 21, 26, 33, 44), while for AAD4 and YML131W, the cofactor preference is not known (Saccharomyces Genome Database at http://www.yeastgenome.org). This is consistent with our finding that central carbon metabolism doubles the NADPH-generating pentose phosphate pathway fluxes upon 25 mM furfural challenge. Hence, we speculate that all these enzymes could be involved in the reduction of furfural.
FIG. 4.
Effect of furfural on transcript levels. (A) Hierarchical clustering of probe set intensities for all conditions and strains. (B) Gene ontology displaying important processes upon furfural concentration increase from 15 and 25 mM.
Upon a further furfural increase from 25 mM to 36 mM that only the evolved strain could support, the differential expression analysis reveals another increase in transcription of the oxireductase genes GRE2, OYE2, OYE3, YML131W, and ADH7, all of which were also induced upon furfural increase from 15 to 25 mM. There is a linear correlation between the observed reduction activities at 15, 25, and 36 mM furfural and the increase in transcript levels of these five genes. This suggests that these enzymes are directly involved in the reduction of furfural. The oxireductase genes GCY1, YKL124W, and AAD4, whose expression was induced from 15 to 25 mM, had only slightly higher expression upon furfural increase from 25 to 36 mM furfural. They either are not directly involved in furfural reduction or are already fully expressed at 25 mM. At this higher furfural challenge, the pentose phosphate pathway mRNAs for ZWF1, GND1, SOL3, and TKL1 also were more abundant.
Besides the genes coding for oxireductases, the two ORFs YKL071W and YLL056C that encode proteins of yet unknown function were expressed at significantly higher levels at 25 and 36 mM furfural in both strains. For YKL071W, a protein homology search in the RCSB Protein Data Bank (http://www.rcsb.org) revealed striking similarities with the Sniffer carbonyl reductases of Drosophila melanogaster (P value = 1.0 × 10−14) and Caenorhabditis elegans (P value = 1.3 × 10−10), both preferring NADPH as their cofactor. Moreover, a search for structural elements reveals a Rossmann fold (37), a protein structural element serving as the NAD(P)H binding site. These are strong indications that the ORF YKL071W encodes an enzyme with reduction activity and suggest that it is involved in the reduction of furfural and thereby for resistance. The homology search for the ORF YLL056C showed that the protein sequence has homologies with the NADH-dependent dehydratases DesIV from Streptomyces venezuelae and RMLB from Streptococcus suis, with P values of 6.4 × 10−6 and 1.0 × 10−4, respectively. Since the deduced protein sequence of YLL056C contains NAD(P)H binding sites, we have some indication that YLL056C also encodes a protein involved in the reduction process.
Other cellular processes involved in resistance.
Next, we asked whether additional cellular processes are relevant for the furfural response beyond the oxireductases and increased NADPH supply. For this purpose, we performed a functional gene ontology search on the transcript data with the 100 most upregulated genes for both strains upon furfural increase from 15 to 25 mM (Fig. 4B). The outcome of this gene ontology search led to the same results for parent and evolved strains. Important functional groups/processes included in this set were the already-mentioned oxireductases, endopeptidase activity, and siderophore iron transmembrane transport. Genes in the latter functional category were the most upregulated genes in both strains at the 15 to 25 mM furfural and the 25 to 36 mM furfural steps (FIT2, FIT3, SIT1, and ARN1). A possible explanation could be a higher demand for iron upon furfural stress, and indeed, we find slightly higher expression of genes (0.1- to 0.5-fold) containing iron-sulfur clusters (MXR1, NAR1, and GRX6) or involved in the biogenesis of iron-sulfur enzymes (CFD1, NFS1, IBA57, ISU2, and DRE2). Not surprisingly, some of these proteins are involved in response to oxidative stress. An alternative, previously raised explanation (2) could be damage to membranes that would result in iron leakage. However, typical genes involved in membrane synthesis and function (such as the ERG and GPI genes) were not differentially expressed in our data set, such that membrane damage appears unlikely. We also observe that mRNAs for heat shock proteins and DNA damage repair are more abundant at 36 mM furfural rather than at 25 mM furfural, which might reflect the general stress at this furfural concentration.
Transcriptional differences between parent and evolved strains.
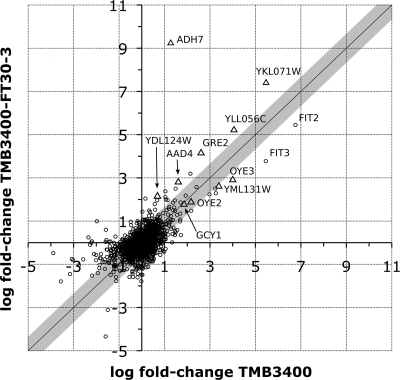
Having revealed general mechanisms and processes associated with furfural challenge, we next asked whether we could identify genes that are important for the higher furfural resistance of the evolved strain. A differential expression analysis at 15 mM furfural between parent and evolved strains showed only slight differences that could not be associated with any particular process or function (data not shown). We therefore compared the differential expression upon furfural concentration increase from 15 to 25 mM for both strains (Fig. 5). Most striking was the more than 9-fold overexpression of ADH7 in the evolved strain compared to only 1.3-fold overexpression in the parent. ADH7 encodes a NADPH-dependent, medium-chain-length alcohol dehydrogenase with a broad substrate specificity that includes aromatic compounds. It is a member of the cinnamyl family of alcohol dehydrogenases and is suspected to be involved in the buildup of fusel alcohols (26). Nothing is known about the molecular function or the regulation of ADH7. Together with the fact that ADH7 has been shown to increase yeast reduction capacity in vitro (28), we hypothesize that ADH7 is responsible for the reduction of furfural and thereby for the resistance in the evolved strain.
FIG. 5.
Differential gene expression upon increase of furfural concentration from 15 to 25 mM between evolved and parent strains. Oxireductases are depicted as ▵ (YKL071W and YLL056C were assumed to be dehydrogenases). The gray diagonal bar indicates the distance interval from the diagonal, in which 98% of the data points are populated. Hence, it gives a visual impression of the likelihood that the dehydrogenases are overexpressed more strongly in the evolved strain.
In addition, transcript levels for the oxireductases GRE2, AAD4, YDL124W, OYE3, YML131W, OYE2, and GCY1 were generally higher in the evolved strain, with a t test P value of 0.1, than those for a random sample from the data set (Fig. 5). In the parent, in contrast, more genes of the general stress response were upregulated (most notably, genes encoding pleiotropic drug resistance, heat shock, and DNA damage repair proteins), indicating that the parent strain is more stressed than the evolved strain at 25 mM furfural.
Inverse metabolic engineering of furfural resistance.
Knowing the precise genetic basis of the furfural resistance would be a prerequisite for inverse metabolic engineering (3) of this phenotype into other strains. One important parameter in this context is the number of alterations that are causal for the mutant resistance. To address these issues, we first estimated the genetic complexity of the evolved resistance trait. For this purpose, we forced the diploid strain to undergo meiosis, separated the haploid segregants, and analyzed them for furfural resistance. A total of 16 of the 17 sporulated diploid cells showed a 2:2 inheritance ratio of the resistant trait, which strongly indicated that the phenotype is caused by a single alteration in the genome (30). The most striking difference between the strains is the highly expressed ADH7 in the evolved strain. Using DNA sequencing, however, we could not detect any alterations in the 900-base-pair DNA region between ADH7 and the preceding ORF (YCR104W) or in the structural ADH7 gene itself. Thus, increased ADH7 expression in the evolved strain might be explained by an alteration in the transcription factor of ADH7 or other regulatory mechanisms.
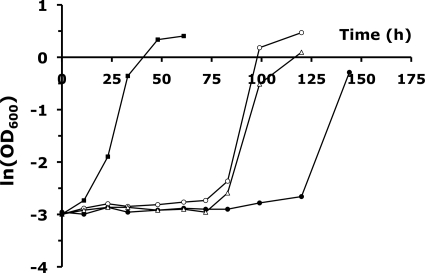
To verify that increased ADH7 expression is indeed responsible for furfural resistance, we overexpressed the gene from the constitutive GPD promoter in the parent strain TMB3400. Furthermore, we also chose to overexpress YKL071W to test its involvement in resistance, since it was the second most differentially expressed gene upon furfural increase. The most pronounced characteristic of furfural stress in batch cultures is long lag phases, during which yeast reduces furfural to a level permissive for growth (2, 18, 24). The duration of these lag phases allows a good quantification of resistance level, whereas chemostat experiments allow only a rough binary quantification in the form of washout. We therefore grew the cells in batch cultures in the presence of 11 mM furfural, which is slightly higher than the maximal residual concentration of 8.8 mM furfural in our 36 mM furfural chemostat in which only the evolved strain survived. Both the ADH7- and YKL071W-overexpressing strains showed highly significant reduction of the furfural-induced lag phase of about 30%, compared to that of the control strain (Fig. 6). We therefore conclude that both genes are directly involved in reducing furfural. The evolved strain, however, showed an about 80% reduction of lag phase. The NADPH-dependent furfural reduction activities in crude cell extracts of ADH7 and YKL071W overexpression strains were only about 30% higher than those in the control with the empty plasmid, whereas the evolved strain shows 50% higher reduction activity (data not shown), which might explain the discrepancy in the observed lag-phase reduction.
FIG. 6.
Growth characteristics of TMB3400 overexpressing ADH7 (○) and YKL071W (▵), TMB3400 carrying the empty plasmid (•), and the evolved strain TMB3400-FT30-3 (▪) in 11 mM furfural containing minimal media. OD600, optical density at 600 nm.
DISCUSSION
By combining 13C-based metabolic flux and global gene expression analysis in microaerobic, glucose-limited chemostats, we revealed key features of yeasts' normal response to furfural. While low concentrations of furfural (<15 mM) can be reduced by NADH-dependent oxireductases, the main physiological response to higher concentrations of furfural (>15 mM) was increased in pentose phosphate pathway flux to provide sufficient NADPH for the reduction of furfural. In wild-type yeast, upregulated expression of several genes coding for NADPH-dependent oxireductases indicates their involvement in the reduction of furfural. Moreover, our transcript data show that genes for iron transmembrane transport are upregulated in response to furfural. By comparing the transcript levels of an evolved, furfural-resistant strain to those of its parent, we demonstrated the key molecular mechanism of the evolved furfural resistance to be the upregulation of the NADPH-dependent dehydrogenase ADH7 and the uncharacterized ORF YKL071W.
Previous chemostat studies showed that the cofactor for furfural reduction is NADH, but only at low concentrations of furfural, around 6 mM (20, 40). These strains were also much less resistant than the semi-industrial strain TMB3400 used here, such that washout already occurred at 8.6 mM furfural. These results are consistent with our flux data at the furfural concentration of 15 mM, where the NADH supply also seems to suffice to quantitatively reduce furfural. At 25 and 36 mM furfural, however, our redox balance data show clearly that the in vivo available NADH would not suffice for furfural reduction. Therefore, we suggest the following model. Upon furfural exposure, yeast preferentially ceases glycerol production as a first means to free NADH to carry out the reduction (40). As this source is exhausted, pentose phosphate pathway activity is increased as a second mechanism to deliver more reduction equivalents in the form of NADPH. Consistent with this model, most of the upregulated genes in our data set at high furfural challenge have a preference for NADPH, whereas previously identified NADH-dependent genes (ADH1 and ADH5) were not upregulated. These enzymes are possibly already fully expressed at 15 mM furfural. Our finding is consistent with the study of Gorsich et al. (17) that demonstrated that a knockout of pentose phosphate pathway genes leads to an increased sensitivity at a high concentration of 50 mM furfural on agar plates. The overexpression of ZWF1 provides no growth advantage at low concentrations of furfural but enables growth at otherwise lethal concentrations of 50 mM furfural in liquid culture.
Lin and coworkers found that in batch cultures with a very high concentration of 177 mM furfural, the yeast response consists of an increase in enzymes catalyzing the reaction of the tricarboxylic acid cycle and upregulation of mainly NADH-dependent dehydrogenases and ADH6 as a NADPH-dependent dehydrogenase (27). The conditions were fundamentally different from our chemostat experiments for the following two main reasons. First, the yeast was cultivated under aerobic conditions, which would in principle allow an increase in tricarboxylic acid activity. Second, cell growth stopped completely after the addition of furfural, which strongly affects metabolic activity (43, 50). Furthermore, since pentose phosphate pathway activity is regulated mainly at the posttranslational level, a potential increase could not be detected by their method.
We attribute the increased resistance of our evolved strain to the observed strong overexpression of ADH7 and YKL071W as well as to the generally higher levels of a further four oxireductases. The higher expression of these genes leads to the demonstrated higher maximal reduction capacity that ensures growth at high concentrations of furfural. By separate, plasmid-based overexpression of ADH7 and YKL071W in the parent strain, we demonstrate the relevance of both events in furfural resistance. Neither of them alone, however, could fully restore the observed resistance level of our evolved strain. We contribute this difference mainly to the 50% higher in vitro reduction activity of the evolved strain compared to only 30% higher in vitro reduction activity of the plasmid-based overexpression strains. Moreover, several further dehydrogenase genes are upregulated upon furfural increase in the evolved strain, cumulating in a higher total reduction activity, which might allow the strain to attain a higher level of resistance.
The inheritance pattern of the resistant trait indicates a single genome alteration that would be causal for the observed phenotype. Since several dehydrogenases were upregulated and neither ADH7 nor YKL071W overexpression alone was sufficient to restore the evolved phenotype, we speculate that a regulatory mutation is the basis for the altered expression level of these genes that conjointly lead to furfural resistance.
Supplementary Material
Acknowledgments
This work was financially supported by the EU Framework VI NILE project within European Commission Framework VI (http://www.nile-bioethanol.org).
We thank the Functional Genomics Center Zurich for their support on the transcript analysis, Karel Novy for segregant growth tests, and the lab of M. Peter for providing p426GPD.
Footnotes
Published ahead of print on 23 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alexandre, H., V. Ansanay-Galeote, S. Dequin, and B. Blondin. 2001. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. FEBS Lett. 498:98-103. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, J. R. M., T. Modig, A. Petersson, B. Hahn-Hagerdal, G. Liden, and M. F. Gorwa-Grauslund. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82:340-349. [Google Scholar]
- 3.Bailey, J. E., A. Sburlati, V. Hatzimanikatis, K. Lee, W. A. Renner, and P. S. Tsai. 2002. Inverse metabolic engineering: a strategy for directed genetic engineering of useful phenotypes. Biotechnol. Bioeng. 79:568-579. [DOI] [PubMed] [Google Scholar]
- 4.Blank, L. M., and U. Sauer. 2004. TCA cycle activity in Saccharomyces cerevisiae is a function of the environmentally determined specific growth and glucose uptake rates. Microbiology 150:1085-1093. [DOI] [PubMed] [Google Scholar]
- 5.Boyle, E. I., S. Weng, J. Gollub, H. Jin, D. Botstein, J. M. Cherry, and G. Sherlock. 2004. GO::TermFinder—open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 20:3710-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Q., T. A. Griest, T. M. Harter, and J. M. Petrash. 2007. Functional studies of aldo-keto reductases in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. N., L. Porubleva, G. Shearer, M. Svrakic, L. G. Holden, J. L. Dover, M. Johnston, P. R. Chitnis, and D. H. Kohl. 2003. Associating protein activities with their genes: rapid identification of a gene encoding a methylglyoxal reductase in the yeast Saccharomyces cerevisiae. Yeast 20:545-554. [DOI] [PubMed] [Google Scholar]
- 8.Cockerill, S., and C. Martin. 2008. Are biofuels sustainable? The EU perspective. Biotechnol. Biofuels 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daran-Lapujade, P., J. M. Daran, A. J. van Maris, J. H. de Winde, and J. T. Pronk. 2009. Chemostat-based micro-array analysis in baker's yeast. Adv. Microb. Physiol. 54:257-311. [DOI] [PubMed] [Google Scholar]
- 10.Delgenes, J. P., R. Moletta, and J. M. Navarro. 1996. Effects of lignocellulose degradation products on ethanol fermentations of glucose and xylose by Saccharomyces cerevisiae, Zymomonas mobilis, Pichia stipitis, and Candida shehatae. Enzyme Microb. Technol. 19:220-225. [Google Scholar]
- 11.Farrell, A. E., R. J. Plevin, B. T. Turner, A. D. Jones, M. O'Hare, and D. M. Kammen. 2006. Ethanol can contribute to energy and environmental goals. Science 311:506-508. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, E., and U. Sauer. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270:880-891. [DOI] [PubMed] [Google Scholar]
- 13.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentleman, R., V. Carey, W. Huber, R. Irizarry, and S. Dudoit (ed.). 2005. Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, NY.
- 15.Gietz, R. D., and R. H. Schiestl. 2007. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2:35-37. [DOI] [PubMed] [Google Scholar]
- 16.Goldemberg, J. 2007. Ethanol for a sustainable energy future. Science 315:808-810. [DOI] [PubMed] [Google Scholar]
- 17.Gorsich, S. W., B. S. Dien, N. N. Nichols, P. J. Slininger, Z. L. Liu, and C. D. Skory. 2006. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 71:339-349. [DOI] [PubMed] [Google Scholar]
- 18.Heer, D., and U. Sauer. 2008. Identification of furfural as the key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb. Biotechnol. 1:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirasawa, T., Y. Nakakura, K. Yoshikawa, K. Ashitani, K. Nagahisa, C. Furusawa, Y. Katakura, H. Shimizu, and S. Shioya. 2006. Comparative analysis of transcriptional responses to saline stress in the laboratory and brewing strains of Saccharomyces cerevisiae with DNA microarray. Appl. Microbiol. Biotechnol. 70:346-357. [DOI] [PubMed] [Google Scholar]
- 20.Horvath, I. S., M. J. Taherzadeh, C. Niklasson, and G. Liden. 2001. Effects of furfural on anaerobic continuous cultivation of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75:540-549. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara, K., H. Yamamoto, K. Mitsuhashi, K. Nishikawa, S. Tsuboi, H. Tsuji, and N. Nakajima. 2004. Purification and characterization of alpha-keto amide reductase from Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 68:2306-2312. [DOI] [PubMed] [Google Scholar]
- 22.Jouhten, P., E. Rintala, A. Huuskonen, A. Tamminen, M. Toivari, M. Wiebe, L. Ruohonen, M. Penttila, and H. Maaheimo. 2008. Oxygen dependence of metabolic fluxes and energy generation of Saccharomyces cerevisiae CEN.PK113-1A. BMC Syst. Biol. 2:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinke, H. B., L. Olsson, A. B. Thomsen, and B. K. Ahring. 2003. Potential inhibitors from wet oxidation of wheat straw and their effect on ethanol production of Saccharomyces cerevisiae: wet oxidation and fermentation by yeast. Biotechnol. Bioeng. 81:738-747. [DOI] [PubMed] [Google Scholar]
- 24.Klinke, H. B., A. B. Thomsen, and B. K. Ahring. 2004. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 66:10-26. [DOI] [PubMed] [Google Scholar]
- 25.Lange, H. C., and J. J. Heijnen. 2001. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75:334-344. [DOI] [PubMed] [Google Scholar]
- 26.Larroy, C., X. Pares, and J. A. Biosca. 2002. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADHVII), a member of the cinnamyl alcohol dehydrogenase family. Eur. J. Biochem. 269:5738-5745. [DOI] [PubMed] [Google Scholar]
- 27.Lin, F. M., B. Qiao, and Y. J. Yuan. 2009. Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl. Environ. Microbiol. 75:3765-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Z. L., J. Moon, B. J. Andersh, P. J. Slininger, and S. Weber. 2008. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81:743-753. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Z. L., P. J. Slininger, and S. W. Gorsich. 2005. Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl. Biochem. Biotechnol. 121:451-460. [PubMed] [Google Scholar]
- 30.Madigan, T., J. Martinko, P. Dunla, P. D. Clark, and T. D. Brock (ed.). 2002. Brock biology of microorganisms. Prentice Hall International, Englewood Cliffs, NJ.
- 31.Margeot, A., B. Hahn-Hagerdal, M. Edlund, R. Slade, and F. Monot. 2009. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 20:372-380. [DOI] [PubMed] [Google Scholar]
- 32.Martin, C., M. Marcet, O. Almazan, and L. J. Jonsson. 2007. Adaptation of a recombinant xylose-utilizing Saccharomyces cerevisiae strain to a sugarcane bagasse hydrolysate with high content of fermentation inhibitors. Bioresour. Technol. 98:1767-1773. [DOI] [PubMed] [Google Scholar]
- 33.Niino, Y. S., S. Chakraborty, B. J. Brown, and V. Massey. 1995. A new old yellow enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 270:1983-1991. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson, A., M. F. Gorwa-Grauslund, B. Hahn-Hagerdal, and G. Liden. 2005. Cofactor dependence in furan reduction by Saccharomyces cerevisiae in fermentation of acid-hydrolyzed lignocellulose. Appl. Environ. Microbiol. 71:7866-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmqvist, E., H. Grage, N. Q. Meinander, and B. Hahn-Hagerdal. 1999. Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol. Bioeng. 63:46-55. [DOI] [PubMed] [Google Scholar]
- 36.Petersson, A., J. R. Almeida, T. Modig, K. Karhumaa, B. Hahn-Hagerdal, M. F. Gorwa-Grauslund, and G. Liden. 2006. A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455-464. [DOI] [PubMed] [Google Scholar]
- 37.Rao, S. T., and M. G. Rossmann. 1973. Comparison of super-secondary structures in proteins. J. Mol. Biol. 76:241-256. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld, E., B. Beauvoit, B. Blondin, and J. M. Salmon. 2003. Oxygen consumption by anaerobic Saccharomyces cerevisiae under enological conditions: effect on fermentation kinetics. Appl. Environ. Microbiol. 69:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Sarvari Horvath, I., C. J. Franzen, M. J. Taherzadeh, C. Niklasson, and G. Liden. 2003. Effects of furfural on the respiratory metabolism of Saccharomyces cerevisiae in glucose-limited chemostats. Appl. Environ. Microbiol. 69:4076-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauer, U. 2006. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smyth, G. K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed] [Google Scholar]
- 43.Sonderegger, M., M. Schümperli, and U. Sauer. 2004. Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70:2892-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stott, K., K. Saito, D. J. Thiele, and V. Massey. 1993. Old Yellow Enzyme. The discovery of multiple isozymes and a family of related proteins. J. Biol. Chem. 268:6097-6106. [PubMed] [Google Scholar]
- 45.Taherzadeh, M. J., L. Gustafsson, C. Niklasson, and G. Liden. 2000. Inhibition effects of furfural on aerobic batch cultivation of Saccharomyces cerevisiae growing on ethanol and/or acetic acid. J. Biosci. Bioeng. 90:374-380. [DOI] [PubMed] [Google Scholar]
- 46.Taherzadeh, M. J., and K. Karimi. 2008. Pretreatment of lignocellulosic wastes to improve ethanol and biogas production: a review. Int. J. Mol. Sci. 9:1621-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. Van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 48.Wahlbom, C. F., W. H. van Zyl, L. J. Jonsson, B. Hahn-Hagerdal, and R. R. Otero. 2003. Generation of the improved recombinant xylose-utilizing Saccharomyces cerevisiae TMB 3400 by random mutagenesis and physiological comparison with Pichia stipitis CBS 6054. FEMS Yeast Res. 3:319-326. [DOI] [PubMed] [Google Scholar]
- 49.Wolinsky, H. 2009. The economics of biofuels. EMBO Rep. 10:551-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, S., T. J. Tschaplinski, N. L. Engle, S. L. Carroll, S. L. Martin, B. H. Davison, A. V. Palumbo, M. Rodriguez, Jr., and S. D. Brown. 2009. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics 10:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaldivar, J., A. Martinez, and L. O. Ingram. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68:524-530. [DOI] [PubMed] [Google Scholar]
- 52.Zamboni, N., S. M. Fendt, M. Ruhl, and U. Sauer. 2009. (13)C-based metabolic flux analysis. Nat. Protoc. 4:878-892. [DOI] [PubMed] [Google Scholar]
- 53.Zamboni, N., E. Fischer, and U. Sauer. 2005. FiatFlux—a software for metabolic flux analysis from 13C-glucose experiments. BMC Bioinformatics 6:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.