Abstract
To understand the prevalence of Cryptosporidium infection in rodents in China and to assess the potential role of rodents as a source for human cryptosporidiosis, 723 specimens from 18 rodent species were collected from four provinces of China and examined between August 2007 and December 2008 by microscopy after using Sheather's sugar flotation and modified acid-fast staining. Cryptosporidium oocysts were detected in 83 specimens, with an overall prevalence of 11.5%. Phodopus sungorus, Phodopus campbelli, and Rattus tanezumi were new reported hosts of Cryptosporidium. The genotypes and subtypes of Cryptosporidium strains in microscopy-positive specimens were further identified by PCR and sequence analysis of the small subunit rRNA and the 60-kDa glycoprotein (gp60) genes. In addition to Cryptosporidium parvum, C. muris, C. andersoni, C. wrairi, ferret genotype, and mouse genotype I, four new Cryptosporidium genotypes were identified, including the hamster genotype, chipmunk genotype III, and rat genotypes II and III. Mixed Cryptosporidium species/genotypes were found in 10.8% of Cryptosporidium-positive specimens. Sequence analysis of the gp60 gene showed that C. parvum strains in pet Siberian chipmunks and hamsters were all of the subtype IIdA15G1, which was found previously in a human isolate in The Netherlands and lambs in Spain. The gp60 sequences of C. wrairi and the Cryptosporidium ferret genotype and mouse genotype I were also obtained. These findings suggest that pet rodents may be potential reservoirs of zoonotic Cryptosporidium species and subtypes.
Cryptosporidium spp. are protozoan parasites that infect a wide range of vertebrates, including humans. Cryptosporidiosis is acute and self-limiting in immunocompetent hosts but life threatening in immunocompromised individuals (48). Humans and animals can acquire Cryptosporidium infection through direct contact with infected individuals or contaminated fomites or by consumption of contaminated food or water (16, 47). Rodents, which are abundant and widespread, have been considered reservoirs of cryptosporidiosis in humans and farm animals. Previous studies based on oocyst morphology showed that many wild rodents might serve as hosts of Cryptosporidium parvum-like and C. muris-like parasites (4, 8, 42). The reported prevalence rates of Cryptosporidium in rodents ranged from 5.0% to 39.2% (11-13). Nearly 40 rodent species belonging to 11 families (Sciuridae, Muridae, Cricetidae, Castoridae, Geomyidae, Hystricidae, Erethizontidae, Myocastoridae, Caviidae, Hydrochoeridae, and Chinchillidae) have been reported as hosts of Cryptosporidium spp. (10, 12, 30, 53).
Recently, PCR-based genotyping and subtyping tools have been used in assessing the human-infective potential of Cryptosporidium spp. in animals and the extent of cross-species transmission of cryptosporidiosis in animals (47, 49, 51). Five Cryptosporidium species and nearly 20 Cryptosporidium genotypes of uncertain species status have been identified in rodents worldwide in recent studies (3, 6, 12, 13, 18-20, 23, 26, 30, 31, 36, 39, 52, 53). Among them, C. parvum, C. meleagridis, cervine genotype, C. muris, C. andersoni, chipmunk genotype I, and skunk genotype have been associated with cryptosporidiosis in humans although the last four species and genotypes are each responsible for only one or a few cases (47). Subtyping based on sequence analysis of the 60-kDa glycoprotein (gp60) gene has been used in tracking the transmission of six Cryptosporidium species and genotypes, including C. hominis, C. parvum, C. meleagridis, C. fayeri, and the rabbit and horse genotypes (7, 37). There are at least 10 gp60 subtype families of C. parvum, two (IIa and IId) of which are involved in zoonotic transmission. In rodents, natural C. parvum infection is rare (11), and only one C. parvum subtype (IIaA15G2R1) has been reported in capybaras (Hydrochoerus hydrochaeris) in Brazil (30).
Until recently there has been no genetic characterization of Cryptosporidium spp. in rodents in China. Worldwide, there are also hardly any genetic data on Cryptosporidium spp. from pet rodents. The purpose of this study was to determine the prevalence of Cryptosporidium in some wild, laboratory, and pet rodents in China and to assess the zoonotic potential of Cryptosporidium spp. from rodents.
MATERIALS AND METHODS
Specimens.
A total of 723 fecal specimens were collected from 18 species of rodents in several areas of China from August 2007 to December 2008. The sampled animals included wild, laboratory, and pet rodents. Relevant information on animal species, sex, and age and clinical signs were recorded at the time of sample collection.
(i) Wild or captive rodents.
Fresh fecal specimens were collected from eight species of captive or wild rodents. The former included one woodchuck (Marmota sibirica) in the Wild Animals Rescue Centre of Henan Province and one nutria and eight porcupines in the Zhengzhou Zoo; the wild rodents included five species of wild rats and mice trapped in different areas (Table 1). Brown rats were trapped mostly from the field, granary, and the zoo in Zhengzhou City, with some from the field in Shangqiu City, both in Henan Province. Asian house rats (Rattus tanezumi) were trapped in a rice shop (17/33) and on a pig farm (16/33) in Longyan City, Fujian Province. House mice were trapped in residences in Linzhou and Kaifeng City, Henan Province. Striped field mice (Apodemus agrarius) and Himalayan field rats (Rattus nitidus) were trapped in the fields of six cities (Dujiangyan, Beichuan, Anxian, Pengzhou, Mianzhu, and Shijin) in Sichuan Province after the massive earthquake in 2008. The trapped animals were killed and transported in coolers with ice packs to the laboratory within 48 h.
TABLE 1.
Prevalence of Cryptosporidium species/genotypes in wild, laboratory, and pet rodents in China
| Host | Location of animal(s) | Total no. of samples | No. of positive samples | Prevalence (%) | No. and/or name of Cryptosporidium species/genotype(s) (no. of samples)c |
|---|---|---|---|---|---|
| Wild rodents | 147 | 10 | 6.8 | 3 genotypes | |
| Hystrix hodgsoni (porcupine) | Zhengzhou Zoo | 8 | 0 | 0.0 | |
| Myocastor coypus (nutria, coypu) | Zhengzhou Zoo | 1 | 0 | 0.0 | |
| Marmota sibirica (woodchuck) | WARCHPa | 1 | 0 | 0.0 | |
| Rattus norvegicus (brown rat) | Zhengzhou and Shangqiu City | 64 | 4 | 6.3 | Mouse genotype I (3), mouse genotype I plus rat genotype III (1) |
| Rattus tanezumi (Asian house rat) | Longyan City, Fujian Province | 33 | 6 | 18.2 | Mouse genotype I (1), rat genotype II (1), rat genotype III (1), mouse genotype I plus rat genotype II (1), mouse genotype I plus rat genotype III (1) |
| Mus musculus (house mouse) | Linzhou and Kaifeng City | 12 | 0 | 0.0 | |
| Rattus nitidus (Himalayan field rat) | Sichuan province | 2 | 0 | 0.0 | |
| Apodemus agrarius (striped field mouse) | Sichuan province | 26 | 0 | 0.0 | |
| Laboratory rodents | 264 | 5 | 1.9 | 1 genotype | |
| Mus musculus (laboratory mouse) | LACHPb | 229 | 4 | 1.7 | Mouse genotype I (4) |
| Rattus norvegicus (laboratory rat) | 25 | 1 | 4.0 | Mouse genotype I (1) | |
| Meriones unguiculataus (Mongolian gerbil) | Capital Medical University | 10 | 0 | 0.0 | |
| Pet rodents | 312 | 68 | 21.8 | 7 genotypes | |
| Mesocricetus auratus (Golden hamster) | Pet market in Zhengzhou City | 50 | 16 | 32.0 | C. muris (6), C. andersoni (5), C. parvum (2), C. muris + C. parvum (1), C. andersoni + C. parvum (1) |
| Phodopus sungorus (Siberian hamster) | 51 | 4 | 7.8 | C. muris (1), C. parvum (1), C. andersoni + C. parvum (1), hamster genotype (1) | |
| Phodopus campbelli (Campbell hamster) | 30 | 3 | 10.0 | C. parvum (1), C. andersoni (1) C. muris + C. parvum (1) | |
| Phodopus roborovskii (desert hamster) | 5 | 0 | 0.0 | ||
| Sciurus vulgaris (red squirrel) | 19 | 5 | 26.3 | Ferret genotype (5) | |
| Tamias sibiricus (Siberian chipmunk) | 20 | 6 | 30.0 | Ferret genotype (3), ferret genotype + C. parvum (1), C. muris + C. parvum + chipmunk genotype III (1) | |
| Chinchilla laniger (Chinchilla) | 96 | 0 | 0.0 | ||
| Cavia porcellus (Guinea pig) | 40 | 34 | 85.0 | C. wrairi (30) | |
| Pteromys sp. | 1 | 0 | 0.0 | ||
| Total | 723 | 83 | 11.5 | 10 genotypes |
WARCHP, Wild Animals Rescue Centre of Henan Province.
LACHP, Laboratory Animal Center of Henan Province.
Plus signs indicate mixed infections.
(ii) Laboratory rodents.
Ten 22-day-old laboratory Mongolian gerbils were bought from the animal facility of the Capital Medical University, and fresh fecal specimens were collected from the animals upon arrival. Laboratory white mice (the KM strain) and rats of 3 to 14 weeks in age were bought from the Laboratory Animal Center of Henan Province and Henan Academy of Agricultural Sciences, respectively, and euthanized upon arrival in the laboratory. Rectal contents from wild, captive, and laboratory rodents were taken for parasite detection.
(iii) Pet rodents.
All pet-derived specimens were from seven pet shops in a pet market in Zhengzhou City, Henan Province. Fresh fecal specimens were collected in the pet shops from red squirrels, Siberian chipmunks, chinchillas, and Pteromys that occupied individual wire cages with removable trays for cleaning. Because hamsters of the same species and age were housed together in glass containers and guinea pigs were housed together in large wire cages, they were purchased from the pet shops and euthanized upon arrival in the laboratory. Rectal contents were taken from the four species of hamsters and one species of guinea pigs killed (Table 1).
Cryptosporidium oocyst detection.
Fecal specimens, rectal contents, and mucosal scrapings from the intestine were examined for the presence of various parasite ova, cysts, and oocysts, including Cryptosporidium. Cryptosporidium oocysts were detected by microscopy after sugar flotation and modified acid-fast staining. The sizes of Cryptosporidium oocysts of various Cryptosporidium species/genotypes were measured using a Nikon light microscope (model Eclipse E200) at a magnification of ×1,000.
DNA extraction.
Oocysts from microscopy-positive specimens were purified by the sucrose density gradient centrifugation as described previously (41). Genomic DNA was extracted from the purified Cryptosporidium oocysts using a Mag Extractor Genome kit (Toyobo Co. Ltd., Osaka, Japan), based on chaotropic extraction and absorption onto silica-coated magnetic beads. The DNA was kept at −20°C before being used in PCRs.
Cryptosporidium genotyping and subtyping.
Nested PCR analyses of the small subunit (SSU) rRNA and gp60 genes were done on DNA of microscopy-positive specimens using primers and procedures previously described by Jiang et al. and Alves et al., respectively (2, 21). Positive and negative controls were included in each analysis. Two-directional sequencing of positive PCR products was done by SinoGenoMax Co., Ltd (Beijing, China), on an ABI Prism 3730 XL DNA analyzer (Applied Biosystems). Restriction fragment length polymorphism (RFLP) analysis of the SSU rRNA PCR products with endonucleases SspI and VspI was performed only on Cryptosporidium specimens from guinea pigs prior to sequencing.
Sequence analysis.
Nucleotide sequences were aligned with each other and with reference sequences in the GenBank using ClustalX, version 1.81 (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/), and the default setting, with manual adjustment. Two neighbor-joining trees based on the SSU rRNA and the gp60 genes and a maximum-parsimony tree based on the SSU rRNA gene were generated using the software Mega 4 (http://www.megasoftware.net/). An SSU rRNA sequence of Monocystis agilis (GenBank accession no. AF457127) was used as the outgroup of the tree based on the SSU rRNA gene. The reliability of branches in two trees was assessed by bootstrap analysis using 1,000 replicates. The degree of similarity among sequences was determined using MegAlign, version 5.0 (a tool in the software DNAStar).
Statistical analysis.
Chi-square analysis was performed to assess the correlation between the prevalence of Cryptosporidium and the age or sex of the host using SPSS, version 11.5 (Statistical Package for the Social Sciences), for Windows (SPSS Inc., Chicago, IL).
Nucleotide sequence accession numbers.
Unique partial Cryptosporidium SSU rRNA and gp60 gene sequences obtained from rodents in this study were deposited in the GenBank database under accession numbers GQ121017 to GQ121030.
RESULTS
Prevalence of Cryptosporidium in rodents.
Cryptosporidium oocysts were detected by microscopy in 83 (11.5%) specimens from 9 of the 18 rodent species. In wild rodents, only Asian house rats and brown rats were positive, with prevalence rates of 6.3% and 18.2%, respectively. In laboratory animals, the prevalence rate of Cryptosporidium in mice and rats was 1.7% and 4.0%, respectively. In pet rodents, guinea pigs had the highest prevalence (85.0%) of Cryptosporidium, followed by sciurid animals (28.2% in squirrels and chipmunks) and hamsters (16.9%). Cryptosporidium oocysts were not detected in other laboratory and pet rodent species (Table 1). The prevalence rate of Cryptosporidium was 12.5% (74/594) and 6.9% (9/129) in young and adult animals and 10.7% (42/392) and 12.4% (41/331) in females and males, respectively. These differences in Cryptosporidium prevalences between the two age or sex groups were not significant (for age, χ2 = 3.13 and P > 0.05; for sex, χ2 = 0.49 and P > 0.05).
Cryptosporidium species/genotypes.
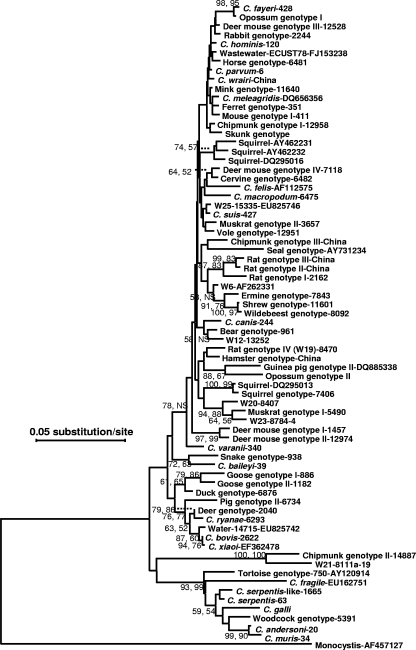
PCR analysis of the SSU rRNA gene generated the expected products in 76 of the 83 specimens. RFLP analysis of the positive PCR products was done with DNA only from guinea pig specimens, which showed only one banding pattern, indicative of the presence of C. wrairi. DNA sequencing of PCR products from six specimens confirmed the identification of C. wrairi. Overall, nucleotide sequence and phylogenetic analysis of the SSU rRNA gene PCR products identified a total of 10 Cryptosporidium species/genotypes, including six species and genotypes previously reported in rodents, such as C. parvum, C. muris, C. andersoni, C. wrairi, and the Cryptosporidium ferret genotype and mouse genotype I. In addition, one Cryptosporidium genotype in rats, the rat genotype II, had sequence almost identical to a 483-bp sequence (AY898790) from a genotype previously reported in a sheep (40) (see the supplemental material). The only difference between the two was an insertion of TT in a poly(T) region in the sequence obtained from the sheep specimen. Three new Cryptosporidium genotypes were found, including the hamster genotype in a Siberian hamster, chipmunk genotype III in a Siberian chipmunk, and rat genotype III in Asian house rats (Table 1).
Sequence and phylogenetic analyses of the partial SSU rRNA gene indicated that the Cryptosporidium hamster genotype was most closely related to the W19 genotype from a New York watershed, with 97.7% similarity. Chipmunk genotype III exhibited highest sequence similarity (97.7 to ∼97.9%) to Sbey03b and Sbey05a from California ground squirrels but formed a clade with the seal genotype that was not supported by bootstrap analysis (Fig. 1). Rat genotypes II and III exhibited 97.1% and 96.6% sequence similarity, respectively, to their closest relative, rat genotype I, with a 98.8% similarity to each other. These three genotypes formed a well-supported clade separated from other common intestinal Cryptosporidium spp. (Fig. 1). The phylogenetic relatedness of the novel Cryptosporidium genotypes to others remains to be substantiated by analysis of the full SSU rRNA and other genes. Sequence differences in the partial SSU rRNA gene among the four new Cryptosporidium genotypes in the present study and their relatives are shown in Table 2.
FIG. 1.
Phylogenetic relationships among various Cryptosporidium species/genotypes from rodents in China and some known Cryptosporidium species and genotypes, as inferred by a neighbor-joining analysis of the partial (∼735 positions in the final alignment) SSU rRNA sequences based on distances calculated using the Tamura-Nei 93 model and adjusted with a gamma distribution (shape parameter, 1). Bootstrap values (in percentages) above 50 from 1,000 pseudoreplicates are shown for both the neighbor-joining (the first value) and maximum-parsimony analyses (the second value). ns, node with bootstrap values lower than 50%. Sequences from brown rats in Japan (23) are not included because of the lack of data from the 5′ end of the fragment under analysis. The multiple-sequence alignment of partial SSU rRNA sequences used in the generation of the phylogenetic tree is presented elsewhere (see the supplemental material).
TABLE 2.
Nucleotide sequence diversity in the partial SSU rRNA gene of new Cryptosporidium genotypes obtained in this study in comparison with related reference sequences in the GenBank
| Organism or genotype (accession no.) | Nucleotide sequence at the indicated position(s)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 473-483 | 632-682 | 693-717 | 750-757 | 770 | 785-793 | 805-826 | 871 | 908 | |
| C. parvum (AF093490) | ACTTTT-TGG | TAATTTATATAAAATATT--TTGATGAATATTTATATAATATTAACATA | CTATATA-TTTTAGTATATGAAATT | GCATAT- | C | ATTAAA-GA | CTT-ATTGGTTCTAAGATAAGA | C | A |
| Hamster genotype | ....A.-... | ..G...-....TTGATAA--.ATT.TGT....A....G........... | ..-...T-.A.G............. | ...AT.- | . | ......-.. | ...-...............GA. | . | . |
| W19 (AY737587) | ....AA-.T. | ..-...-....TT..TAA--..T..T.T....A.............G.. | ..-.T.T-.-.A............. | ....T.- | . | ......-.. | ...-...............GA. | . | . |
| Chipmunk genotype III | ......AACA. | ........G..T......--..CG.-..............G........ | .....A--...........G.... | ......AT | . | ..A..GA.. | T..-................A. | . | . |
| Sbey03b (AY462232) | ......AA.A. | ........G..C......--..A.A-......GT............... | ......--................ | ......AT | . | .CA...-.. | T..-.........GG.....A. | . | . |
| Sbey05a (DQ295017) | .....-A... | ...........C......--..C.A-..............T........ | ....G.--..-............. | ..G...- | . | .CA..G-.. | T..-..........G.....A. | . | . |
| Rat genotype I (FJ205699) | ..C...T-C.. | .TGC..-....T...T.ATGCT...C..C...........G.......... | ......TT..C............C. | ..T.T.- | T | .A....A.. | ...T...............GA. | T | G |
| Rat genotype II | ..C...-C.. | .T.C..-....T...C.-GCAC.T-.TG..G.......G.......... | ....--................. | ..T.T.- | T | .A.T..A.. | ...-................A. | T | G |
| Rat genotype III | ..C...AA... | .T.C..-....T...T..-ACAC.TTC.TG..G.......G.......... | ....--................. | ..T.T.- | T | .A.T..A.. | ...-................A. | T | G |
Dots indicate nucleotide identity to the C. parvum sequence (AF093490), and dashes represent nucleotide deletions.
Subtypes of C. parvum, C. wrairi, and the Cryptosporidium ferret genotype and mouse genotype I.
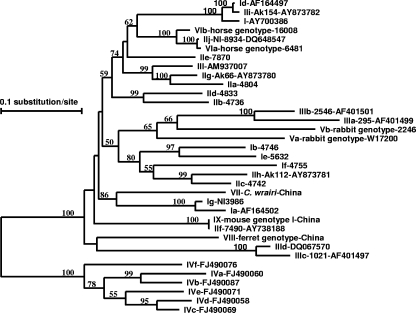
Sequences of the gp60 gene were obtained from C. parvum, C. wrairi, ferret genotype, and mouse genotype I in this study. These sequences were aligned with reference sequences downloaded from the GenBank. Only one subtype was identified in each of the four Cryptosporidium species/genotypes. Sequences of C. parvum isolates from chipmunks and hamsters were all identified as subtype IIdA15G1. The sequence of C. wrairi had the highest similarity to sequences of C. hominis subtypes IgA24 and IaA19R3 at 77.2% and 77.1%, respectively. The sequence of the Cryptosporidium ferret genotype was most closely related to a sequence from C. meleagridis, with a similarity of only 71.7%. The sequence of the Cryptosporidium mouse genotype I was most closely related to C. parvum subtype IIfA6, with 97.2% similarity (Fig. 2). Numbers of serine-coding trinucleotide repeats (TCA, TCG, and TCT) in various gp60 subtypes obtained in this study are shown in Table 3.
FIG. 2.
Phylogenetic relationship of gp60 nucleotide sequences of C. parvum, C. wrairi, and the Cryptosporidium ferret genotype and mouse genotype I obtained in this study and known Cryptosporidium subtype families, as inferred by a neighbor-joining analysis based on distance calculated using the Kimura two-parameter model. Bootstrap values greater than 50% from 1,000 pseudoreplicates are shown. The name of each subtype family starts with the Cryptosporidium species or genotype designation, with C. hominis, C. parvum, C. meleagridis, C. fayeri, rabbit genotype, horse genotype, C. wrairi, ferret genotype, and mouse genotype I represented by I, II, III, IV, V, VI, VII, VIII, and IX, respectively. The sequence of the C. parvum subtype family IIk (AB237137) is not shown because of its short length.
TABLE 3.
The number of serine-coding trinucleotide repeats in various gp60 subtypes obtained in this study
| Species or genotype | No. of samples subtyped | No. of subtypes | Subtype family | No. of serine-coding trinucleotide repeats |
||
|---|---|---|---|---|---|---|
| TCA | TCG | TCT | ||||
| C. parvum | 10 | 1 | IId | 15 | 1 | 0 |
| C. wrairi | 6 | 1 | VII | 17 | 0 | 1 |
| Ferret genotype | 5 | 1 | VIII | 5 | 2 | 0 |
| Mouse genotype I | 5 | 1 | IX | 6 | 0 | 0 |
Morphometrics of Cryptosporidium oocysts.
The sizes of oocysts of eight Cryptosporidium species and genotypes obtained in the study were measured under a light microscope. The morphometric measurements of oocysts of C. parvum, C. wrairi, C. andersoni, and C. muris were within the reported ranges, with those of C. andersoni and C. muris significantly larger and more elongated than those of C. parvum and C. wrairi (Table 4). Other Cryptosporidium genotypes from rodents had oocyst morphometric measurements similar to those of C. parvum and C. wrairi although the mouse genotype I had slightly smaller oocysts (Table 4).
TABLE 4.
Morphometric measurements of some Cryptosporidium species and genotypes in this study
| Cryptosporidium species or genotypea | Animal source | No. of oocysts | Parametera |
|||||
|---|---|---|---|---|---|---|---|---|
| Length (μm) |
Width (μm) |
Shape index (length/width) |
||||||
| Mean | 95% CL | Mean | 95% CL | Mean | 95% CL | |||
| C. muris | Golden hamster | 50 | 8.10 | 8.00-8.20 | 6.03 | 5.98-6.08 | 1.34 | 1.32-1.36 |
| C. andersoni | Golden hamster | 50 | 7.85 | 7.78-7.92 | 5.90 | 5.84-5.96 | 1.33 | 1.32-1.34 |
| C. wrairi | Guinea pig | 50 | 5.25 | 5.22-5.28 | 4.72 | 4.68-4.76 | 1.11 | 1.10-1.12 |
| C. parvum | Siberian hamster | 50 | 5.42 | 5.36-5.48 | 4.90 | 4.85-4.95 | 1.11 | 1.10-1.12 |
| Ferret genotype | Red squirrel | 50 | 5.31 | 5.26-5.36 | 4.98 | 4.92-5.04 | 1.07 | 1.06-1.08 |
| Mouse genotype I | Laboratory mouse | 50 | 4.83 | 4.79-4.87 | 4.07 | 4.02-4.12 | 1.19 | 1.18-1.20 |
| Rat genotype II | Asian house rat | 20 | 5.14 | 5.06-5.22 | 4.42 | 4.35-4.49 | 1.16 | 1.13-1.19 |
| Rat genotype III | Asian house rat | 20 | 5.21 | 5.14-5.28 | 4.40 | 4.31-4.49 | 1.16 | 1.13-1.19 |
The morphometric measurements of oocysts from the hamster genotype and chipmunk genotype III were not obtained because of low oocyst numbers or the presence of mixed infections. CL, confidence limit.
Clinical signs.
Most of the animals with Cryptosporidium infection showed no apparent clinical signs of disease at the time of specimen collection. However, five hamsters, including one golden hamster positive for Giardia, one golden hamster positive for both Giardia and C. parvum, and one golden hamster and two Siberian hamsters with no identifiable parasites, had fecal materials on their tails.
DISCUSSION
In this study, the overall Cryptosporidium prevalence in 723 rodents in China was 11.5%, which was within the reported range of 5.0% to 39.2% (11-13). The infection rate (21.8%) in pet rodents was apparently higher than infection rates in wild (6.8%) and laboratory rodents (1.9%). This might have been caused by the overcrowded living conditions and poor management in pet stores; chinchillas were mostly housed singly or paired and were negative for Cryptosporidium spp. in this study. Cryptosporidium oocysts were not detected in the majority of wild rodents examined in this study, which might have partially resulted from the small sample sizes. We reported Cryptosporidium infection in Phodopus sungorus, Phodopus campbelli, and R. tanezumi for the first time.
Ten Cryptosporidium species/genotypes were found by DNA sequencing in this study. Concurrent infections with mixed Cryptosporidium species/genotypes were seen in 10.8% of the positive animals, as reported in a few previous studies (12, 42). The oocyst shedding intensity varied by hosts; golden hamsters shed higher numbers of C. muris-like oocysts than the other two hamster species (hundreds versus a few per slide), and red squirrels shed more Cryptosporidium oocysts than chipmunks.
Among the Cryptosporidium spp. found, C. parvum is the only species commonly found in humans (47). Calves are major recognized reservoirs for C. parvum although other ruminants (sheep, goats, and deer) (38, 39), alpacas (43), horses (9, 17), and occasionally carnivores (dogs, gray wolves, and raccoon dogs) (15, 29, 35), pigs (27, 54), and rodents (mice, hamsters, squirrels, eastern chipmunks, nutrias, and capybaras) (6, 12, 30, 31, 36, 39) are also hosts. In the present study, C. parvum was identified in pet rodents for the first time.
Cryptosporidium muris has been found in many rodents (mice, wood mice, rats, bank voles, Syrian hamsters, desert hamsters, squirrels, and Siberian chipmunks), and some marsupials (bilbies) (45), other mammals (Bactrian camels, mountain goats, reticulated giraffe, ringed seals, cats, rock hyraxes, cynomolgus monkeys, dogs, and pigs) (11, 24, 25, 28), and birds (tawny frogmouth) (33). It has also been identified in a few humans in developing countries (14, 32, 34). It was found in pet hamsters (6.6%) and pet Siberian chipmunks (5.0%) in the present study.
In this study, C. andersoni was unexpectedly identified in pet hamsters, and the morphology and size of its oocysts were similar to those of C. muris (Table 4). Its prevalence (5.9%) in pet hamsters was similar to that of C. muris (6.6%). In addition to ruminants (cattle, sheep, Bactrian camels, and European wisnets), natural C. andersoni infection has been found in a few rodents (Marmota bobac) (39), birds (wood partridge) (33), and humans.
A high prevalence (85.0%) of Cryptosporidium was found in pet guinea pigs (mostly <300 g), but no clinical signs of cryptosporidiosis were observed, which is consistent with reports in the literature (44). Only C. wrairi was detected in guinea pigs; the other host-adapted Cryptosporidium sp., guinea pig genotype II (19), was not found in this study.
The Cryptosporidium ferret genotype has been found in ferrets and red squirrels (Sciurus vulgaris) (26, 50). In the present study, 26.3% of red squirrels and 25.0% of Siberian chipmunks harbored the genotype, a result similar to that observed (21.4%) in red squirrels (S. vulgaris) in Italy (26). The Cryptosporidium chipmunk genotype I, previously reported in red squirrels, eastern squirrels, eastern chipmunks, and deer mice in the United States and Italy (12, 26), was not detected in this study.
The Cryptosporidium mouse genotype I was previously identified mostly in house mice (Mus musculus) and other small rodents (yellow-necked mice, common voles, and bank voles) and occasionally in prairie bisons, takins, red pandas, and leopards (1, 5, 22). In this study, it was found in 2.0% of laboratory mice and rats, 6.3% of wild brown rats, and 9.1% of Asian house rats.
Several other novel Cryptosporidium genotypes were identified in rodents in this study, including the hamster genotype, chipmunk genotype III, and rat genotypes II and III although the rat genotype II is likely the same parasite previously found in a sheep in Australia (40). Two common Cryptosporidium genotypes previously found in wild brown rats in Japan, the rat genotype I and a genotype phylogenetically related to the W19 genotype in a New York watershed (23), were not found in this study. Because a high sequence heterogeneity in the SSU rRNA gene is known for W19 (21), we consider the BR3, BR4, BR12, BR16, BR20, BR40, and BR44 sequences of the partial SSU rRNA gene from the Japanese study to belong to the Cryptosporidium W19 genotype and propose to name this group of parasites the Cryptosporidium rat genotype IV. Although also related to W19, the Cryptosporidium hamster genotype has nucleotide substitutions in both the hypervariable and semiconserved regions and thus should be considered a separate genotype.
The gp60 subtyping technique is useful in tracking infection sources of Cryptosporidium spp. Cryptosporidium parvum isolates in this study were all identified as IIdA15G1, belonging to the zoonotic C. parvum subtype family IId, which is not as common as the major zoonotic subtype family IIa (47). The subtype IIdA15G1 was previously reported in humans and lambs in southwest Europe (38, 46). Thus far, there have been no confirmed C. parvum cases in humans in China, and the IId subtype family has not been found in farm animals in the country. Therefore, the source of C. parvum in pet rodents is not clear. Because C. parvum was detected only in pet rodents in this study and belonged to one subtype, it could have originated from other animals kept by the pet shop. The IId subtype family of C. parvum was previously known mostly as a parasite of sheep and goats in southern Europe (38). More samples from rodents and other animals should be examined to understand the distribution of C. parvum and its subtypes in China. In addition to C. parvum, partial gp60 nucleotide sequences of C. wrairi, ferret genotype, and mouse genotype I were obtained for the first time.
The results of this study indicate that a high genetic diversity of Cryptosporidium spp. exists in rodents in China. However, this observation is based on sampling of limited animal species in limited regions. Nevertheless, data obtained thus far suggest that pet rodents may play a potential role in the zoonotic transmission of Cryptosporidium spp. With the rapid increase in the number of pet rodents in recent years, more studies should be conducted to assess the public health importance of Cryptosporidium spp. from rodents and the role of these animals in the transmission of human cryptosporidiosis.
Supplementary Material
Acknowledgments
This study was supported in part by the National Natural Science Foundation of China (grants 30871863 and 30928019), Henan Province Great Special Fund of Public Welfare (Henan Province financial administration grant 2008-145), and Ministry of Health Special Funds of Public Sector Research (grant number 200808012).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 9 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alves, M., L. Xiao, V. Lemos, L. Zhou, V. Cama, M. B. da Cunha, O. Matos, and F. Antunes. 2005. Occurrence and molecular characterization of Cryptosporidium spp. in mammals and reptiles at the Lisbon zoo. Parasitol. Res. 97:108-112. [DOI] [PubMed] [Google Scholar]
- 2.Alves, M., L. Xiao, I. Sulaiman, A. A. Lal, O. Matos, and F. Antunes. 2003. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 41:2744-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atwill, E. R., R. Phillips, M. D. Pereira, X. Li, and B. McCowan. 2004. Seasonal shedding of multiple Cryptosporidium genotypes in California ground squirrels (Spermophilus beecheyi). Appl. Environ. Microbiol. 70:6748-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajer, A., M. Bednarska, A. Pawelczyk, J. M. Behnke, F. S. Gilbert, and E. Sinski. 2002. Prevalence and abundance of Cryptosporidium parvum and Giardia spp. in wild rural rodents from the Mazury Lake District region of Poland. Parasitology 125:21-34. [DOI] [PubMed] [Google Scholar]
- 5.Bajer, A., S. Caccio, M. Bednarska, J. M. Behnke, N. J. Pieniazek, and E. Sinski. 2003. Preliminary molecular characterization of Cryptosporidium parvum isolates of wildlife rodents from Poland. J. Parasitol. 89:1053-1055. [DOI] [PubMed] [Google Scholar]
- 6.Berrilli, F., D. Di Cave, C. D. Orazi, B. Bongiovanni, P. Scaramozzino, F. Scholl, and C. De Liberato. 2006. Giardia and Cryptosporidium in kennel dogs, pet rabbits and hamsters from Rome (Italy). Parassitologia 48:261. [Google Scholar]
- 7.Chalmers, R. M., G. Robinson, K. Elwin, S. J. Hadfield, L. Xiao, U. Ryan, D. Modha, and C. Mallaghan. 2009. Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerg. Infect. Dis. 15:829-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalmers, R. M., A. P. Sturdee, S. A. Bull, A. Miller, and S. E. Wright. 1997. The prevalence of Cryptosporidium parvum and C. muris in Mus domesticus, Apodemus sylvaticus and Clethrionomys glareolus in an agricultural system. Parasitol. Res. 83:478-482. [DOI] [PubMed] [Google Scholar]
- 9.Chalmers, R. M., A. L. Thomas, B. A. Butler, and M. C. Morel. 2005. Identification of Cryptosporidium parvum genotype 2 in domestic horses. Vet. Rec. 156:49-50. [DOI] [PubMed] [Google Scholar]
- 10.Fayer, R. 2007. General biology, p. 1-42. In R. Fayer and L. Xiao (ed.), Cryptosporidium and cryptosporidiosis, 2nd ed. CRC Press, Boca Raton, FL.
- 11.Feng, Y. 27 November 2008, posting date. Cryptosporidium in wild placental mammals. Exp. Parasitol. doi: 10.1016/j.exppara.2008.11.005. [DOI] [PubMed]
- 12.Feng, Y., K. A. Alderisio, W. Yang, L. A. Blancero, W. G. Kuhne, C. A. Nadareski, M. Reid, and L. Xiao. 2007. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73:6475-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foo, C., J. Farrell, A. Boxell, I. Robertson, and U. M. Ryan. 2007. Novel Cryptosporidium genotype in wild Australian mice (Mus domesticus). Appl. Environ. Microbiol. 73:7693-7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatei, W., C. N. Wamae, C. Mbae, A. Waruru, E. Mulinge, T. Waithera, S. M. Gatika, S. K. Kamwati, G. Revathi, and C. A. Hart. 2006. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am. J. Trop. Med. Hyg. 75:78-82. [PubMed] [Google Scholar]
- 15.Giangaspero, A., R. Iorio, B. Paoletti, D. Traversa, and G. Capelli. 2006. Molecular evidence for Cryptosporidium infection in dogs in Central Italy. Parasitol. Res. 99:297-299. [DOI] [PubMed] [Google Scholar]
- 16.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajdusek, O., O. Ditrich, and J. Slapeta. 2004. Molecular identification of Cryptosporidium spp. in animal and human hosts from the Czech Republic. Vet. Parasitol. 122:183-192. [DOI] [PubMed] [Google Scholar]
- 18.Hikosaka, K., and Y. Nakai. 2005. A novel genotype of Cryptosporidium muris from large Japanese field mice, Apodemus speciosus. Parasitol. Res. 97:373-379. [DOI] [PubMed] [Google Scholar]
- 19.Huber, F., S. da Silva, T. C. Bomfim, K. R. Teixeira, and A. R. Bello. 2007. Genotypic characterization and phylogenetic analysis of Cryptosporidium sp. from domestic animals in Brazil. Vet. Parasitol. 150:65-74. [DOI] [PubMed] [Google Scholar]
- 20.Hurkova, L., O. Hajdusek, and D. Modry. 2003. Natural infection of Cryptosporidium muris (Apicomplexa: Cryptosporiidae) in Siberian chipmunks. J. Wildl. Dis. 39:441-444. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, J., K. A. Alderisio, and L. Xiao. 2005. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl. Environ. Microbiol. 71:4446-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karanis, P., J. Plutzer, N. A. Halim, K. Igori, H. Nagasawa, J. Ongerth, and M. Liqing. 2007. Molecular characterization of Cryptosporidium from animal sources in Qinghai province of China. Parasitol. Res. 101:1575-1580. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, A., A. Edagawa, K. Okada, A. Takimoto, S. Yonesho, and P. Karanis. 2007. Detection and genotyping of Cryptosporidium from brown rats (Rattus norvegicus) captured in an urban area of Japan. Parasitol. Res. 100:1417-1420. [DOI] [PubMed] [Google Scholar]
- 24.Kodadkova, A., M. Kvac, O. Ditrich, B. Sak, and L. Xiao. 2009. Cryptosporidium muris in a reticulated giraffe (Giraffa camelopardalis reticulata). J. Parasitol. 17:1. [DOI] [PubMed] [Google Scholar]
- 25.Kvac, M., D. Hanzlikova, B. Sak, and D. Kvetonova. 2009. Prevalence and age-related infection of Cryptosporidium suis, C. muris and Cryptosporidium pig genotype II in pigs on a farm complex in the Czech Republic. Vet. Parasitol. 160:319-322. [DOI] [PubMed] [Google Scholar]
- 26.Kvac, M., L. Hofmannova, S. Bertolino, L. Wauters, G. Tosi, and D. Modry. 2008. Natural infection with two genotypes of Cryptosporidium in red squirrels (Sciurus vulgaris) in Italy. Folia Parasitol. (Praha) 55:95-99. [PubMed] [Google Scholar]
- 27.Kvac, M., B. Sak, D. Hanzlikova, J. Kotilova, and D. Kvetonova. 2009. Molecular characterization of Cryptosporidium isolates from pigs at slaughterhouses in South Bohemia, Czech Republic. Parasitol. Res. 104:425-428. [DOI] [PubMed] [Google Scholar]
- 28.Lupo, P. J., R. C. Langer-Curry, M. Robinson, P. C. Okhuysen, and C. L. Chappell. 2008. Cryptosporidium muris in a Texas canine population. Am. J. Trop. Med. Hyg. 78:917-921. [PubMed] [Google Scholar]
- 29.Matsubayashi, M., N. Abe, K. Takami, I. Kimata, M. Iseki, T. Nakanishi, H. Tani, K. Sasai, and E. Baba. 2004. First record of Cryptosporidium infection in a raccoon dog (Nyctereutes procyonoides viverrinus). Vet. Parasitol. 120:171-175. [DOI] [PubMed] [Google Scholar]
- 30.Meireles, M. V., R. M. Soares, F. Bonello, and S. M. Gennari. 2007. Natural infection with zoonotic subtype of Cryptosporidium parvum in capybara (Hydrochoerus hydrochaeris) from Brazil. Vet. Parasitol. 147:166-170. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, U. M., A. P. Sturdee, G. Singleton, M. S. Gomez, M. Gracenea, J. Torres, S. G. Hamilton, D. P. Woodside, and R. C. Thompson. 1999. The Cryptosporidium “mouse” genotype is conserved across geographic areas. J. Clin. Microbiol. 37:1302-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muthusamy, D., S. S. Rao, S. Ramani, B. Monica, I. Banerjee, O. C. Abraham, D. C. Mathai, B. Primrose, J. Muliyil, C. A. Wanke, H. D. Ward, and G. Kang. 2006. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J. Clin. Microbiol. 44:632-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng, J., I. Pavlasek, and U. Ryan. 2006. Identification of novel Cryptosporidium genotypes from avian hosts. Appl. Environ. Microbiol. 72:7548-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer, C. J., L. Xiao, A. Terashima, H. Guerra, E. Gotuzzo, G. Saldias, J. A. Bonilla, L. Zhou, A. Lindquist, and S. J. Upton. 2003. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg. Infect. Dis. 9:1174-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paziewska, A., M. Bednarska, H. Nieweglowski, G. Karbowiak, and A. Bajer. 2007. Distribution of Cryptosporidium and Giardia spp. in selected species of protected and game mammals from north-eastern Poland. Ann. Agric. Environ. Med. 14:265-270. [PubMed] [Google Scholar]
- 36.Perz, J. F., and S. M. Le Blancq. 2001. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl. Environ. Microbiol. 67:1154-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Power, M. L., C. Cheung-Kwok-Sang, M. Slade, and S. Williamson. 2009. Cryptosporidium fayeri: diversity within the GP60 locus of isolates from different marsupial hosts. Exp. Parasitol. 121:219-223. [DOI] [PubMed] [Google Scholar]
- 38.Quilez, J., E. Torres, R. M. Chalmers, S. J. Hadfield, E. Del Cacho, and C. Sanchez-Acedo. 2008. Cryptosporidium genotypes and subtypes in lambs and goat kids in Spain. Appl. Environ. Microbiol. 74:6026-6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan, U., L. Xiao, C. Read, L. Zhou, A. A. Lal, and I. Pavlasek. 2003. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microbiol. 69:4302-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan, U. M., C. Bath, I. Robertson, C. Read, A. Elliot, L. McInnes, R. Traub, and B. Besier. 2005. Sheep may not be an important zoonotic reservoir for Cryptosporidium and Giardia parasites. Appl. Environ. Microbiol. 71:4992-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suresh, P., and J. E. Rehg. 1996. Comparative evaluation of several techniques for purification of Cryptosporidium parvum oocysts from rat feces. J. Clin. Microbiol. 34:38-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres, J., M. Gracenea, M. S. Gomez, A. Arrizabalaga, and O. Gonzalez-Moreno. 2000. The occurrence of Cryptosporidium parvum and C. muris in wild rodents and insectivores in Spain. Vet. Parasitol. 92:253-260. [DOI] [PubMed] [Google Scholar]
- 43.Twomey, D. F., A. M. Barlow, S. Bell, R. M. Chalmers, K. Elwin, M. Giles, R. J. Higgins, G. Robinson, and R. M. Stringer. 2008. Cryptosporidiosis in two alpaca (Lama pacos) holdings in the south-west of England. Vet. J. 175:419-422. [DOI] [PubMed] [Google Scholar]
- 44.Vetterling, J. M., H. R. Jervis, T. G. Merrill, and H. Sprinz. 1971. Cryptosporidium wrairi sp. n. from the guinea pig Cavia porcellus, with an emendation of the genus. J. Protozool. 18:243-247. [DOI] [PubMed] [Google Scholar]
- 45.Warren, K. S., R. A. Swan, U. M. Morgan-Ryan, J. A. Friend, and A. Elliot. 2003. Cryptosporidium muris infection in bilbies (Macrotis lagotis). Aust. Vet. J. 81:739-741. [DOI] [PubMed] [Google Scholar]
- 46.Wielinga, P. R., A. de Vries, T. H. van der Goot, T. Mank, M. H. Mars, L. M. Kortbeek, and J. W. van der Giessen. 2008. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int. J. Parasitol. 38:809-817. [DOI] [PubMed] [Google Scholar]
- 47.Xiao, L., and R. Fayer. 2008. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int. J. Parasitol. 38:1239-1255. [DOI] [PubMed] [Google Scholar]
- 48.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao, L., and Y. Feng. 2008. Zoonotic cryptosporidiosis. FEMS Immunol. Med. Microbiol. 52:309-323. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, W., P. Chen, E. N. Villegas, R. B. Landy, C. Kanetsky, V. Cama, T. Dearen, C. L. Schultz, K. G. Orndorff, G. J. Prelewicz, M. H. Brown, K. R. Young, and L. Xiao. 2008. Cryptosporidium source tracking in the Potomac River watershed. Appl. Environ. Microbiol. 74:6495-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, L., R. Fayer, J. M. Trout, U. M. Ryan, F. W. Schaefer III, and L. Xiao. 2004. Genotypes of Cryptosporidium species infecting fur-bearing mammals differ from those of species infecting humans. Appl. Environ. Microbiol. 70:7574-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ziegler, P. E., S. E. Wade, S. L. Schaaf, D. A. Stern, C. A. Nadareski, and H. O. Mohammed. 2007. Prevalence of Cryptosporidium species in wildlife populations within a watershed landscape in southeastern New York State. Vet. Parasitol. 147:176-184. [DOI] [PubMed] [Google Scholar]
- 54.Zintl, A., D. Neville, D. Maguire, S. Fanning, G. Mulcahy, H. V. Smith, and T. De Waal. 2007. Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitology 134:1575-1582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.