Abstract
Studying the interactions between enteric pathogens and their environment is important to improving our understanding of their persistence and transmission. However, this remains challenging in large part because of difficulties associated with tracking pathogens in their natural environment(s). In this study, we report a fluorescent labeling strategy which was applied to murine norovirus (MNV-1), a human norovirus surrogate, and hepatitis A virus (HAV). Specifically, streptavidin-labeled Quantum dots (Q-Dots) were bound to biotinylated capsids of MNV-1 and HAV (bio-MNV-1 and bio-HAV); the process was confirmed by using a sandwich-type approach in which streptavidin-bound plates were reacted with biotinylated virus followed by a secondary binding to Q-Dots with an emission range of 635 to 675 nm (Q-Dots 655). The assay demonstrated a relative fluorescence of 528 ± 48.1 and 112 ± 8.6 for bio-MNV-1 and control MNV-1, respectively. The biotinylation process did not impact virus infectivity, nor did it interfere with the interactions between the virus and host cells or model produce items. Using fluorescent microscopy, it was possible to visualize both bio-HAV and bio-MNV-1 attached to the surfaces of permissive mammalian cells and green onion tissue. The method provides a powerful tool for the labeling and detection of enteric viruses (and their surrogates) which can be used to track virus behavior in situ.
Over the last 2 decades, production and consumption of fresh fruits and vegetables have increased dramatically, with an accompanying rise in the number of produce-associated food-borne disease outbreaks (12). It is clear from review of the CDC Food-borne Disease Outbreak database (1996 to 2006; available at: http://www.cdc.gov/foodborneoutbreaks/outbreak_data.htm) that the human noroviruses (HuNoV) are the leading cause of produce-associated food-borne disease. Although most HuNoV outbreaks are small and limited in scope, vegetable salads and berries are notable vehicles (8, 11, 17, 18). Hepatitis A virus (HAV) is the other human enteric virus of concern in fresh produce, and several high-profile outbreaks have occurred in association with HAV-contaminated strawberries and green onions in the last decade (9, 21).
Little is known about the behavior of the enteric viruses in fresh produce items although virus persistence in such products is well documented (reviewed by Papafragkou et al. [14]). It is likely that a complex set of physicochemical interactions governs such persistence, which may be associated with attachment and/or internalization of the virus in the produce item(s). Unfortunately, there are no known means by which to visualize the HuNoV and HAV in situ in order to study such purported virus-surface interactions or other mechanisms by which these viruses persist in fresh produce. In the only study of its kind, Chancellor et al. (5) used fluorescent microspheres as a surrogate to examine the uptake of HAV into green onions. The fluorophores were observed microscopically on the surface and within the green onion tissue over a 60-day period. However, the fluorophores employed in that study ranged from 1 μm to 10 μm in diameter, much larger than a HAV particle (28 to 33 nm) (20), calling into question the relevance of these observations (5).
Quantum dots, or Q-Dots, are small, 10- to 20-nm fluorescent semiconductor nanocrystals composed of a cadmium selenide core coated in a zinc sulfide shell. Q-Dots absorb photons over a wide range of wavelengths but have a very narrow range of emission; in addition, the fluorescence emitted by Q-Dots is both bright and photostable. The fluorescent wavelength of the Q-Dot is determined by the size of the nanocrystal, which moves from red to blue as the Q-Dots decrease in size. A wide range of Q-Dots are available with different emission wavelengths, ranging from 525 nm (green) to 800 nm (blue). Properties such as these make Q-Dots excellent candidates for use in virus labeling for study of surface interactions. For example, Kampani et al. (10) described a Q-Dot-based fluorescence assay that was used to study the attachment of human T-cell leukemia virus type 1 to host cells. In this case, the viral envelope was biotinylated, and this step was followed by attachment of streptavidin-coated Q-Dots. The fluorescently labeled viruses could be visualized binding to the appropriate host cell line using fluorescent microscopy, and data were interpreted both qualitatively and quantitatively.
As HuNoV cannot be cultivated in vitro and as there is no animal model for propagation, a surrogate must be used to study the environmental behavior of these viruses. Recently, a cultivable murine norovirus (MNV-1) has been isolated, and recent studies indicate that this may be an appropriate surrogate for HuNoV (3). In this paper, we report a method for labeling MNV-1 and HAV with streptavidin-coated Q-Dots, as well as the development of a sandwich-type detection assay. We also demonstrate visualization of the labeled viruses bound to the surface of permissive mammalian cell lines and representative fresh produce items.
MATERIALS AND METHODS
Mammalian cell culture, infectivity assay, and virus propagation.
The MNV-1 P3 strain (obtained courtesy of H. Virgin, Washington University, St. Louis, MO) was cultured in 80 to 90% confluent monolayers of RAW 264.7 cells as previously described (3). HAV clone HM-175 (ATCC, Manassas, VA) was cultured in FRhK-4 cells also as previously described (2). In both instances, virus stocks were prepared from infected cell cultures displaying typical cytopathic effects by three sequential freeze-thaw cycles at −80°C, with cellular debris removed by centrifugation at 2,800 × g for 15 min. MNV-1 stock titers approximated 3.0 × 107 PFU/ml; HAV stock titers were around 3.0 × 106 PFU/ml.
Biotinylation of MNV-1 and HAV.
Prior to application of the biotinylation protocol, the virus stock solutions were concentrated by polyethylene glycol precipitation by the addition of 10% polyethylene glycol 8000 (Sigma Aldrich, St. Louis, MO) to 25 ml of stock, followed by overnight incubation at 4°C. The sample was centrifuged at 27,000 × g for 15 min, the supernatant was discarded, and the resulting pellet was resuspended in 1 ml of phosphate-buffered saline (PBS) supplied with an EZ-Link Sulfo-NHS-LC-Biotinylation kit (Pierce, Rockford, IL). The concentrated virus solution was then biotinylated following the manufacturer's instructions. Briefly, the virus solution was first incubated in the presence of a 20-fold molar excess of sulfo-NHS-LC-biotin (sulfosuccinimidyl-6-biotinamido-hexanoate) at room temperature for 60 min, after which the excess biotin reagent was removed by buffer exchange using a Zeba Desalt spin column. The biotinylated virus was eluted from the column in distilled H2O by centrifugation for 2 min at 1,000 × g and stored at 4°C until use in experiments.
Confirmation of MNV-1 and HAV biotinylation (sandwich-type assay).
A Nunc Immobilizer Streptavidin plate (Nalge Nunc International, Rochester, NY) was prepared following the manufacturer's protocol. The biotinylated virus was diluted 1/10 in PBST (PBS, pH 7.2, and 0.05% Tween 20), and 100-μl aliquots were added to the wells of the plate and incubated at room temperature for 60 min with gentle agitation. The virus was aspirated from the wells, which were then washed three times with 300 μl of PBST. One hundred microliters of 10 or 50 nM streptavidin-coated Q-Dots 655 (Q-Dots with a fluorescence range of 635 to 675 nm) (Invitrogen, Carlsbad, CA) was added to the wells and incubated at room temperature, protected from light with gentle agitation for 30 min. The wells were then washed three times with 300 μl of PBST, after which corresponding fluorescence was read using a Biotek Synergy HT plate reader (Winooski, VT) at an excitation of 360 nm and an emission of 645 nm.
Biotinylated virus detection and binding to mammalian cells and fresh produce.
RAW 264.7 and FRhK-4 cells were seeded into 24-well Corning Costar flat-bottomed cell culture plates (Corning, Corning, NY) (seeding densities of 1 × 106 cells/ml and 5 × 104 cells/ml for RAW 264.7 and FRhK-4 cells, respectively) and grown to confluence at 37°C in 5% CO2 for 24 h (RAW 264.7) or 48 h (FRhK-4). The biotin-labeled virus suspensions were diluted 1/10 or 1/100 and bound to the cells following the protocol described by Kampani et al. (10). Briefly, 40 μl of virus suspension was added to the cells, with incubation at 37°C in 5% CO2 for 1 h with periodic gentle mixing, after which the cells were washed three times with 300 μl of PBS (FRhK-4) or Earle's balanced salt solution (RAW 264.7). Forty microliters of 10 nM streptavidin-labeled Q-Dots 655 was added to the wells and incubated for 30 min at 4°C. The cells were then fixed by the addition of 500 μl of ice-cold 4% formaldehyde, followed by incubation on ice for 5 min and at 4°C for 10 min. After the cells were washed three times with 300 μl of PBS or Earle's balanced salt solution, fluorescence was read as described above at an excitation of 360 nm and an emission of 645 nm.
Real-time RT-qPCR.
Two different reverse transcription-quantitative PCR (RT-qPCR) assays were used in this study, one for MNV-1 and one for HAV. The primer and probe sequences for detection of MNV-1 RNA were as follows: primer G54763F, 5′-TGATCGTGCCAGCATCGA-3′; primer G54863R, 5′-GTTGGGAGGGTCTCTGAGCAT-3′; and probe G54808, 5′-FAM-CTACCCACCAGAACCCCTTTGAGACTC-BHQ1-3′ (where FAM is 6-carboxyfluorescein and BHQ is Black Hole quencher) (sequences provided by J. Vinje, U.S. Centers for Disease Control and Prevention, personal communication). Real-time RT-qPCR was performed utilizing a Qiagen OneStep RT-PCR Kit (Qiagen, Valencia CA). Amplification reagents consisted of a 400 nM concentration of each primer, 200 nM probe, 400 μM of each deoxynucleoside triphosphate, 10 U of RNase inhibitor (Promega, Madison, WI), 2 μl of Qiagen OneStep RT-PCR enzyme mix, 1× Qiagen OneStep RT-PCR buffer, and 2.5 μl of RNA in a final volume of 25 μl. Amplifications were performed using a SmartCycler (Cepheid, Sunnyvale, CA) under conditions of 50°C for 30 min and 95°C for 15 min, followed by 45 cycles of 95°C 10 s, 55°C for 15 s (optics on), and 72°C for 30 s.
HAV RT-qPCR was performed as described by Papafragkou et al. (15) using the following primers and probes: forward primer, 5′-GGTAGGCTACGGGTGAAAC-3′; reverse primer, 5′-AACAACTCACCAATATCCGC-3′; and probe, 5′-FAM-CTTAGGCTAATACTTCTATGAAGAGATGC-BHQ1-3′. The RT-qPCR reaction was performed using a Quantitect Probe RT-PCR kit (Qiagen) following the manufacturer's instructions and contained a 400 nM concentration of each primer, 150 nM probe, and 2.5 μl of RNA in a final volume of 25 μl. All amplifications were done using a SmartCycler II and beginning with 50°C for 30 min and 95°C for 15 min, followed by 45 cycles of 95°C for 10 s, 55°C for 20 s (optics on), and 72°C for 15 s.
Microscopy.
In order to visualize virus binding to host cells, minor methodological modifications were required. Specifically, RAW 264-7 and FRhK-4 cells were propagated as described above in six-well plates (Corning, Corning, NY) containing sterile glass coverslips. After growth to confluence, the cells were exposed to 1/10 diluted biotinylated virus stocks and incubated for 1 h at 37°C. The cells were then washed three times and incubated with 10 nM streptavidin-labeled Q-Dots 655 for 30 min at 4°C. The Q-Dots were removed from the cells, and the cells were washed once before being fixed with formaldehyde as described above. After cells were fixed, the coverslip was placed onto a microscope slide, and slides were viewed using laser confocal microscopy with a Leica TCS SPI confocal microscope (Leica Microsystems Inc., Bannockburn, IL) under a magnification of ×63 at an excitation of 488 nm and emission in the range of 635 nm to 675 nm.
Green onions were used as the model food commodity to demonstrate virus binding to fresh produce. Specifically, the first (outer) layer of onion was removed, and small (approximately 1 cm2) squares of tissue were cut from the onion using an X-Acto knife. The tissue slices were placed into the wells of a six-well sterile plate to which 100 μl of diluted HAV (at a final concentration of approximately 6.5 × 105 PFU/ml) was added to the surface of the onion slice, and cultures were incubated at room temperature for 2 h. The samples were washed three times with either 1 ml of PBS or 1 ml of beef extract buffer (50 mM glycine, 100 mM Tris, 1% [wt/vol] beef extract [pH 9.5]). This was followed by the addition of 100 μl of 10 nM Q-Dots 655, incubation for 30 min at room temperature, and washing the samples three times with 1 ml (each) of PBS. The epidermis was then removed from the piece of green onion and placed on a microscope slide with the waxy cuticle next to the coverslip. The samples were viewed by laser confocal microscopy at a magnification ×20 and an excitation of 488 nm and emission in the range of 635 nm to 675 nm.
Statistical analysis.
When required, statistical analysis was performed using a Tukey-Kramer multiple comparisons test with a P value of ≤0.05 (GraphPad InStat, GraphPad InStat Software Inc., La Jolla, CA).
RESULTS
Confirmation of virus biotinylation.
To ensure that both HAV and MNV-1 had been successfully biotinylated, a sandwich-type assay was designed. This approach consisted of streptavidin labeling of a solid support surface, followed by sequential addition of biotinylated virus, a washing step, and then addition of streptavidin-coated Q-Dots. A positive binding reaction was demonstrated as an increase in fluorescence and indicated successful biotinylation of virus stocks. The controls, which were designed to monitor degree of background fluorescence as well as nonspecific binding, consisted of PBST alone, Q-Dots alone, and biotinylated HAV (bio-HAV) or bio-MNV-1 alone. Results, shown in Table 1, clearly demonstrate a marked and statistically significant increase in the relative fluorescence of the bio-HAV and bio-MNV-1 viruses compared to controls. Initial experiments were performed utilizing 10 nM or 50 nM Q-Dot concentrations. However, in the absence of a statistically significant difference between results at the 10 and 50 nM concentrations, all further experiments were done using a 10 nM Q-Dot concentration.
TABLE 1.
Demonstration of biotinylation of HAV and MNV-1 using Q-Dot binding assay
| Sample | Relative fluorescence of the indicated virusa |
|
|---|---|---|
| HAV | MNV-1 | |
| Wells only | 22 ± 5.6A | 24 ± 1.8A |
| PBST only | 19.5 ± 1.0A | 20.75 ± 1.3A |
| 10 nM Q-Dots only | 21 ± 2.3A | 22 ± 2.8A |
| 50 nM Q-Dots only | 21 ± 2.7A | 20.75 ± 1.5A |
| Bio-virus only | 22.5 ± 1.7A | 23.25 ± 1.3A |
| Virus only | 22.25 ± 2.2A | 21.25 ± 2.2A |
| Bio-virus and 10 nM Q-Dots | 1,337 ± 68.8B | 1,049 ± 48.4B |
| Bio-virus and 50 nM Q-Dots | 1,434 ± 72.3C | 1,419 ± 55.8C |
| Virus and 10 nM Q-Dots | 22.5 ± 4.4A | 23.5 ± 2.9A |
| Virus and 50 nM Q-Dots | 26.25 ± 3.6A | 26.75 ± 4.0 |
Results are an average of four readings. Different superior letters indicate a statistically significant difference between the results (P ≤ 0.05).
Effect of biotinylation on virus binding/attachment properties.
In an effort to ensure that the biotinylation process did not otherwise impact virus infectivity or binding efficiency, several validation experiments were undertaken. There was no statistically significant difference in the titers of parental virus stocks and their biotinylated counterparts, as evaluated by plaque assay (data not shown). In another set of experiments, biotinylated and nonbiotinylated viruses were applied to the surface of a model food (raspberries for MNV-1 and green onions for HAV), allowed to adsorb, and then eluted from the food surface. The eluate was extracted for RNA, and the titer of eluted virus was approximated using RT-qPCR. Again, there were no statistically significant differences between the resulting threshold cycle values of biotinylated and nonbiotinylated viruses (data not shown). Taken together, these data demonstrate that the biotinylation process did not appear to influence virus infectivity or binding characteristics, at least for the food matrices studied in these experiments.
Binding and visualization of biotinylated viruses to permissive host cells.
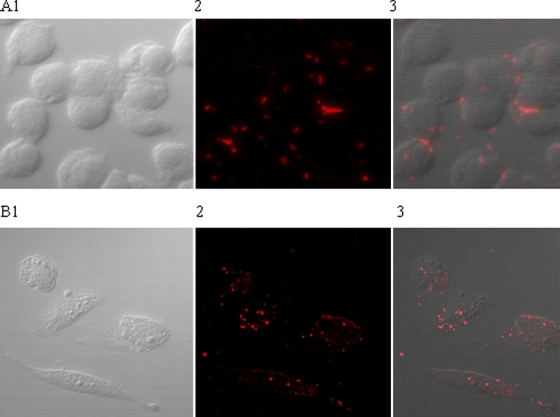
To further compare the behavior of bio-HAV and bio-MNV-1 relative to the wild-type stocks, 24-well plates with confluent FRhK-4 or RAW 264.7 cells were exposed to biotinylated virus. This was followed by the addition of streptavidin-coated Q-Dots 655. Table 2 shows that increased relative fluorescence was observed only in the presence of both Q-Dots 655 and bio-HAV or bio-MNV-1, suggesting that the biotinylated viruses were able to adhere to their host cells. These data are supported by fluorescent microscopy experiments which illustrate clear visualization of biotinylated virus (and no matrix-associated background fluorescence) after attachment to host cells and subsequent exposure to streptavidin-coated Q-Dots (Fig. 1). Negative control samples (cells alone, cells challenged with nonbiotinylated virus but without subsequent Q-Dots 655 treatment, or cells challenged with nonbiotinylated virus) showed no fluorescence (data not shown).
TABLE 2.
Demonstration of bio-HAV and bio-MNV-1 binding to their relevant host cell lines
| Sample | Relative fluorescence of the indicated cellsa |
|
|---|---|---|
| HAV-FRhk-4 | MNV-1-RAW 264.7 | |
| Cells only | 138 ± 1.7A | 133 ± 6.2A |
| 10 nM Q-Dots only | 134 ± 4.6A | 153 ± 12.6A |
| Bio-virus only (10−1) | 144 ± 9.2A | 137 ± 11.0A |
| Bio-virus only (10−2) | 133 ± 5.1A | 137 ± 17.8A |
| Virus only 10−1 | 132 ± 6.2A | 145 ± 3.7A |
| Virus only (10−2) | 133 ± 7.1A | 137 ± 8.8A |
| Bio-virus (10−1) and 10 nM Q-Dots | 450 ± 65.7B | 455 ± 58.4B |
| Bio-virus (10−2) and 10 nM Q-Dots | 216 ± 6.8C | 260 ± 33.4C |
| Virus (10−1) and 10 nM Q-Dots | 135 ± 4.4A | 163 ± 10.1A |
| Virus (10−2)and 10 nM Q-Dots | 142 ± 1.0A | 156 ± 14.5A |
Results are an average of four readings. Different superior letters indicate a statistically significant difference between the results (P ≤ 0.05).
FIG. 1.
Visualization of biotinylated viruses binding to host cells after exposure to Q-Dots 655. Panel A corresponds to the RAW 264.7 cells and bio-MNV-1, and panel B shows FRhK-4 cells and bio-HAV. Image 1 shows the host cells only, image 2 shows the fluorescently labeled viruses after challenge with biotinylated virus and subsequent addition of Q-Dots 655, and image 3 is an overlay of images 1 and 2.
Visualization of HAV bound to the surface of green onion tissue using biotinylated virus and streptavidin-coated Q-Dots.
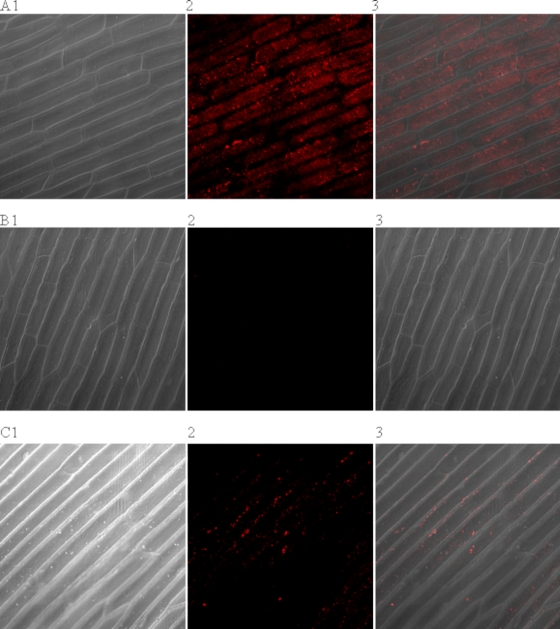
When dissected green onion tissue was exposed to a high titer (approximately 6.5 × 105 PFU/ml) of bio-HAV, it was possible to visualize the virus, with no matrix-associated background fluorescence, using confocal microscopy (Fig. 2A). Rinsing the contaminated green onion tissue with 1% beef extract, a commonly used virus eluant, greatly reduced the number of virus-Q-Dot conjugates present on the onion epidermis although, as expected, some virus remained (Fig. 2C). Control experiments performed with nonbiotinylated HAV and Q-Dots (data not shown) and Q-Dots on their own (Fig. 2 B) did not produce any visible fluorescence on the surface of the onion.
FIG. 2.
Visualization of biotinylated virus binding to the surface of onion epidermis. (A) Onion epidermis after exposure to bio-HAV and Q-Dots 655. (B) Onion treated with the Q-Dots only. (C) Onion epidermis after the bio-HAV had been eluted from the surface with beef extract buffer before the addition of the Q-Dots. Image 1 shows the onion epidermis only under light microscopy, image 2 shows the fluorescence from the Q-Dots, and image 3 is an overlay of images 1 and 2.
DISCUSSION
With increasing recognition of the importance of enteric viruses in the overall burden of produce-associated food-borne disease, there remains a need to develop novel techniques which can be used to study the interactions of these agents with product tissue. Typically, these sorts of studies have entailed the spotting of virus suspensions on the surface of the food product, followed by washing and enumeration using mammalian cell culture infectivity assays or real-time RT-PCR. This approach is both time-consuming and inefficient, yielding results which may be difficult to interpret. Only one study to date has used fluorescent fluorophores to follow the migration of virus (HAV) in green onion tissue (5), but the method was limited because the fluorophores were used as a proxy for infectious virus; hence, direct comparisons between virus behavior and that of the fluorescent particles were not possible. Also, the fluorophores utilized in that study were considerably larger (1 to 10 μm diameter) than virus particles (approximately 20 to 30 nm diameter). The Q-Dot method outlined in this paper provides a more relevant model for in situ visualization of enteric viruses.
The utilization of Q-Dots has many advantages over current techniques for the study of virus-food interactions. For example, although fluorescent in situ hybridization is commonly used to facilitate visualization of bacteria and, to a lesser extent, viruses on foods (6, 7, 13), the fluorescent dyes used in these techniques are generally not photostable and tend to bleach quickly, requiring immediate imaging of the sample (4, 16). Also, auto-fluorescence of the plant tissue is particularly problematic when fluorescent in situ hybridization is used with produce samples due to the wide emission ranges of the dyes involved (4, 16). Both of these issues can be overcome using Q-Dot technology as Q-Dots are very photostable (1), and, due to their narrow range of wavelength emission, Q-Dots can be chosen with consideration of matrix effects such that auto-fluorescence associated with plant chlorophyll does not interfere with experimental design or interpretation. In this work we used Q-Dots 655, which fluoresce maximally at 655 nm (range 635 to 675 nm). Indeed, we initially used Q-Dots 525 for the cell culture experiments but observed auto-fluorescence that was easily overcome by substituting Q-Dots 655. In addition, the narrow emission range of these Q-Dots fell outside of the chlorophyll auto-fluorescence wavelength (685 nm), making this an optimal choice for plant-based products.
We also demonstrated that biotinylation of the virus stock solutions did not appear to affect their behavior when they were exposed to permissive mammalian cells or candidate food matrices. Specifically, we showed that the biotinylated viruses were able to both bind and infect host cells at an efficiency which did not differ from that of nonbiotinylated virus. Although biotin is small molecule (224 Da) and can bind to proteins without affecting their biological function, the physicochemical interactions involved with viral adherence to produce are complex, and a demonstration that the biotinylation process did not otherwise influence virus attachment is critical for the practical use of this approach to study virus-matrix interactions. In addition, the biotinylation process did not appear to impact HAV or MNV-1 infectivity, which is somewhat different from the results of Skulstad et al. (19), who reported that the infectivity of herpes simplex virus type 1 was reduced by 40% when the virus is biotinylated. However, herpes simplex virus is an enveloped virus, whereas both HAV and MNV-1 are nonenveloped, which may explain why their infectivity was not affected by the biotinylation process.
In this work, we have established proof-of-concept that human enteric viruses (HAV and MNV-1, a culturable HuNoV surrogate) could be biotinylated and used in conjunction with fluorescent Q-Dots for in situ investigation of their interactions with the external environment. Clearly, the virus biotinylation process did not negatively impact virus infectivity or binding characteristics, and virus interactions could be clearly visualized by fluorescent microscopy. This approach provides a powerful tool to study interactions between nonenveloped enteric viruses and their external environments, which should improve our ability to characterize the importance of virus attachment, internalization, and persistence in fresh produce items which may be at risk for human enteric virus contamination.
Acknowledgments
We thank Eva Johannes at the North Carolina State Cellular and Molecular Imaging Facility for all her help with the confocal microscopy work.
This study was supported in part by a grant from the U.S. Department of Agriculture, Cooperative State Research, Education and Extension Service, National Research Initiative, Competitive Grants Program, Biological Approaches to Food Safety, Project 2007-35201-18373.
The use of trade names in this publication does not imply endorsement by the North Carolina Agricultural Research Service or criticism of similar ones not mentioned.
The manuscript is designated FSR09-34 in the journal series of the Department of Food, Bioprocessing, and Nutrition Sciences, North Carolina State University.
Footnotes
Published ahead of print on 23 October 2009.
REFERENCES
- 1.Arya, H., Z. Kaul, R. Wadhwa, K. Taira, T. Hirano, and S. C. Kaul. 2005. Quantum dots in bio-imaging: revolution by the small. Biochem. Biophys. Res. Commun. 329:1173-1177. [DOI] [PubMed] [Google Scholar]
- 2.Bidawid, S., J. M. Farber, S. A. Sattar, and S. Hayward. 2000. Heat inactivation of hepatitis A virus in dairy foods. J. Food Prot. 63:522-528. [DOI] [PubMed] [Google Scholar]
- 3.Cannon, J. L., E. Papafragkou, G. W. Park, J. Osborne, L. A. Jaykus, and J. Vinje. 2006. Surrogates for the study of norovirus stability and inactivation in the environment: a comparison of murine norovirus and feline calicivirus. J. Food Prot. 69:2761-2765. [DOI] [PubMed] [Google Scholar]
- 4.Chan, W. C., and S. Nie. 1998. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281:2016-2018. [DOI] [PubMed] [Google Scholar]
- 5.Chancellor, D. D., S. Tyagi, M. C. Bazaco, S. Bacvinskas, M. B. Chancellor, V. M. Dato, and F. de Miguel. 2006. Green onions: potential mechanism for hepatitis A contamination. J. Food Prot. 69:1468-1472. [DOI] [PubMed] [Google Scholar]
- 6.Fang, Q., S. Brockmann, K. Botzenhart, and A. Wiedenmann. 2003. Improved detection of Salmonella spp. in foods by fluorescent in situ hybridization with 23S rRNA probes: a comparison with conventional culture methods. J. Food Prot. 66:723-731. [DOI] [PubMed] [Google Scholar]
- 7.Harders, J., N. Lukacs, M. Robert-Nicoud, T. M. Jovin, and D. Riesner. 1989. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hjertqvist, M., A. Johansson, N. Svensson, P. E. Åbom, C. Magnusson, M. Olsson, K. O. Hedlund, and Y. Andersson. 2006. Four outbreaks of norovirus gastroenteritis after consuming raspberries, Sweden, June-August 2006. Eurosurveillance 11:E060907. [DOI] [PubMed] [Google Scholar]
- 9.Hutin, Y. J. F., V. Pool, E. H. Cramer, O. V. Nainan, J. Weth, I. T. Williams, S. T. Goldstein, K. F. Gensheimer, B. P. Bell, C. N. Shapiro, M. J. Alter, H. S. Margolis, and the National Hepatitis A Investigation Team. 1999. A multistate, foodborne outbreak of hepatitis A. N. Engl. J. Med. 340:595-602. [DOI] [PubMed] [Google Scholar]
- 10.Kampani, K., K. Quann, J. Ahuja, B. Wigdahl, Z. K. Khan, and P. Jain. 2007. A novel high throughput quantum dot-based fluorescence assay for quantitation of virus binding and attachment. J. Virol. Methods 141:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Guyader, F. S., C. Mittelholzer, L. Haugarreau, K.-O. Hedlund, R. Alsterlund, M. Pommepuy, and L. Svensson. 2004. Detection of noroviruses in raspberries associated with a gastroenteritis outbreak. Int. J. Food Microbiol. 97:179-186. [DOI] [PubMed] [Google Scholar]
- 12.Leon, J. S., L. Jaykus, and C. Moe. 2009. Microbiology of fruits and vegetables, p. 255-290. In N. Heredia, I. Wesley, and S. Garcia (ed.), Microbiologically safe foods. John Wiley and Sons, Inc., Hoboken, NJ.
- 13.Ootsubo, M., T. Shimizu, R. Tanaka, T. Sawabe, K. Tajima, M. Yoshimizu, Y. Ezura, T. Ezaki, and H. Oyaizu. 2002. Oligonucleotide probe for detecting Enterobacteriaceae by in situ hybridization. J. Appl. Microbiol. 93:60-68. [DOI] [PubMed] [Google Scholar]
- 14.Papafragkou, E., D. H. D'Souza, and L. A. Jaykus. 2006. Foodborne transmitted viruses: prevention and control, p. 289-330. In S. Goyal (ed.), Viruses in foods. Springer, New York, NY.
- 15.Papafragkou, E., M. Plante, K. Mattison, S. Bidawid, K. Karthikeyan, J. M. Farber, and L. A. Jaykus. 2008. Rapid and sensitive detection of hepatitis A virus in representative food matrices. J. Virol. Methods 147:177-187. [DOI] [PubMed] [Google Scholar]
- 16.Parak, W. J., T. Pellegrino, and C. Plank. 2005. Labelling of cells with quantum dots. Nanotechnology 16:R9-R25. [DOI] [PubMed] [Google Scholar]
- 17.Schmid, D., H. Stüger, I. Lederer, A. M. Pichler, G. Kainz-Arnfelser, E. Schreier, and F. Allerberger. 2007. A foodborne norovirus outbreak due to manually prepared salad, Austria 2006. Infection 35:232-239. [DOI] [PubMed] [Google Scholar]
- 18.Showell, D., T. Sundkvist, M. Reacher, and J. Gray. 2007. Norovirus outbreak associated with canteen salad in Suffolk, United Kingdom. Eurosurveillance 12:E071129.6.. [DOI] [PubMed] [Google Scholar]
- 19.Skulstad, S., E. Rodahl, K. Jakobsen, N. Langeland, and L. Haarr. 1995. Labeling of surface proteins of herpes simplex virus type 1 using a modified biotin-streptavidin system. Virus Res. 37:253-270. [DOI] [PubMed] [Google Scholar]
- 20.Volkin, D. B., C. J. Burke, K. E. Marfia, C. B. Oswald, B. Wolanski, and C. R. Middaugh. 1997. Size and conformational stability of the hepatitis A virus used to prepare VAQTA, a highly purified inactivated vaccine. J. Pharm. Sci. 86:666-673. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler, C., T. M. Vogt, G. L. Armstrong, G. Vaughan, A. Weltman, O. V. Nainan, V. Dato, G. Xia, K. Waller, J. Amon, T. M. Lee, A. Highbaugh-Battle, C. Hembree, S. Evenson, M. A. Ruta, I. T. Williams, A. E. Fiore, and B. P. Bell. 2005. An outbreak of hepatitis A associated with green onions. N. Engl. J. Med. 353:890-897. [DOI] [PubMed] [Google Scholar]