Abstract
Campylobacter jejuni, a gram-negative, microaerophilic bacterium, is a predominant cause of bacterial gastroenteritis in humans. Although considered fragile and fastidious and lacking many classical stress response mechanisms, C. jejuni exhibits a remarkable capacity for survival and adaptation, successfully infecting humans and persisting in the environment. Consequently, understanding the physiological and genetic properties that allow C. jejuni to survive and adapt to various stress conditions is crucial for therapeutic interventions. Of importance is polyphosphate (poly-P) kinase 1 (PPK1), which is a key enzyme mediating the synthesis of poly-P, an essential molecule for survival, mediating stress responses, host colonization, and virulence in many bacteria. Therefore, we investigated the role of PPK1 in C. jejuni pathogenesis, stress survival, and adaptation. Our findings demonstrate that a C. jejuni Δppk1 mutant was deficient in poly-P accumulation, which was associated with a decreased ability to form viable-but-nonculturable cells under acid stress. The Δppk1 mutant also showed a decreased frequency of natural transformation and an increased susceptibility to various antimicrobials. Furthermore, the Δppk1 mutant was characterized by a dose-dependent deficiency in chicken colonization. Complementation of the Δppk1 mutant with the wild-type copy of ppk1 restored the deficient phenotypes to levels similar to those of the wild type. Our results suggest that poly-P plays an important role in stress survival and adaptation and might contribute to genome plasticity and the spread and development of antimicrobial resistance in C. jejuni. These findings highlight the potential of PPK1 as a novel target for therapeutic interventions.
Campylobacter jejuni, a gram-negative, microaerophilic bacterium, occurs as a commensal among the intestinal microflora of various animals, especially chickens and cattle (6, 73). However, C. jejuni can infect human hosts, invading the intestinal mucosa and causing watery and/or bloody diarrhea (9). C. jejuni is transmitted to humans primarily through the consumption of contaminated chicken products, raw milk, or water (2, 3). Currently, C. jejuni is considered a leading bacterial cause of human food-borne gastroenteritis (3, 61) and has also been associated with a plethora of symptoms, including acute neuromuscular paralysis (Guillain-Barré syndrome) (26). Since an appropriate vaccine for human campylobacteriosis has yet to be introduced, it has been suggested that C. jejuni infections might be alternatively controlled by reducing colonization in food animals (73). Consequently, determining the physiological and genetic properties that allow the survival of C. jejuni and its colonization of animal hosts, pathogenicity, and adaptation to various stresses is of critical importance.
The mechanisms underlying C. jejuni adaptation and survival under stresses imposed by its environment and host are not well understood. High variability between different C. jejuni strains and the unavailability of appropriate genetic tools and animal models have contributed to the lack of knowledge regarding its stress tolerance and pathogenicity. However, it is suggested that the capacity of C. jejuni to form viable-but-nonculturable (VBNC) cells under stress (14) and its readiness for natural transformation (68) and acquiring resistance to antibiotics (39) are among the strategies that promote stress adaptation and survival. Although little is known about the genetics underlying these processes, recent advances in C. jejuni genomics show that this bacterium carries several important genes that might play key roles in mediating stress adaptation and survival. Of particular interest are genes encoding polyphosphate (poly-P) kinases, ppk1 (CJJ81176_1361) and ppk2 (CJJ81176_0633), that were predicted to be involved in the metabolism of poly-P (22, 25, 47), an intracellular granule that impacts several physiological properties in many bacterial species, including pathogenicity, host colonization, adaptation to different environments, and survival (28, 31, 46).
Poly-P kinase 1 (PPK1) is encoded by ppk1, which mediates the synthesis of all or most of the poly-P in the cell (33), while ppk2 encodes an enzyme (PPK2) that synthesizes GTP from poly-P (27). Both ppk genes have been associated with the metabolism of poly-P, which consists of phosphate residues that are linked by high-energy phosphoanhydride bonds and is widely distributed in bacterial species (60). Previous reports showed that poly-P plays important roles in bacterial survival and stress tolerance, including ATP production (8), entry of DNA through membrane channels (13, 54), capsule composition (67), maintaining nutritional requirements during starvation (34), motility, biofilm formation, and resistance to oxidative, osmotic, heat, acid and alkaline stresses, and stationary-phase survival (28, 31, 46, 48, 50, 52, 65). Because of their importance in many bacterial species, it is not surprising to assume a role for PPK and poly-P in C. jejuni survival, colonization, and stress tolerance (8).
Interestingly, PPK1 has been shown to be important for C. jejuni stress responses and pathogenicity (10). However, the role of ppk1 in key metabolic and physiological responses of C. jejuni still needs further analysis. For instance, it has been proposed that during starvation, poly-P might act as a reservoir for phosphorus and energy (7). Subsequently, poly-P would be crucial for maintaining viability/metabolism in stressed cells. This has been observed in H. pylori, where the occurrence of poly-P correlated with culturability and structurally intact cells (45). Poly-P-containing nonculturable H. pylori showed a capacity for ATP and mRNA synthesis after a nutrient stimulus (45). Consequently, poly-P might be an important factor for the formation of VBNC cells by stressed bacteria, including C. jejuni. Furthermore, natural transformation is perhaps one of the most important mechanisms in the adaptation of C. jejuni, and poly-P has been reported to play a role in the entry of DNA through membrane channels (13, 54). It follows that poly-P might be important for natural transformation, adaptation, and acquisition of antibiotic resistance genes in C. jejuni. Poly-P can further impact the survival and adaptation in C. jejuni by modulating antibiotic resistance properties. For example, poly-P interacted with Escherichia coli ribosomes (42), which are known targets of several antibiotics. These observations suggest that ppk1 might be linked to important physiology and functions such as VBNC cell formation, natural transformation, and antimicrobial resistance in C. jejuni. Therefore, in the present study, we determined the contribution of PPK1 to C. jejuni stress responses and adaptation, including the ability to form VBNC cells under acid stress, natural transformation, and antimicrobial resistance. Furthermore, we assessed the impact of ppk1 deletion on in vivo chicken colonization. Our findings highlight the importance of PPK1 in C. jejuni survival, adaptation to different environmental stresses, and in vivo colonization. These findings also indicate the suitability of PPK1 as a potential target for controlling the proliferation of this pathogen.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All of the bacterial strains and plasmids used in this study are described in Table 1. C. jejuni 81-176 is a highly invasive strain originally isolated from diarrheic patients (32). C. jejuni strains were cultured on Mueller-Hinton (MH) medium under microaerophilic conditions (85% N2, 10% CO2, 5% O2) at 42°C for 24 h. For isolation of C. jejuni from chicken feces and organs, MH agar plates were supplemented with Campylobacter selective supplement (SR117E; Oxoid, Lenexa, KS). E. coli strain DH5α was used for plasmid propagation and cloning purposes. E. coli strains were grown on Luria-Bertani medium at 37°C overnight. Chloramphenicol (20 μg/ml for E. coli and 10 μg/ml for Campylobacter), kanamycin (30 μg/ml), and zeocin (50 μg/ml) were added to the media where necessary.
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant description | Source or reference |
|---|---|---|
| Strains | ||
| C. jejuni | ||
| 81-176 | Wild type | Qijing Zhang |
| Δppk1/DG001 | 81-176 derivative with deletion in ppk1 gene; ppk1::Kan | This study |
| Δppk1c/DG002 | DG001 harboring pDG3, Cm | This study |
| 81-176 (dcuA::tetO) | Carries tetracycline resistance marker for natural transformation; Tet | Qijing Zhang |
| E. coli DH5α | Used for cloning | Invitrogen |
| Plasmids | ||
| pZErO-1 | Cloning vector for making suicide vector; Zeo | Invitrogen |
| pRY111 | E. coli-Campylobacter shuttle vector for complementation | 71 |
| pRK2013 | Helper plasmid for conjugation | 1 |
| pUC4K | Source for kanamycin | Amersham |
| pDG1 | pZErO-1 containing ppk1 region plus 1 kb upstream and downstream of 81-176; Zeo | This study |
| pDG2 | A portion of ppk1 replaced with kanamycin resistance region from pUC4K in pDG1; Zeo Kan | This study |
| pDG3 | pRY111 containing ppk1 coding region and the upstream promoter sequence for complementation; Cm | This study |
Kan, kanamycin; Cm, chloramphenicol; Zeo, zeocin; tetO, tetracycline resistance gene.
Construction of the C. jejuni ppk1 mutant and the complemented strain.
Genomic DNA was extracted from C. jejuni strain 81-176 with a Masterpure DNA purification kit (Epicentre, Madison, WI) according to the manufacturer's instructions. Oligonucleotides were designed from the published genome sequence of C. jejuni strain 81-176 (GenBank accession no. NC_008787) with Vector NTI software and commercially synthesized by Integrated DNA Technologies (Skokie, IL). All of the oligonucleotides used in this study are listed in Table 2. Appropriate restriction sites were included in the primers to facilitate digestion and ligation into cloning vectors. Restriction enzymes were purchased from Promega (Madison, WI). To generate the deletion mutant, ppk1 was amplified by using the PPK1 F and PPK1 R primers from the C. jejuni genomic DNA along with approximately 1 kb of DNA sequences upstream and downstream of the target gene. The PCR products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA). Purified PCR products were ligated into zeocin-resistant pZErO-1 (Invitrogen, Carlsbad, CA), a zero background cloning vector, with a Fast-Link DNA ligation kit (Epicentre) and transformed to Library Efficiency DH5α E. coli competent cells (Invitrogen) to generate the plasmid pDG1. Plasmid (pDG1) DNA was isolated with a QIAprep spin mini prep kit (Qiagen), and the whole plasmid, except the target gene, was amplified by inverse PCR with the PPK1 INV F and PPK1 INV R primers. Purified inverse PCR products were ligated to a kanamycin resistant cassette from pUC4K, and the resulting suicide vector was named pDG2. pDG2 was electroporated into C. jejuni as described by Wilson et al. (69). Briefly, C. jejuni strains were grown on MH agar plates at 42°C for 24 h under microaerophilic conditions and the cells were harvested in 1 ml sterile, chilled, ice-cold wash buffer (15% glycerol, 272 mM sucrose) and pelleted (13,000 rpm for 5 min at 4°C). The cells were then washed three times in ice-cold wash buffer and resuspended in 500 μl of ice-cold wash buffer, and 50 μl of cell suspension was used for electroporation. One microgram of pDG2 plasmid DNA was added to 50 μl of the cell suspension and mixed thoroughly by pipetting. The cell-DNA mixture was transferred into a prechilled 0.2-cm-wide cuvette and electroporated at 2.5 kV, 200 Ω, and 25 μF with a Gene Pulser Xcell (Bio-Rad, Hercules, CA). One hundred fifty microliters of SOC medium (Invitrogen) was added to stabilize the cells, and the suspension was spotted onto a nonselective MH agar plate and incubated overnight at 42°C. Following incubation, the cells were harvested, plated onto MH agar with kanamycin (30 μg/ml), and incubated at 42°C for 2 to 3 days under microaerophilic conditions. The kanamycin-resistant colonies were patched and streak purified, and the ppk1 deletion was confirmed by PCR with the PPK1 F and PPK1 R primers. One of the PCR-confirmed mutant clones (DG001) was selected for further studies.
TABLE 2.
Primers used in this study
| Name | Sequence (5′-3′)a | Use |
|---|---|---|
| PPK1 F | ATAAAAGGTACCATTTAGCCATAAAACTCCCG | Amplification of 81-176 ppk1 region |
| PPK1 R | AAAAAACTGCAGGCGATAGGTTTGAACCTTTA | |
| PPK1 INV F | ATAAAAGGATCCTACAGATCAAAGTCGTGCAA | Amplification of pDG1 except ppk1 |
| PPK1 INV R | AAAAAAGGATCCGAACATTGGTCTAAAACACG | |
| PPK1 COMP F | AAATAACTGCAGGTCCTAAACCACTTTGCGCT | Amplification of ppk1 gene for complementation |
| PPK1 COMP R | AAATAAGGTACCCCTCGCTTCCACCAGTTTTA | |
| CsrA F | TTATCGGAGAAGGTATAG | Real-time quantitative PCR amplification of cDNA from ppk2, csrA, spoT, phosR, cmeC, pstS, pstC, and CJJ81176_0750 from 81-176 strain |
| CsrA R | TTTCTAAGTATCATAAGGG | |
| SpoT F | GTAACCACTCGCACAATATC | |
| SpoT R | GATGTCGCAGTTTATTCTCC | |
| PhosR F | GCAAACATAATCATCACAACCAC | |
| PhosR R | GAGAGCAAGGATACAAAGAAGC | |
| CmeC F | GCTGCTGCTCAATTAGGTATAG | |
| CmeC R | GCTTCATAATCATACTCACTTGC | |
| PstS F | CCTTATACAAACTGGAATCAAATC | |
| PstS R | GACACATCACTCATTACAAGC | |
| PstC F | CGCTTATGCTTTAGGTATGAC | |
| PstC R | GCTGCCATCACCACTATC | |
| CJJ81176_0750 F | GGTCTTGTTGCCTTATTG | |
| CJJ81176_0750 R | GTATCGCTATGTTCTATGC | |
| PPK2 F | ATCTAATACTCCAACTTGTC | |
| PPK2 R | TTCTTCTTCTCCACTACG |
Underlining indicates restriction sites added to the 5′ end of each primer.
For construction of the complemented strain (the Δppk1c mutant), the ppk1 gene, along with the potential upstream promoter sequence, was amplified from strain 81-176 chromosomal DNA with the PPK1 COMP F and PPK1 COMP R primers. Appropriate restriction sites for PstI and KpnI were incorporated in the oligonucleotides to facilitate ligation into digested pRY111, an E. coli-Campylobacter shuttle vector (71). The resulting complementation plasmid (pDG3) was introduced into the C. jejuni Δppk1 mutant by conjugation as described previously (44). The resulting complemented strain (the Δppk1c mutant) was designated DG002.
TEM.
Transmission electron microscopy (TEM) procedures were used as described previously (23, 46). Briefly, bacterial cells grown to the mid-log phase were adjusted to an optical density at 600 nm (OD600) of 0.5 and fixed in 2% paraformaldehyde-2.5% glutaraldehyde, embedded in 1% agar, and postfixed in 1% osmium tetroxide. The samples were then dehydrated in a graded ethanol series and embedded in Spurr's resin (Ted Pella Inc., Redding, CA), and 80- to 90-nm sections were cut with a diamond microtome knife. The specimens were then stained with 2% uranyl acetate and lead citrate and examined under a Hitachi 7500 transmission electron microscope (Pleasanton, CA) at 80 kV at the Molecular and Cellular Imaging Center (Ohio Agricultural Research and Development Center [OARDC]; http://www.oardc.ohio-state.edu/mcic).
Induction and enumeration of VBNC cells of C. jejuni.
C. jejuni VBNC cells were induced as described by Chaveerach et al. (14). Briefly, 1 ml of a culture grown overnight containing 5 × 108 bacterial cells/ml (OD600 of 0.5) was added to 4 ml of MH broth with the pH adjusted to 4.0 with formic acid. The cultures were incubated under microaerophilic conditions at 42°C for 3 h. The induction of VBNC cells was confirmed by determining the culturable cell counts by the spread plate method and viable cell counts by fluorescence microscopy with 5-cyano-2,3-ditolyl tetrazolium chloride (CTC)-4′,6-diamidino-2-phenylindole (DAPI) staining.
To determine culturable cell counts, 100-μl volumes of culture at 0, 0.5, 1, 2, and 3 h posttreatment with formic acid were serially diluted (10-fold) and plated onto MH agar in triplicate. The plates were incubated at 42°C under microaerophilic conditions, and the number of CFU per milliliter (culturable count) was calculated.
At the aforementioned time intervals, 1 ml of culture was stained with CTC-DAPI for actively respiring and total C. jejuni cell counts as described by Cappelier et al. (12). Briefly, CTC (Polysciences, Inc., Warrington, PA) was added to a final concentration of 5 mM to 1 ml of formic acid-treated cells and incubated in the dark for 1 h at room temperature. The cells were then counterstained with 5 μg/ml of DAPI for 30 min. The cells were subsequently pelleted by centrifuging at 13,000 rpm for 4 min, and the pellet was resuspended in 100 μl of phosphate-buffered saline (PBS) and fixed with formaldehyde. Twenty microliters of the cell suspension was placed on a clean, oil-free slide, and an 18-mm coverslip was placed on the sample and sealed with nail polish. The slides were observed with a Zeiss (Thornwood, NY) Axiophot upright fluorescence microscope equipped with AxioCam HRc and Axiovision 2.05 software. The cells were subjected to fluorescent light with an excitation filter of 405 nm and a 455-nm dichroic mirror, which facilitates the simultaneous visualization of both DAPI (blue) and CTC (red). Viable cells convert CTC into insoluble formazan crystals, which accumulate in the cell and appear red under fluorescence microscopy, while both viable and dead cells are stained by DAPI. Ten fields were counted for each sample, with an average of 90 bacteria in each field. The percentage of viable cells was calculated as follows: % viability = (viable cell count/total cell count) ×100. Total cell counts were determined by adding the viable (red) and dead (blue) cell counts. The results represent the mean of three independent experiments. We also recorded the OD600 at the specified time points after CTC staining in order to observe the temporal change in intensity of tetrazolium reduction and further confirm the viability results generated from the fluorescence microscopy counts.
Flow cytometry.
Flow cytometry analysis of the bacterial samples was performed after formic acid treatment to determine possible change in cell morphology and granularity (the shifts in forward scatter [FSC] and side scatter [SSC]) with a FACScalibur (BD Biosciences, San Jose, CA) as described previously (66). Briefly, 100 μl of the bacterial samples at 0, 0.5, 1, 2, and 3 h posttreatment were diluted to 1 ml with 1× PBS (pH 7.4) and the samples were analyzed at an excitation wavelength of 488 nm. All parameters were read as logarithmic values, and 100,000 events were recorded for each sample. The assay was repeated two times.
Natural transformation frequency.
Natural transformation was performed as described previously (29). Briefly, cultures of the C. jejuni wild-type, Δppk1, and Δppk1c strains grown overnight on MH agar plates were resuspended in fresh MH broth to an OD600 of 0.05. The bacterial suspension (0.5 ml) was then incubated in sterile tubes at 42°C with shaking at 200 rpm under microaerophilic conditions for 3 h. After adding 1 μg of donor DNA from the dcuA::tetO mutant of C. jejuni 81-176 (tetracycline resistant), the bacterial suspension was incubated for another 4 h under the conditions described above. Transformants and total bacterial counts were obtained by plating on MH agar with or without tetracycline (5 μg/ml), respectively. Transformation frequency was expressed as the number of transformants from 1 μg of DNA divided by the total bacterial count. The experiment was repeated three times with triplicate transformation reactions each time.
Antibiotic susceptibility testing.
The susceptibility of the wild-type, Δppk1, and Δppk1c strains to various antimicrobials was determined by the standard microtiter broth dilution method as described previously (38, 49). The bacterial cultures were grown to mid-log phase and adjusted to an OD600 of 0.05 in MH broth. One hundred microliters of culture was then added to serially diluted (twofold) antimicrobials on a 96-well microtiter plate, mixed by pipetting, and incubated at 42°C under microaerophilic conditions for 2 days. The MIC was determined by recording the lowest concentration of an antimicrobial showing complete inhibition of bacterial growth. The antimicrobials and compounds used in this study were ciprofloxacin, erythromycin, tetracycline, rifampin (rifampicin), polymyxin B, cefotaxime, deoxycholic acid, taurocholic acid (Sigma Chemical Co., St. Louis, MO), sodium dodecyl sulfate (SDS; EM Science, Gibbstown, NJ), cholic acid (ACROS Organics, Belgium), and ethidium bromide (Amresco, Solon, OH). The susceptibility testing was repeated three times, and the mean MIC (μg/ml) was calculated for each antimicrobial.
Survival under nutrient downshift.
The role of PPK1 in C. jejuni survival under nutrient downshift was assessed as described previously (10, 49). Briefly, mid-log-phase cultures of the wild-type, Δppk1, and Δppk1c strains were pelleted and washed twice with minimal essential medium (MEM). The pellets were resuspended in MEM, and the OD600 was adjusted to 0.05. The suspensions were then incubated under microaerophilic conditions at 42°C with shaking at 200 rpm. One hundred-microliter volumes of the culture at different time points were serially diluted (10-fold) in MEM and plated onto MH agar in triplicate. The plates were incubated under microaerophilic conditions, and the number of CFU/ml was calculated. The experiment was repeated three times.
Reverse transcription (RT)-PCR analysis.
To study differential gene expression in the wild-type and mutant strains with and without formic acid treatment, we targeted key genes involved in phosphate uptake (phosR, CJJ81176_0899; pstS, CJJ81176_0642; pstC, CJJ81176_0643; and the periplasmic substrate binding protein-encoding gene, CJJ81176_0750) (53, 70), the stringent response (spoT, CJJ81176_1288) (24), and multidrug resistance (cmeC, CJJ81176_0388) (38), as well as posttranscriptional global regulator csrA (CJJ81176_1121) (20). The real-time quantitative PCR primers used in this study were designed with Beacon Designer 7.0 software (Premier Biosoft International, Palo Alto, CA). The wild-type and Δppk1 mutant strains were grown to mid-log phase at 42°C in MH broth under microaerophilic conditions, and 1 ml of culture with an OD600 of 0.5 was added to 4 ml of MH broth with or without pH adjustment to 4.0 with formic acid and incubated under microaerophilic conditions at 42°C for 30 min. Formic acid-treated, as well as untreated, wild-type and mutant strains were used for total RNA extraction with an RNeasy Mini kit (Qiagen), and cDNA was synthesized with SuperScript III First-Strand Synthesis SuperMix (Invitrogen) according to the manufacturer's protocol. Quantitative RT-PCR was performed with a SensiMixPlus SYBR RT-PCR kit (Quantace, Norwood, MA) in a Mastercycler ep realplex2 thermal cycler (Eppendorf, Westbury, NY). The relative expression of different genes in the wild-type and mutant strains was normalized with the rpoA (CJJ81176_1582) gene amplified from the corresponding sample. The difference in the expression of the genes was determined by the threshold cycle (CT) method, and the assay was repeated three times with two replicates each time for each sample.
Since PPK2 is also involved in the metabolism of poly-P (27), we performed quantitative RT-PCR analysis of ppk2 expression in the wild-type and Δppk1 mutant strains as described above.
Chicken colonization studies.
Day-old broiler chicks were obtained from a local hatching facility at the Food Animal Health Research Program (OARDC, Wooster, OH), and the chicks were confirmed to be negative for Campylobacter by culturing of cloacal swabs on MH agar with Campylobacter selective supplement (SR117E; Oxoid). The chicks were divided randomly into six groups with five chicks in each group. Groups 1, 2, and 3 were inoculated orally with 103, 104, and 105 CFU/chick of the wild-type bacterium, respectively, while groups 4, 5, and 6 received 103, 104, and 105 CFU/chick of the Δppk1 mutant in 200 μl of 1× PBS (pH 7.4), respectively. The chicks were given feed and water ad libitum and cared for in accordance with the guidelines of Association for the Assessment and Accreditation of Laboratory Animal Care. The chicks from each group were euthanized on day 8 postinoculation. Cecal contents, feces, and bursae were collected aseptically, weighed, and homogenized in 1× PBS (pH 7.4). The suspensions were serially diluted (10-fold) and plated onto MH agar with Campylobacter selective supplement. The plates were incubated at 42°C under microaerophilic conditions, and the number of CFU per gram of cecal contents, feces, or bursas was calculated. The data represent the average from five chicks.
Statistical analysis.
All of the data generated in this study were analyzed by one-way analysis of variance, followed by Tukey's honestly significant difference test and were expressed as mean ± SE (standard error). A P value of <0.01 or 0.05 (α level) was considered statistically significant for all of our assays.
RESULTS
The Δppk1 mutant is defective in the accumulation of poly-P granules.
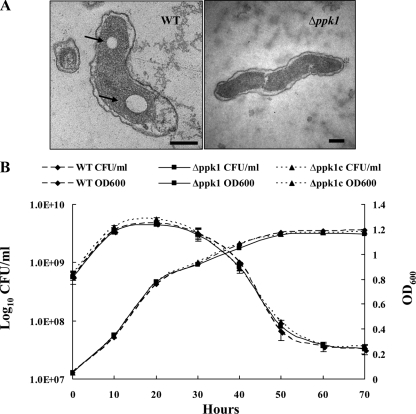
The ability to accumulate poly-P has been demonstrated in several bacteria, and PPK mutants have been shown to have low levels of poly-P (4, 10, 46, 65). TEM of the C. jejuni wild-type and Δppk1 mutant strains revealed the presence of large putative poly-P granules in the wild-type bacterial sections, while such granules were either absent or present as relatively smaller and indistinct granules in the Δppk1 mutant (Fig. 1A). Specifically, large putative poly-P granules were found in 30% or more of the examined wild-type sections, and these granules were absent in the majority of Δppk1 mutant sections, with only 3% or less of the sections showing comparatively small and indistinct granules. Our results corroborate the previous finding showing decreased levels of poly-P in the ppk1 mutant strain compared to the wild type with the toluidine blue O staining assay (10).
FIG. 1.
(A) Transmission electron micrographs of the wild-type and Δppk1 mutant C. jejuni strains. The TEM sections show large, distinct poly-P granules (arrows) in the wild type but not in the Δppk1 mutant, as observed with other bacteria (46, 65). Representative images are shown. Bars, 200 nm. (B) Growth curves of the wild-type, Δppk1, and Δppk1c strains. The growth curves were assessed by growing the bacteria in MH broth and direct plating on MH agar in triplicate or by measuring the OD600 at different time points. Data represent the mean ± SE of two independent experiments. WT, wild type.
We also monitored the growth of the wild-type, Δppk1, and Δppk1c strains in MH broth by plating 100 μl of the culture at 0, 10, 20, 30, 40, 50, 60, and 70 h after inoculation to determine if these strains have any growth defects under standard incubation conditions. All of the strains exhibited similar growth patterns in MH broth (Fig. 1B).
Role of poly-P in the formation of VBNC cells of C. jejuni.
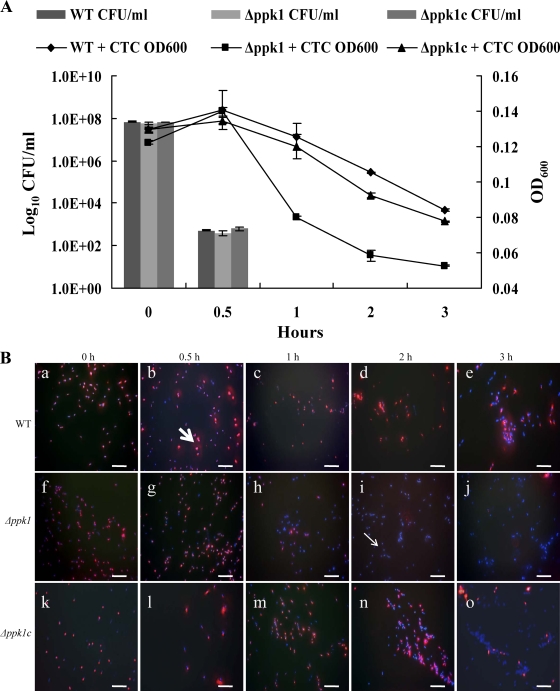
Since VBNC cell formation has been suggested as a mechanism for C. jejuni survival under environmental stress, we investigated the contribution of poly-P to VBNC cell formation. VBNC cells were induced with formic acid as described previously (11, 14), and direct culturable, total, and viable cell counts were determined at different time points. At 0 h, the wild-type and Δppk1 and Δppk1c mutant strains showed a high number of culturable cells (5 × 108 CFU/ml). However, the culturability of these strains decreased drastically (5 × 102 CFU/ml) at 30 min posttreatment with acid, which was not significantly different among the three strains. Interestingly, cell culturability was completely lost at 1 h in all three strains (Fig. 2A). Despite the loss of culturability, CTC-DAPI staining showed the presence of viable cells (i.e., stained with CTC) of the wild-type, Δppk1, and Δppk1c strains after formic acid treatment, suggesting the formation of VBNC cells. The wild-type strain had a significantly (P < 0.01) higher number of VBNC cells than did the Δppk1 mutant strain (Fig. 2B). The number of viable cells of the wild-type strain decreased gradually over time, while the number of these cells of the Δppk1 mutant drastically dropped at 1, 2, and 3 h posttreatment (Table 3). In the wild-type strain at 1 h, 96% of the cells were viable but none were culturable. In contrast, cells of the mutant strain showed only 36% viability while none were culturable. A similar trend was seen at 2 h with 94% of the cells maintaining viability in the wild type compared to 4% in the Δppk1 mutant strain. At 3 h posttreatment, there was a significant decline in the viable cell counts of the wild-type strain (42%) but still the viable counts were significantly higher than that of the Δppk1 mutant (3%). Complementation of the mutant with the wild-type copy of ppk1 restored its viability to levels similar to those of the wild type. We also recorded the OD600 at specified time points after CTC staining, and we found that the viability followed a trend similar to that observed by fluorescence microscopy (Fig. 2A). The difference between the viable counts of the wild-type and Δppk1 mutant strains was significant (P < 0.01) at 1, 2, and 3 h posttreatment with acid.
FIG. 2.
Effect of ppk1 deletion on the formation of VBNC cells of C. jejuni. (A) Culturability of the wild-type, Δppk1, and Δppk1c strains after formic acid treatment as determined by CFU enumeration and OD600 measurements after CTC staining. The data represent the mean ± SE of three independent experiments. (B) Fluorescence microscopy images of CTC-DAPI-stained C. jejuni cells showing viable (CTC-stained red cells, thick arrow) and dead (DAPI-stained blue cells, thin arrow) cells at different time points after formic acid treatment. Microscopy images a, b, c, d, and e (wild-type strain); f, g, h, i, and j (Δppk1 mutant strain); and k, l, m, n, and o (complemented strain) are the representative images of three independent experiments treated with formic acid for 0, 0.5, 1, 2, and 3 h, respectively. Note the drop in viable cells (red cells) of the Δppk1 mutant at 1, 2, and 3 h posttreatment compared to the wild-type strain. Bars, 10 μm. WT, wild type.
TABLE 3.
Percentages of viable cells as determined by CTC-DAPI staininga
| Strain | % of viable cells after: |
||||
|---|---|---|---|---|---|
| 0 h | 0.5 h | 1 h | 2 h | 3 h | |
| Wild type | 100 | 98.4 | 96.7 | 94.5 | 42.4 |
| Δppk1 mutant | 100 | 97.7 | 30.5 | 3.5 | 2.8 |
| Δppk1c mutant | 100 | 98.9 | 95.2 | 46.9 | 27.6 |
A total of 10 fields were counted for each sample, with an average of 90 bacteria in each field. The percentage of viable cells was calculated as follows: % viability = (viable cell count/total cell count) ×100. Total cell counts were determined by adding viable (red) and dead (blue) cell counts. The results represent the mean of three independent experiments.
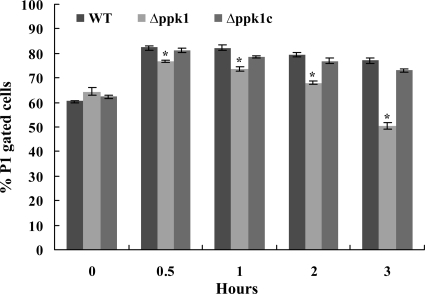
We performed fluorescence-activated cell sorter analysis to determine if bacterial cells undergo changes in morphology and granularity after formic acid treatment. Our results showed a change in cell size (FSC) and granularity (SSC) in the mutant strain compared to the wild-type strain from 1 to 3 h posttreatment. Separation of the total (P1) cell population based on their size and granularity demonstrated that there was a significant (P < 0.01) drop in P1 gated cells of the Δppk1 mutant from 1 to 3 h posttreatment with formic acid compared to those of the wild-type strain (Fig. 3). Complementation of the mutant with the wild-type copy of ppk1 restored cell size and granularity to wild-type levels.
FIG. 3.
Two-parameter FSC/SSC flow cytometry analysis of C. jejuni strains with and without formic acid treatment. FSC represents cell size, while reverse scatter is for cell granularity. A gate (P1) was set on FSC versus SSC to select the cell population for determining the change in cell size/granularity. The percent changes in P1-gated cells of the wild-type and Δppk1 and Δppk1c mutant strains at different time points after formic acid treatment are shown. The bars represent the mean ± SE of two independent experiments. *, P < 0.01. WT, wild type.
The Δppk1 mutant exhibits a decrease in transformation frequency.
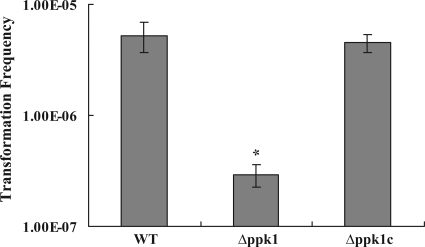
To determine if ppk1 has a role in natural transformation, we assessed the transformation frequency in the wild-type, Δppk1, and Δppk1c strains. Since the C. jejuni 81-176 strain used in the present study is pTet (confers tetracycline resistance) plasmid cured, we used genomic DNA from a dcuA::tetO mutant of the 81-176 strain (tetracycline resistant) as the donor for determining the transformation frequency. The Δppk1 mutant showed a significant decrease (18-fold, P < 0.01) in natural transformation frequency compared to the wild-type strain (Fig. 4). Complementation of the mutant with the wild-type copy of ppk1 restored the transformation frequency to wild-type levels.
FIG. 4.
Effect of ppk1 deletion on C. jejuni natural transformation. The donor DNA contained a tetracycline resistance (tetO) marker. The data represent the mean transformation frequency ± SE of three independent experiments with triplicate transformation reactions in each experiment. *, P < 0.01. WT, wild type.
The Δppk1 mutant is sensitive to various antimicrobials and bile acids.
In order to determine the contribution of ppk1 to antimicrobial resistance, we tested the susceptibility of the 81-176 wild-type, Δppk1, and Δppk1c strains to various antimicrobials and compounds. Our results showed that the Δppk1 mutant strain was more susceptible to several antimicrobials (Table 4) than was the wild-type strain. For example, the Δppk1 mutant showed a significant increase in susceptibility to erythromycin (128-fold, P < 0.01), cefotaxime (66-fold, P < 0.01), and ciprofloxacin (16-fold, P < 0.05). The Δppk1 mutant strain was also sensitive to rifampin (twofold), polymyxin B (fourfold), and tetracycline (eightfold). Interestingly, the Δppk1 mutant showed susceptibility to cholic acid (8-fold), taurocholic acid (8-fold), and deoxycholic acid (16-fold). In addition, the Δppk1 mutant demonstrated 31-fold and 4-fold increases in sensitivity to ethidium bromide and SDS, respectively. Complementation of the mutant with the wild-type copy of ppk1 either partially or completely restored resistance to wild-type levels.
TABLE 4.
Susceptibility of C. jejuni to antimicrobials, bile acids, and other compounds
| Agent | Inhibitory concn(μg/ml) |
||
|---|---|---|---|
| Wild type | Δppk1 mutant (fold difference)a | Δppk1c mutant | |
| Ciprofloxacin | 0.26 | 0.016 (16) | 0.13 |
| Erythromycin | 0.5 | 0.003 (128) | 0.12 |
| Cefotaxime | 2.0 | 0.03 (66) | 1.0 |
| Rifampin | 103 | 51.5 (2) | 51.5 |
| Polymyxin | 4.0 | 1.0 (4) | 1.0 |
| Tetracycline | 2.06 | 0.26 (8) | 2.06 |
| Ethidium bromide | 1.25 | 0.04 (31) | 1.25 |
| SDS | 375 | 93.75 (4) | 375 |
| Cholic acid | 5,156 | 645 (8) | 5,156 |
| Taurocholic acid | 36,000 | 4,500 (8) | 36,000 |
| Deoxycholic acid | 16,500 | 1,031 (16) | 8,250 |
Fold difference in susceptibility between the wild type and the Δppk1 mutant.
Transcription of the ppk2, phosR, pstS, pstC, cmeC, csrA, spoT, and CJJ81176_0750 genes was affected in the Δppk1 mutant.
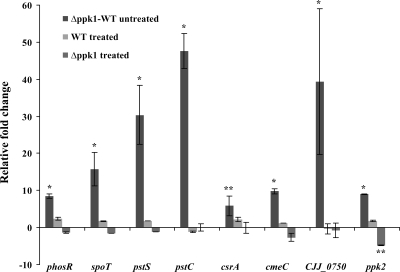
In order to understand the mechanism behind PPK1-mediated stress responses and adaptation, quantitative RT-PCR analysis was performed to examine the expression of phosphate regulon genes (phosR, pstS, pstC, and the periplasmic substrate binding protein-encoding gene, CJJ81176_0750), PPK2 (ppk2), the multidrug resistance efflux pump gene (cmeC), the global posttranscriptional regulator (csrA), and the stringent response regulator (spoT). Interestingly, the aforementioned genes were found to be significantly (P < 0.01) upregulated (fivefold or more) in the Δppk1 mutant relative to the wild-type strain (Fig. 5). Of particular interest, pstC, CJJ81176_0750, pstS, phosR, and ppk2 were upregulated 48-fold, 40-fold, 30-fold, 9-fold, and 9-fold, respectively. Gene expression was also analyzed after formic acid treatment and during the induction of VBNC cells. Our results showed that, with the exception of ppk2, the expression of the aforementioned genes was not affected in the acid-treated Δppk1 mutant. The ppk2 gene was significantly downregulated (fivefold) (P < 0.05) in the Δppk1 mutant after acid treatment. However, the expression of ppk2 and the aforementioned genes was not affected in the wild type before and after acid treatment.
FIG. 5.
Histogram showing the relative change in the expression of target genes before and after formic acid treatment. The relative change (2−ΔΔCT) in gene expression was calculated from the ΔΔCT value after normalization. Δppk1-WT (wild type) untreated indicates the fold change in gene expression of the Δppk1 mutant strain relative to the wild-type strain. WT treated indicates the fold change in gene expression in the wild-type strain treated with formic acid relative to the untreated wild type. Δppk1 treated indicates the relative fold change in gene expression of the Δppk1 mutant treated with formic acid relative to the untreated Δppk1 mutant. Genes with a twofold or greater (P < 0.01 or 0.05) relative change in expression were considered upregulated or downregulated. Each bar represents the mean ± SE of the relative fold change in expression from three independent experiments with triplicate reactions for each sample. *, P < 0.01; **, P < 0.05.
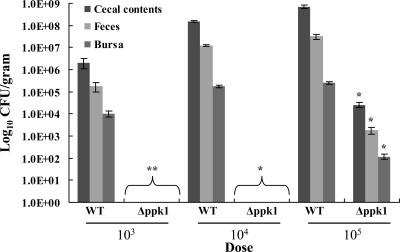
Poly-P is required for dose-dependent chicken colonization.
We assessed the ability of the Δppk1 mutant to colonize the cecum and bursa and its shedding in the feces of 2-day-old chicks. The dose dependency of colonization was studied by inoculating the chicks with three doses of inoculum separately, 103, 104, and 105 CFU/chick (n = 5). Our results demonstrated that the Δppk1 mutant was significantly defective (P < 0.05) in chicken colonization at all inoculum doses for all organs and feces compared to the wild type (Fig. 6). Chicks infected with 103 or 104 CFU/chick of the Δppk1 mutant had no detectable bacteria in any of the samples tested. However, chicks infected with 105 CFU/chick of the Δppk1 mutant were colonized with approximately 4 logs, 3 logs, and 2 logs fewer bacteria in the cecal contents, feces, and bursae, respectively, compared to the wild-type strain.
FIG. 6.
Effect of ppk1 deletion on chicken colonization. Chicks were randomly assigned to six groups of five. The first three groups were inoculated with 103, 104, and 105 CFU/chick of the wild-type strain, while the other three groups received 103, 104, and 105 CFU/chick of the Δppk1 mutant strain. At 8 days postinoculation, the chicks were euthanized and the numbers of CFU/gram of cecal contents, feces, and bursae were calculated. Each bar represents the mean ± SE of five chicks. *, P < 0.01; **, P < 0.05. WT, wild type.
DISCUSSION
To better understand the role of poly-P in stress survival and environmental adaptation mechanisms in an important enteric pathogen, C. jejuni, we constructed a deletion mutant by targeting PPK1 (Δppk1), an enzyme that mediates poly-P synthesis in the cell. Although the C. jejuni Δppk1 mutant showed growth patterns similar to those of the wild-type strain, as expected, the mutant was deficient in the accumulation of poly-P and exhibited defects in key survival and adaptation responses. Significantly, the Δppk1 mutant showed reduced abilities to form VBNC cells under stress, acquire DNA through natural transformation, and resist antimicrobials. The impairment of these physiological and stress responses, along with other observed deficiencies, confirmed that poly-P plays an essential role in the survival and adaptation of C. jejuni.
The absence of ppk genes was associated with defects in growth, responses to stress and starvation, and viability in several bacterial species (31, 40, 45, 58). This is also true of C. jejuni survival and stress tolerance, as our results showed that the Δppk1 mutant, after losing culturability (Fig. 2A), exhibited reduced viability (Table 3) and an inability to form VBNC cells under acid stress (Fig. 2B). Furthermore, flow cytometry analysis showed a significant change in cell size and granularity of the Δppk1 mutant during acid treatment (Fig. 3), probably indicating an increase in dead cells (35) and corroborating the inability to form VBNC cells, which have been reported to occur in spiral (noncoccoidal) forms in C. jejuni (19). Poly-P particles have been found in starved and morphologically altered Vibrio parahaemolyticus (15), while Nilsson et al. (45) showed that poly-P accumulated in structurally intact coccoid forms of starved H. pylori, probably for storing energy during starvation. Despite this, no direct association between poly-P and PPK1 and the formation of VBNC cells has been reported previously. This is important, as VBNC cell formation has been proposed as a strategy for surviving environmental stresses in many bacterial species, including enteric pathogens such as C. jejuni (55, 56). While usually undetectable by standard culturing techniques, in many cases, VBNC cells were reported to maintain pathogenicity, further increasing the public health importance of these forms. Although VBNC cell forms have been reported for C. jejuni strains that were exposed to starvation and other stresses (14, 37, 55), the significance of these forms in the epidemiology of C. jejuni has been a subject of debate (43, 57, 63), probably due to variations in the animal model, dose of VBNC cells, and strains used (14). However, it appears that VBNC C. jejuni cells were successfully resuscitated after passage in embryonated eggs (12, 14) and the strains recovered their capacity to adhere to HeLa cells, an indication of regained pathogenicity (12).
PPK1 is responsible for most of the poly-P synthesis in bacterial cells (8), which was also inferred from our electron microscopy images that showed deficient accumulation of poly-P granules in the C. jejuni Δppk1 mutant (Fig. 1A). Analysis of ppk mutants of other bacteria has also revealed defective poly-P granules (46, 65). Since poly-P acts as an energy store allowing cells to tolerate stress (45), we propose that poly-P- and PPK1-deficient bacteria lose their ability to enter a VBNC state, which impacts their capacity to reduce metabolism and potentially tolerate and adapt to environmental stresses and the possibility for recovery when conditions are favorable (41). With the exception of ppk2, which was downregulated under formic acid treatment, our quantitative RT-PCR data did not show any significant changes in the expression of selected genes involved in phosphate uptake and stress tolerance in the Δppk1 mutant (Fig. 5). Since ppk2 mediates a poly-P-driven reverse reaction, synthesizing GTP from GDP in a greater magnitude (75-fold) than the forward reaction (i.e., poly-P synthesis from GTP) (51), the downregulation of ppk2 can be interpreted as an additional effort by the cell to decrease energy (GTP/ATP) expenditure and maintain its already decreased poly-P reserves, which might be needed to form VBNC cells and resist stress. Unlike PPK1, PPK2 is stimulated and becomes stable in the presence of poly-P (51). As a result, the absence of or decrease in poly-P accumulation (Fig. 1A) might have prompted the overexpression of ppk2 in the non-acid-treated Δppk1 mutant compared to the wild type (Fig. 5), possibly compensating for the deficiency in poly-P-dependent PPK2 stability and partially maintaining its functions. Interestingly, the upregulation of ppk2 in the non-acid-treated Δppk1 mutant was accompanied by overexpression of the genes related to phosphate regulation and uptake, possibly for maintaining intracellular phosphate levels (51). This does not contradict the gene expression profiles of the acid-treated Δppk1 mutant, since VBNC cell formation might require a certain decrease in some metabolic and cellular processes, which would conserve energy and phosphate utilization and consequently limit the need for the overexpression of the aforementioned genes. However, it should be emphasized that the precise mechanism(s) by which poly-P influences the formation of VBNC cells requires further investigation.
An important mechanism for adaptation to environmental changes and antibiotic stress is the acquisition of important genetic material through natural transformation (16, 29, 30). Natural transformation plays an important role in mediating genetic diversity and the acquisition of antibiotic resistance in C. jejuni while also facilitating genetic manipulations in this naturally competent bacterium (17, 29, 68). Interestingly, poly-P is involved in the formation of cell membrane channels that allow DNA uptake (13, 54). However, to our knowledge, the impact of poly-P and its associated enzyme (PPK1) on natural transformation has not been investigated before. Our results show that the Δppk1 mutant was significantly deficient in the frequency of natural transformation (Fig. 4) compared to the wild-type and complemented strains. While it is true that the Δppk1 mutant showed an increased susceptibility to tetracycline (Table 4), it should be emphasized that the transformation experiments with the tetracycline gene occurred under nonselective conditions. Therefore, the acquisition of the tetracycline resistance gene by the mutant should have not been affected by its susceptibility to tetracycline. We suggest that PPK1 and poly-P possibly impact genetic diversity, acquisition of antibiotic resistance genes, and adaptation in C. jejuni by indirectly impacting the entry of DNA from the environment. This is important, as ppk-deficient bacteria, especially those with relatively smaller genomes, like C. jejuni, might be incapable of accessing collective gene pools (“supragenome”), which greatly restricts natural transformation and adaptation to environments (5).
Poly-P increases the activity of the stationary-phase RNA polymerase RpoS (59), possibly leading to adaptive mutations (21), including those that induce resistance to antibiotics (59). However, certain stress response factors, such as RpoS, are not found in C. jejuni. The Δppk1 mutant was significantly more susceptible to a variety of antibiotics and other antimicrobials than was the wild-type strain (Table 4). It follows that poly-P and its associated enzyme (PPK1) can affect the antibiotic resistance of C. jejuni through various other pathways that are evidently RpoS independent. For example, poly-P accumulation regulates the synthesis of guanosine tetraphosphate (ppGpp) (59), an important molecule in stringent responses, which at high levels, in turn, induces the accumulation of poly-P (34). Gaynor et al. (24) showed that the stringent response mediated the survival of C. jejuni under exposure to rifampin, probably through the activation of RelA and/or SpoT, enzymes that affect the synthesis of ppGpp, which alters transcription of genes that facilitate survival under stress. This was further confirmed by our observation that the Δppk1 mutant indeed showed an increased susceptibility to rifampin (Table 4). However, spoT was upregulated in the Δppk1 mutant (Fig. 5), probably in an attempt to induce ppGpp-dependent accumulation of poly-P (through the inhibition of exopolyphosphatase) (34) in the deficient mutant, which also highlights the importance of previously suggested interactions of poly-P with stringent response factors (36). The resistance of C. jejuni to macrolides (erythromycin) and fluoroquinolones (ciprofloxacin) is mediated through changes in the ribosome and mutations in DNA gyrase (encoded by gyrA), respectively (18). Interestingly, an E. coli ppk mutant showed a 40% decrease in induced mutagenesis to nalidixic acid resistance (64), which is a result of base mutations in DNA gyrase genes (72), while poly-P has been reported to interact with ribosomes, supporting translation fidelity in E. coli (42). Furthermore, ppk deletion mutations in Salmonella enterica serovar Typhimurium and S. enterica serovar Dublin showed an increased susceptibility to polymyxin B (31), corroborating our observations that the C. jejuni Δppk1 mutant was susceptible to polymyxin (Table 4) and emphasizing the role of poly-P in mediating antibiotic resistance. Surprisingly, cmeC, a component of a multidrug efflux pump that mediates resistance to antibiotics and other important compounds such as bile, which induces virulence gene expression (38), was upregulated in the Δppk1 mutant under no stress (Fig. 5), possibly in response to poly-P deficiency. However, overexpression of cmeC did not appear to facilitate the tolerance of the Δppk1 mutant when it was challenged with antibiotics or bile. Although our results indicate a role for PPK1 and poly-P in mediating the resistance of C. jejuni to different antibacterial compounds, the precise mechanism of the observed susceptibility and/or resistance remains largely unknown and requires further investigation. However, this might emphasize the importance of poly-P and its associated enzymes as potential targets for antibiotics and/or other compounds.
Differential-expression analysis of C. jejuni genes involved primarily in phosphate uptake, transport, and metabolism showed that ppk2 (regulates the reversible synthesis of poly-P from GTP or ATP), phosS/phosR (a two-component system that activates the transcription of the phosphate regulon), pstSC (involved in phosphate transport), and a gene for a periplasmic solute-binding protein (CJJ81176_0750) that is part of an operon encoding a putative ABC transporter for phosphate uptake (20, 53, 70) were expectedly upregulated under normal conditions in the Δppk1 mutant compared to the wild type (Fig. 5). It has been previously suggested that the Pho regulon is associated with stress responses (36), as the Pho regulon was suppressed in E. coli mutant strains (e.g., relA and spoT mutants) that were deficient in ppGpp accumulation (62). Consequently, direct and/or indirect interactions have been proposed to occur between pathways involved in phosphate regulation, poly-P synthesis, and the stringent response (36). In C. jejuni, the phosS/phosR two-component system triggers transcription of the phosphate regulon, including CJJ81176_0750 and the pstS and pstC genes, both of which are also transcribed during phosphate limitation (70). Consequently, the upregulation of ppk2 and other genes that are involved in phosphate uptake and transport in the Δppk1 mutant appears to constitute a compensatory response by the bacterium in an attempt to accumulate poly-P. These observations further highlight the role of poly-P and its associated metabolic enzymes in adaptation and survival responses of C. jejuni.
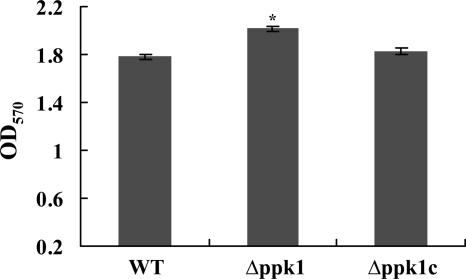
Consistent with previous findings, C. jejuni motility and resistance to oxidative stress were not impacted in the Δppk1 mutant (data not shown; 10), which surprisingly showed enhanced biofilm formation (Fig. 7). Interestingly, csrA (putative global posttranscriptional regulator), which contributes to biofilm formation and oxidative resistance in C. jejuni (20), was upregulated in the Δppk1 mutant (Fig. 5), possibly explaining the tolerance of this mutant to oxidative stress and its enhanced capacity for biofilm formation. Furthermore, our results indicated that the Δppk1 mutant was susceptible to osmotic stress and nutrient downshift (Fig. 8A and B) and exhibited a dose-dependent chicken colonization deficiency (Fig. 6), while the thermal stress response and resistance to heavy metals were not impacted (data not shown). With the exception of thermal stress and resistance to heavy metals, the tolerance to osmotic and nutrient stresses was previously investigated, and our results were consistent with the earlier report (10). Similar to the ppk1 mutant, a C. jejuni spoT mutant was deficient in poly-P accumulation during growth in a rich medium despite the presence of an intact ppk1 gene (10). In our Δppk1 mutant, poly-P did not accumulate despite the overexpression of spoT and the phosphate regulon genes, further confirming that functional PPK1 is required for poly-P accumulation. Additionally, this suggested that spoT might affect the activity of ppk1, which could also be inferred from E. coli spoT mutants that failed to accumulate ppGpp and pppGpp, as well as poly-P, under nutrient deprivation (34).
FIG. 7.
Relative amounts of biofilm formation in the wild-type, Δppk1, and Δppk1c strains. Static biofilm formation in borosilicate tubes was assessed by inoculating 100 μl of culture with an OD600 of 0.05 into 1 ml of MH broth and incubating it at 42°C under microaerophilic conditions for 2 days without shaking. Biofilms were visualized by staining with 250 μl of 1% crystal violet for 15 min. Biofilms were quantified by measuring the absorbance at 570 nm after dissolving them in 1 ml of dimethyl sulfoxide for 24 to 48 h. Each bar represents the mean ± SE of two independent experiments. *, P < 0.01. WT, wild type.
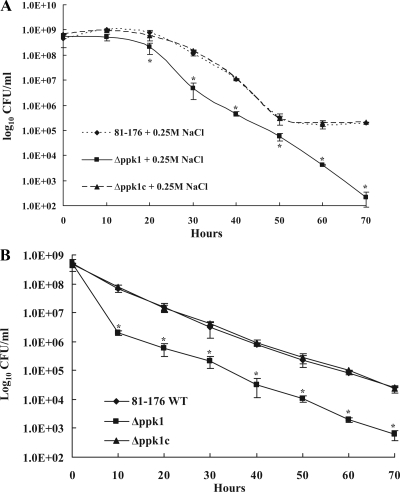
FIG. 8.
(A) Survival of the wild-type, Δppk1, and Δppk1c strains in MH broth with 0.25 M NaCl. Bacterial strains were grown into mid-log phase and adjusted to an OD600 of 0.05 in MH broth with and without 0.25 M NaCl and incubated under microaerophilic conditions at 42°C for 70 h with shaking at 200 rpm. At different time points, samples were serially diluted (10-fold) and plated onto MH agar in triplicate. The plates were incubated under microaerophilic conditions, and the number of CFU/ml was calculated. The assay was repeated three times, and the mean number of CFU/ml was reported. (B) C. jejuni survival under nutritional downshift in MEM. Each data point represents the mean ± SE of three independent experiments. *, P < 0.01. WT, wild type.
In summary, our observations confirm the importance of PPK1 in the adaptation, stress tolerance, and survival of C. jejuni. Furthermore, we elucidate for the first time the role of poly-P in influencing VBNC cell formation, natural transformation, and resistance to antimicrobial compounds and bile acids, while impacting the expression of genes that are important in these mechanisms. With limited options (e.g., absence of RpoS) for survival under various stresses, poly-P and its associated enzymes might be critical for C. jejuni survival, both in vivo and during transmission. In addition, poly-P might contribute to the plasticity of the C. jejuni genome, as well as the spread and development of antibiotic resistance, by mediating natural transformation and antimicrobial resistance. Further studies are warranted to understand the molecular mechanisms of poly-P regulation of stress responses and pathogenicity and the interplay of poly-P, phosphate regulation genes, SpoT, and ppGpp in C. jejuni pathophysiology.
Acknowledgments
We thank Qijing Zhang and Byeonghwa Jeon for help with the natural transformation study and for critically reading the manuscript. We also thank Juliette Hanson for assistance with chicken colonization studies, Tea Meulia for help with electron microscopy, and Aradhya Gourapura for help with flow cytometry.
The research in G. Rajashekara's laboratory is supported by USDA grant 2007-35201-18414 and by the OARDC, The Ohio State University.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Akiba, M., J. Lin, Y. W. Barton, and Q. Zhang. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57:52-60. [DOI] [PubMed] [Google Scholar]
- 2.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 3.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni: an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayraud, S., B. Janvier, A. Labigne, C. Ecobichon, C. Burucoa, and J. L. Fauchere. 2005. Polyphosphate kinase: a new colonization factor of Helicobacter pylori. FEMS Microbiol. Lett. 243:45-50. [DOI] [PubMed] [Google Scholar]
- 5.Baltrus, D. A., K. Guillemin, and P. C. Phillips. 2008. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62:39-49. [DOI] [PubMed] [Google Scholar]
- 6.Beery, J. T., M. B. Hugdahl, and M. P. Doyle. 1988. Colonization of gastrointestinal tracts of chicks by Campylobacter jejuni. Appl. Environ. Microbiol. 54:2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode, G., F. Mauch, H. Ditschuneit, and P. Malfertheiner. 1993. Identification of structures containing polyphosphate in Helicobacter pylori. J. Gen. Microbiol. 139:3029-3033. [DOI] [PubMed] [Google Scholar]
- 8.Brown, M. R., and A. Kornberg. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33:284-290. [DOI] [PubMed] [Google Scholar]
- 9.Butzler, J. P., and M. B. Skirrow. 1979. Campylobacter enteritis. Acta Paediatr. Belg. 32:89-94. [PubMed] [Google Scholar]
- 10.Candon, H. L., B. J. Allan, C. D. Fraley, and E. C. Gaynor. 2007. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 189:8099-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cappelier, J. M., B. Lázaro, A. Rossero, A. Fernandez-Astorga, and M. Federighi. 1997. Double staining (CTC-DAPI) for detection and enumeration of viable but non-culturable Campylobacter jejuni cells. Vet. Res. 28:547-555. [PubMed] [Google Scholar]
- 12.Cappelier, J. M., C. Magras, J. L. Jouve, and M. Federighi. 1999. Recovery of viable but non-culturable Campylobacter jejuni cells in two animal models. Food Microbiol. 16:375-383. [Google Scholar]
- 13.Castuma, C. E., R. Huang, A. Kornberg, and R. N. Reusch. 1995. Inorganic polyphosphates in the acquisition of competence in Escherichia coli. J. Biol. Chem. 270:12980-12983. [DOI] [PubMed] [Google Scholar]
- 14.Chaveerach, P., A. A. H. M. Huurne, L. J. A. Lipman, and F. Knapen. 2003. Survival and resuscitation of ten strains of Campylobacter jejuni and Campylobacter coli under acid conditions. Appl. Environ. Microbiol. 69:711-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, S. Y., W. N. Jane, Y. S. Chen, and H. C. Wong. 2009. Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int. J. Food Microbiol. 129:157-165. [DOI] [PubMed] [Google Scholar]
- 16.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 17.de Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. van Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 18.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Federighi, M., J. L. Tholozan, J. M. Cappelier, J. P. Tissier, and J. L. Jouve. 1998. Evidence of non-coccoid viable but non-culturable Campylobacter jejuni cells in microcosm water by direct viable count, CTC-DAPI double staining, and scanning electron microscopy. Food Microbiol. 15:539-550. [Google Scholar]
- 20.Fields, J. A., and S. A. Thompson. 2008. Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190:3411-3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster, P. L. 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42:373-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraley, C. D., M. H. Rashid, S. S. Lee, R. Gottschalk, J. Harrison, P. J. Wood, M. R. Brown, and A. Kornberg. 2007. A polyphosphate kinase 1 (ppk1) mutant of Pseudomonas aeruginosa exhibits multiple ultrastructural and functional defects. Proc. Natl. Acad. Sci. USA 104:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaynor, E. C., D. H. Wells, J. K. MacKichan, and S. Falkow. 2005. The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56:8-27. [DOI] [PubMed] [Google Scholar]
- 25.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes, R. 2004. Campylobacter jejuni in Guillain-Barré syndrome. Lancet Neurol. 3:644. [DOI] [PubMed] [Google Scholar]
- 27.Ishige, K., H. Zhang, and A. Kornberg. 2002. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. USA 99:16684-16688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahid, I. K., A. J. Silva, and J. A. Benitez. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon, B., W. Muraoka, O. Sahin, and Q. Zhang. 2008. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob. Agents Chemother. 52:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnsborg, O., L. Eldholm, and L. S. Håvarstein. 2007. Natural genetic transformation: prevalence, mechanisms and function. Res. Microbiol. 158:767-778. [DOI] [PubMed] [Google Scholar]
- 31.Kim, K. S., N. N. Rao, C. D. Fraley, and A. Kornberg. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. USA 99:7675-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 34.Kuroda, A., H. Murphy, M. Cashel, and A. Kornberg. 1997. Guanosine tetra and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272:21240-21243. [DOI] [PubMed] [Google Scholar]
- 35.Kusters, J. G., M. M. Gerrits, J. A. G. Van Strijp, and C. M. J. E. Vandenbroucke-Gauls. 1997. Coccoid forms of Helicobacter pylori are the morphologic manifestation of cell death. Infect. Immun. 65:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamarche, M. G., B. L. Wanner, S. Cre′pin, and J. Harel. 2008. The Pho regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32:461-473. [DOI] [PubMed] [Google Scholar]
- 37.Lázaro, B., J. Carcamo, A. Audicana, I. Perales, and A. Fernandez-Astorga. 1999. Viability of DNA maintenance in nonculturable spiral Campylobacter jejuni cells after long-term exposure to low temperatures. Appl. Environ. Microbiol. 65:4677-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multi-drug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luangtongkum, T., B. Jeon, J. Han, P. Plummer, C. M. Logue, and Q. Zhang. 2009. Antibiotic resistance in Campylobacter: emergence, transmission and persistence. Future Microbiol. 4:189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manganelli, R. 2007. Polyphosphate and stress response in mycobacteria. Mol. Microbiol. 65:258-260. [DOI] [PubMed] [Google Scholar]
- 41.McDougald, D., S. A. Rice, D. Weichart, and S. Kjelleberg. 1998. Nonculturability: adaptation or debilitation? FEMS Microbiol. Ecol. 25:1-9. [Google Scholar]
- 42.McInerney, P., T. Mizutani, and T. Shiba. 2006. Inorganic polyphosphate interacts with ribosomes and promotes translation fidelity in vitro and in vivo. Mol. Microbiol. 60:438-447. [DOI] [PubMed] [Google Scholar]
- 43.Medema, G. J., F. M. Schets, A. W. Van de Giessen, and A. Havelaar. 1992. Lack of colonization of one day old chicks by viable, non-culturable Campylobacter jejuni. J. Appl. Bacteriol. 72:512-516. [DOI] [PubMed] [Google Scholar]
- 44.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrel. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nilsson, H. O., J. Blom, W. A. Al-Soud, A. Ljungh, L. P. Andersen, and T. Wadstrom. 2002. Effect of cold starvation, acid stress, and nutrients on metabolic activity of Helicobacter pylori. Appl. Environ. Microbiol. 68:11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa, N., C. M. Tzeng, C. D. Fraley, and A. Kornberg. 2000. Inorganic polyphosphate in Vibrio cholerae: genetic, biochemical, and physiologic features. J. Bacteriol. 182:6687-6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 48.Price-Carter, M., T. G. Fazzio, E. I. Vallbona, and J. R. Roth. 2005. Polyphosphate kinase protects Salmonella enterica from weak organic acid stress. J. Bacteriol. 187:3088-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajashekara, G., M. Drozd, D. Gangaiah, B. Jeon, Z. Liu, and Q. Zhang. 2009. Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog. Dis. 6:935-945. [DOI] [PubMed] [Google Scholar]
- 50.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao, N. N., M. R. Gómez-García, and A. Kornberg. 2009. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78:605-647. [DOI] [PubMed] [Google Scholar]
- 52.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid, A. N., R. Pandey, K. Palyada, L. Whitworth, E. Doukhanine, and A. Stintzi. 2008. Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl. Environ. Microbiol. 74:1598-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reusch, R. N., and H. L. Sadoff. 1988. Putative structure and functions of a poly-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc. Natl. Acad. Sci. USA 85:4176-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rollins, D. M., and R. R. Colwell. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roszak, D. B., D. J. Grimes, and R. R. Colwell. 1984. Viable but nonrecoverable stage of Salmonella enteritidis in aquatic systems. Can. J. Microbiol. 30:334-338. [DOI] [PubMed] [Google Scholar]
- 57.Saha, S. K., S. Saha, and S. C. Sanyal. 1991. Recovery of injured Campylobacter jejuni cells after animal passage. Appl. Environ. Microbiol. 57:388-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seufferheld, M. J., H. M. Alvarez, and M. E. Farias. 2008. Role of polyphosphates in microbial adaptation to extreme environments. Appl. Environ. Microbiol. 74:5867-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shiba, T., K. Tsutsumi, H. Yano, Y. Ihara, A. Kameda, K. Tanaka, H. Takahashi, M. Munekata, N. N. Rao, and A. Kornberg. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. USA 94:11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shiba, T., K. Tsutsumi, K. Ishige, and T. Noguchi. 2000. Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry 65:315-323. [PubMed] [Google Scholar]
- 61.Skirrow, M. B. 1994. Diseases due to Campylobacter, Helicobacter, and related bacteria. J. Comp. Pathol. 111:113-149. [DOI] [PubMed] [Google Scholar]
- 62.Spira, B., N. Silberstein, and E. Yagil. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J. Bacteriol. 177:4053-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stern, N. J., D. M. Jones, I. V. Wesley, and D. M. Rollins. 1994. Colonization of chicks by non-culturable Campylobacter spp. Lett. Appl. Microbiol. 18:333-336. [Google Scholar]
- 64.Stumpf, J. D., and P. L. Foster. 2005. Polyphosphate kinase regulates error-prone replication by DNA polymerase IV in Escherichia coli. Mol. Microbiol. 57:751-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tan, S., C. D. Fraley, M. Zhang, D. Dailidiene, A. Kornberg, and D. E. Berg. 2005. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J. Bacteriol. 187:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tholozan, J. L., J. M. Cappelier, J. P. Tissier, G. Delattre, and M. Federighi. 1999. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl. Environ. Microbiol. 65:1110-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tinsley, C. R., B. N. Manjula, and E. C. Gotschlich. 1993. Purification and characterization of polyphosphate kinase from Neisseria meningitidis. Infect. Immun. 61:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson, D. L., J. A. Bell, V. B. Young, S. R. Wilder, L. S. Mansfield, and J. E. Linz. 2003. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 149:3603-3615. [DOI] [PubMed] [Google Scholar]
- 70.Wösten, M. M., C. T. Parker, A. van Mourik, M. R. Guilhabert, L. van Dijk, and J. P. van Putten. 2006. The Campylobacter jejuni PhosS/PhosR operon represents a non-classical phosphate-sensitive two-component system. Mol. Microbiol. 62:278-291. [DOI] [PubMed] [Google Scholar]
- 71.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 72.Yoshida, H., T. Kojima, J. Yamagishi, and S. Nakamura. 1988. Quinolone resistant mutations of the gyrA gene of Escherichia coli. Mol. Gen. Genet. 211:1-7. [DOI] [PubMed] [Google Scholar]
- 73.Young, K. T., L. M. Davis, and V. J. DiRita. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5:665-679. [DOI] [PubMed] [Google Scholar]