Abstract
The genomes of the two lytic mutant Staphylococcus aureus bacteriophages, vB_SauS-phiIPLA35 (phiIPLA35) and vB_SauS-phiIPLA88 (phiIPLA88), isolated from milk have been analyzed. Their genomes are 45,344 bp and 42,526 bp long, respectively, and contain 62 and 61 open reading frames (ORFS). Enzymatic analyses and sequencing revealed that the phiIPLA35 DNA molecule has 3′-protruding cohesive ends (cos) 10 bp long, whereas phiIPLA88 DNA is 4.5% terminally redundant and most likely is packaged by a headful mechanism. N-terminal amino acid sequencing, mass spectrometry, bioinformatic analyses, and functional analyses enabled the assignment of putative functions to 58 gene products, including DNA packaging proteins, morphogenetic proteins, lysis components, and proteins necessary for DNA recombination, modification, and replication. Point mutations in their lysogeny control-associated genes explain their strictly lytic behavior. Muralytic activity associated with other structural components has been detected in virions of both phages. Comparative analysis of phiIPLA35 and phiIPLA88 genome structures shows that they resemble those of φ12 and φ11, respectively, both representatives of large genomic groupings within the S. aureus-infecting phages.
Staphylococcus aureus is an important etiologic agent of food-borne diseases due to its ability to produce heat-resistant staphylococcal enterotoxins (SEs) when it grows in foods. In fact some S. aureus strains may produce up to 20 serologically distinct SEs, which could be responsible for food poisoning (30). SEs have been divided initially into serological types SEA through SEE, and recently the existence of new types of SEs has also been reported (5).
S. aureus strains harboring enterotoxin genes have been isolated from a variety of foods (38) including dairy products (9, 46, 56). Mastitis caused by this pathogen and poor hygienic processing conditions are the most important sources of dairy product contamination. Growth of enterotoxigenic S. aureus in both raw milk and dairy products poses a potential health hazard to consumers. In this context, new biocontrol strategies to prevent growth of S. aureus, suitable to be applied in the food industry, are being explored.
Currently, there is a renewed interest in exploiting the antimicrobial potential of bacterial viruses for bacterial-control applications in agriculture, aquaculture, and the food industry (11, 18, 23, 49). In fact, the use of phages for the treatment of infectious diseases (or phage therapy) has a long successful history in the countries of Eastern Europe (or former Soviet Union) (50). Specifically, S. aureus bacteriophages have been assayed in the treatment of venous leg ulcers and eye infections (22, 42).
Prior to any phage application, genome analysis is a prerequisite to examine the safety of the phages, specifically, traits which might enhance the virulence of the infected bacterium. In addition, genome analysis might uncover novel antibacterial targets or agents (33) with promising biotechnological applications (6). For example, various lytic phage proteins (endolysins) have shown great potential in veterinary and human medicine for the treatment and prophylaxis of infections (12) and have been applied as biocontrol agents in dairy products (36). Several technologies employing phages and endolysins for pathogen detection and decontamination have also been patented (7).
To date, genomes of over 47 S. aureus phages are available in public databases. The number of known, strictly lytic phages is limited to the close-knit Myoviridae genus of the SPO1-like viruses, containing phages K, Twort, and G1. Apart from this group, a large number of genomes from unclassified Siphoviridae in lysogenic S. aureus strains are available (26, 37). Some temperate bacteriophages may play an important role in the pathogenicity of S. aureus by carrying virulence factors, mediating lateral gene transfer, and even facilitating the adaptation of the pathogen during infection (1, 21, 52).
In previous work, we have characterized phiIPLA35 and phiIPLA88 S. aureus phages (17). These two lytic phages, previously named φ35 and φ88, were selected as mutants of the temperate phages φA72 and φH5, respectively, isolated from raw bovine milk. They belong to the Siphoviridae family of double-stranded DNA bacterial viruses in the order Caudovirales. Remarkably, these phages infect S. aureus of bovine and dairy origin while clinical isolates appear to be resistant. Both phiIPLA35 and phiIPLA88 are very well adapted to the dairy environment and effectively inhibit S. aureus growth in milk and curd-manufacturing processes (17, 20).
In this study, we have sequenced and annotated the genomes of both bacteriophages, elucidated their physical genome structures, and identified peptidoglycan hydrolytic activities. Comparative genome analysis also allowed us to put phiIPLA35 and phiIPLA88 into a phylogenetic context.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
S. aureus Sa9 was used as the host strain of phages phiIPLA35 and phiIPLA88 (17). Escherichia coli strain DH10B (Invitrogen) was used for recombinant DNA work. Both were grown in 2× TY broth (45) at 37°C with vigorous shaking. Plates of 2× TY broth contained 2% (wt/vol) bacteriological agar. For the selection of plasmid-bearing cells, ampicillin was added at 100 μg/ml. Bacterial stocks were stored at −80°C in 2× TY supplemented with 20% (final concentration) glycerol.
Bacteriophage propagation and DNA purification.
Bacteriophages phiIPLA35 and phiIPLA88 were routinely propagated on S. aureus Sa9 as described previously (17). Phage enumeration was performed by the double-layer technique (45) using soft 2× TY medium (0.75% agar plus 10 mM CaCl2 and 10 mM MgSO4) in the upper layer. In order to prepare RNA-free DNA for sequence analysis, the phages were dialyzed against SM buffer (20 mM Tris-HCl, 10 mM MgSO4, 10 mM CaCl2, 100 mM NaCl, pH 7.5), and the DNA was extracted and purified as described previously (15).
One-step growth curve.
One-step growth curves were performed in 2× TY medium using a multiplicity of infection of 1, as previously described (25).
Phage genome sequencing and analysis.
Approximately 10 μg of both phiIPLA35 and phiIPLA88 DNA was sheared by sonication, size selected (2 to 3 kbp), and cloned into pUC18. Individual clones were sequenced and assembled. Trace assembly was done with the phredPhrap package (10). Each sequenced base had at least a fivefold coverage. Primer walking was used to close gaps in the sequence. Open reading frames (ORFs) were predicted with ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/) software and by visual inspection. The primary nucleotide sequence was scanned in all reading frames for start codons with a threshold of 50 codons. BLASTX and BLASTP (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) were used to search for homologous proteins. Structural predictions and motif searches were performed with InterProScan (http://www.ebi.ac.uk/InterProScan/), Pfam (http://pfam.sanger.ac.uk/search?tab=search -SequenceBlock), and YASPIN (http://www.ibi.vu.nl/programs/yaspinwww/). σ70 promoter sequences were identified using Bprom (http://www.softberry.com) and PPP (Prokaryotic Promoter Prediction [http://bioinformatics.biol.rug.nl/websoftware/ppp/ppp_start.php]). Putative terminator sequences were detected with the terminator function of the GCG program (version 10.2). The search for putative tRNA-encoding genes was performed with tRNAscan-SE 1.21 (http://selab.janelia.org/tRNAscan-SE/). Prediction of +1 and −1 frameshifting was carried out with FSFinder 2 (http://wilab.inha.ac.kr/fsfinder2/). Cumulative GC skew (http://mips.gsf.de/services/analysis/genskew) was used to predict the oriC. Pseudoknot predictions were carried out by pknotsRG (41). Dlot plot analyses were carried out by Nucleic Acids Dot Plots (http://www.vivo.colostate.edu/molkit/dnadot/index.html).
Determination of phage genome physical structure.
To test whether the phage genomes have cohesive ends (cos), purified DNA (0.5 μg) was incubated with T4 DNA ligase, followed by digestion with the restriction enzymes BamHI, EcoRI, HindIII, and PstI (Takara, Otsu, Shiga, Japan) and fragment separation by agarose gel electrophoresis. A nonligated DNA sample was equally treated but heated at 70°C for 10 min prior to the electrophoresis. To obtain the sequence of the single-stranded cos extensions, the ends of phiIPLA35 DNA were sequenced directly using primers A (5′-AGTTATACGACACAAGTACACGAGG-3′) and B (5′-TGAGTGACTTGCTCTGCATACCATG-3′) and aligned with the circular phiIPLA35 genome.
To determine if the phiIPLA88 DNA molecule has open ends, phage DNA (5 μg) and phage DNA heated at 70°C for 10 min were incubated with 20 units of the double-stranded DNA exonuclease Bal31 (MBI Fermentas, Vilnius, Lithuania) according to the manufacturer's instructions. The DNA was ethanol precipitated, dissolved in Tris-EDTA buffer, digested with PvuII, and analyzed by agarose gel electrophoresis. As a control, we used lambda DNA (Quimigen, Madrid, Spain) known to have specific ends.
For electron microscopy of phage DNA, phage were dialyzed on a 0.025-μm-pore-size Millipore filter against TBT buffer (100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM MgCl2). The phage were spread with 50% formamide, carbonate buffer, and cytochrome c on a water surface as described earlier (48). Plasmid RSF1010 DNA (47) was added as an internal standard for length comparison with the released phage DNA.
Proteomic analysis of virion proteins.
Phage structural proteins were extracted, purified as described previously (15), and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a Miniprotean III (Bio-Rad, Richmond, CA). Proteins were stained with Coomassie R-250 blue or silver (PlusOne silver staining kit; GE Healthcare, Piscataway, NJ). The bands of interest were excised manually and digested with porcine trypsin (Promega, Madison, WI), and the resulting peptides were analyzed by matrix-assisted laser desorption ionization—time of flight mass spectrometry, essentially as previously described (16). N-terminal amino acid sequences were determined in bands excised from polyvinylidene difluoride membrane blots using an Applied Biosystems 477A automated protein sequencer.
Zymogram analysis.
Purified phage suspensions were dialyzed against SM buffer, mixed with loading buffer (1% SDS, 6% sucrose, 100 mM dithiothreitol, 10 mM Tris, pH 6.8, 0.0625% bromophenol blue) and boiled for 5 min before loading onto 12% SDS-PAGE gels containing 0.2% S. aureus Sa9 autoclaved cells (31). Gels were cast according to Laemmli (28), except that only 0.01% SDS was used to allow protein renaturation. After electrophoresis, gels were washed for 30 min with water and then soaked for 1 day at room temperature in 150 mM sodium phosphate buffer, pH 7.0, containing 0.1% Triton X-100 and 10 mM MgCl2. Zymograms were stained for 3 h with 0.1% methylene blue in 0.001% KOH and washed with water. Peptidoglycan hydrolase activity was detected as a clear zone in a dark blue background of stained peptidoglycan.
Nucleotide sequence accession numbers.
The sequences of phiIPLA35 and phiIPLA88 have been deposited in the GenBank under accession numbers EU861004 and EU861005, respectively.
RESULTS AND DISCUSSION
One-step growth curve of phiIPLA35 and phiIPLA88.
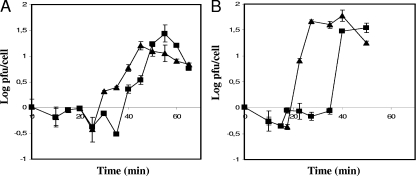
The stages of infection of the phages phiIPLA35 and phiIPLA88 have been followed using a one-step growth curve. We observed that phages behaved slightly differently when propagated on S. aureus Sa9 (Fig. 1). The eclipse phase of phage phiIPLA35, determined by the release of infecting particles after the treatment with chloroform, was longer than that of phiIPLA88 (30 versus 22.5 min). However, in both cases infecting particles were detected in the supernatant after 40 min postinfection. The burst sizes were estimated around 27 and 45 new virions per infecting particle of phiIPLA35 and phiIPLA88, respectively. These values are relatively lower than the burst size reported for φMR11 (34).
FIG. 1.
One-step growth curves of phages phiIPLA35 (A) and phiIPLA88 (B) on exponential cultures of S. aureus Sa9 incubated in 2× YT medium at 37°C under agitation. Cells were chloroform treated (triangles) or left untreated (squares).
Physical structure of the phiIPLA35 and phiIPLA88 genomes.
The phiIPLA35 cos sites were localized using restriction pattern analysis of ligated and unligated DNA samples, revealing typical fragments arising from the ligation of the cos sites (data not shown). Moreover, direct sequencing with appropriate oligonucleotides pointing outwards from the putative cos site and sequence alignment with the circular DNA show that phiIPLA35 DNA has 10-bp single-stranded 3′ extensions with the sequence 5′-CGGCGGGGGC-3′, located upstream from the terminase small subunit (position 19942 to 19951).
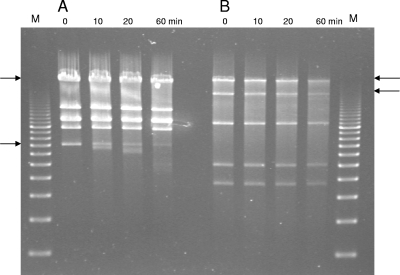
In contrast, no evidence could be found for the presence of cos ends in the phiIPLA88 DNA molecule. Ligation of its DNA did not alter the restriction patterns, which were compatible with a circular map (data not shown). Restriction endonuclease digestions of phiIPLA88 DNA also failed to reveal a “submolar” fragment, which would contain the pac site for initial recognition and subsequent cutting of sequentially packaged phage concatemers by the terminase enzyme (2). Another approach to determine genome ends involved a time-limited treatment of phiIPLA88 DNA with the exonuclease Bal31 (an exonuclease which degrades double-stranded linear DNA from both ends simultaneously), followed by complete digestion with a restriction enzyme (Fig. 2). In this situation, specific degradation of the restriction fragments overlapping the genome ends would be observed. To this end, the lambda phage DNA was used as a positive control. Bal31 treatment followed by digestion with the restriction enzyme EcoRI showed that the two expected EcoRI-specific fragments were degraded (Fig. 2A). Similarly, two specific phiIPLA88 DNA PvuII restriction fragments of 21.5 and 10.7 kbp were simultaneously shortened (Fig. 2B). These fragments would bear the physical genome ends of phiIPLA88. In fact, they are located in the surrounding region of a possible pac site placed upstream of gene product 35 (gp35), the small terminase subunit. This position is where pac sites are usually found in headful packagers (3).
FIG. 2.
Time-limited digestion with Bal31 exonuclease (0 to 60 min, as indicated) of control phage lambda DNA (A) and phiIPLA88 DNA (B), followed by restriction enzyme digestion (EcoRI in panel A and PvuII in panel B). The arrows point to the two sequentially degraded fragments of the linear control DNA and phiIPLA88 DNA. M, molecular size marker (0.5 to 10 kbp). Sampling times are indicated in minutes.
The phiIPLA88 DNA was measured on transmission electron micrographs. The size was 44,466 kb ± 1,025 kb (n = 36), which is significantly longer (+1,940 kb) than that derived from sequencing, and suggested that the phiIPLA88 DNA genome is about 4.5% terminally redundant.
Genome overview of phiIPLA35 and phiIPLA88 phages.
The complete genome sequences of phiIPLA35 and phiIPLA88 were determined. The phiIPLA35 genome comprised 45,344 bp coding for 62 putative ORFs of more than 50 codons, while in the phiIPLA88 genome 42,526 bp and 61 putative ORFs were identified (Tables 1 and 2; Fig. 3 and 4). The G+C content of phiIPLA35 and phiIPLA88 was 33.2% and 34.9%, respectively, which is slightly higher than that of bovine S. aureus strains (32.8%) (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi).
TABLE 1.
Features of bacteriophage phiIPLA35 ORFs, gene products, and functional assignments
| ORF | Position (nt) |
Length (nt) | No. of amino acids | Size (kDa [pI]) | Predictive function | Closest hit (e value) | % Amino acid identity (% similarity) | Accession no. | Predicted domain (protein ID) (e value)a | |
|---|---|---|---|---|---|---|---|---|---|---|
| From | To | |||||||||
| 1 | 1206 | 1 | 1,206 | 401 | 47.2 (10.24) | Integrase | S. aureus MSSA476 (0.0) | 100 (100) | YP_043018.1 | Phage integrase (PF00589) (1.3e−46) |
| 2 | 1333 | 1956 | 624 | 207 | 24.3 (9.53) | Hypothetical protein | Staphylococcus φPVL108 (1e−34) | 100 (100) | YP_918893.1 | Signal peptide; transmembrane regions |
| 3 | 2549 | 2154 | 396 | 131 | 15.3 (4.46) | Hypothetical protein | Staphylococcus φPV83 (3e−70) | 99 (99) | NP_061593.1 | |
| 4 | 3011 | 2577 | 435 | 144 | 15.8 (9.54) | Putative lipoprotein | S. aureus MSSA476 (4e−73) | 93 (97) | YP_043022.1 | Signal peptide |
| 5 | 3029 | 3400 | 372 | 123 | 14.7 (8.78) | Unknown conserved protein | S. aureus MSSA476 (3e−66) | 99 (100) | YP_043023.1 | DUF955 (PF06114) (2.9e−18) |
| 6 | 3833 | 3579 | 255 | 84 | 9.9 (9.99) | Truncated repressor | S. aureus Mu50 (8e−15) | 90 (92) | NP_371375.1 | HTH_3 (PF01381) (1.1e−12) |
| 7 | 3994 | 4185 | 192 | 63 | 7.2 (5.59) | Cro/CI family transcription regulator | S. aureus MSSA476 (1e−28) | 100 (100) | YP_043025.1 | HTH_3 (PF01381) (2.9e−10) |
| 8 | 4273 | 4449 | 177 | 58 | 6.8 (4.63) | Hypothetical protein | S. aureus MSSA476 (2e−26) | 98 (100) | YP_043026.1 | |
| 9 | 4676 | 4446 | 231 | 76 | 8.8 (6.21) | Hypothetical protein | S. aureus USA300 (1e−36) | 100 (100) | YP_494616.1 | |
| 10 | 4732 | 4947 | 216 | 71 | 84 (9.93) | Hypothetical protein | S. aureus MSSA476 (5e−33) | 100 (100) | YP_043028.1 | |
| 11 | 4972 | 5235 | 264 | 87 | 10.4 (9.83) | Hypothetical protein | Staphylococcus φ12 (4e−43) | 97 (98) | NP_803315.1 | |
| 12 | 5247 | 5408 | 162 | 53 | 6.1 (4.16) | Hypothetical conserved protein | Staphylococcus φ47 (2e−21) | 98 (100) | YP_240044.1 | DUF1270 (PF06900) (3.4e−36); transmembrane region; signal peptide |
| 13 | 5487 | 5810 | 324 | 107 | 12.5 (7.85) | Hypothetical protein | Staphylococcus φ47 (1e−55) | 100 (100) | YP_240046.1 | |
| 14 | 5825 | 6187 | 363 | 120 | 13.6 (4.63) | Hypothetical protein | S. aureus MSSA476 (3e−54) | 93 (95) | YP_043032.1 | |
| 15 | 6184 | 7350 | 1,167 | 388 | 44.3 (5.39) | Hypothetical protein | S. aureus MSSA476 (0.0) | 98 (99) | YP_043033.1 | |
| 16 | 7376 | 7933 | 558 | 185 | 20.1 (4.65) | Hypothetical protein | S. aureus φ12 (1e−99) | 99 (100) | NP_803317.1 | |
| 17 | 8001 | 9953 | 1,953 | 650 | 73.6 (6.59) | Phage DNA polymerase A | S. aureus MSSA476 (0.0) | 98 (99) | YP_043035.1 | DNA_pol_A (PF00476) (1.9e−06) |
| 18 | 9966 | 10151 | 186 | 61 | 7.1 (4.19) | Hypothetical protein | Staphylococcus φ12 (9e−27) | 100 (100) | NP_803319.1 | |
| 19 | 10148 | 10552 | 405 | 134 | 16.7 (10.35) | Primase | S. aureus MSSA476 (1e−72) | 99 (100) | YP_043037.1 | PVL_ORF50 (PF07768) (1.7e−90); HTH motif |
| 20 | 10552 | 10809 | 258 | 85 | 9.7 (5.45) | Hypothetical protein | Staphylococcus φ12 (8e−43) | 96 (98) | NP_803321.1 | |
| 21 | 10812 | 11012 | 201 | 66 | 7.5 (10.78) | Hypothetical protein | Staphylococcus φNPH82 (1e−16) | 67 (83) | YP_950654.1 | |
| 22 | 11009 | 11698 | 690 | 229 | 27.1 (4.93) | Putative methyltransferase | Listeria welshimeri SLCC5334 (1e−53) | 49 (68) | YP_849399.1 | N6_Mtase (PF02384) (1.6e−05) |
| 23 | 11712 | 11918 | 207 | 68 | 7.9 (4.68) | Hypothetical protein | S. aureus JH9 (2e−26) | 82 (89) | YP_001245710.1 | |
| 24 | 11921 | 12349 | 429 | 142 | 16.3 (5.16) | Hypothetical protein | Staphylococcus φ71 (9e−24) | 48 (57) | YP_240442.1 | |
| 25 | 12346 | 12540 | 195 | 64 | 7.5 (9.24) | Hypothetical protein | Staphylococcus φ187 (1e−24) | 88 (95) | YP_239554.1 | |
| 26 | 12570 | 12821 | 252 | 83 | 10.1 (8.62) | Hypothetical protein | Staphylococcus φPV83 (1e−35) | 93 (97) | NP_061617.1 | |
| 27 | 12814 | 13062 | 249 | 82 | 9.2 (4.00) | Hypothetical conserved protein | Staphylococcus φ71 (2e−38) | 97 (100) | YP_240444.1 | DUF1024 (PF06260) (2.5e−61) |
| 28 | 13055 | 13615 | 561 | 186 | 22.1 (4.77) | dUTPase | Staphylococcus φX2 (5e−105) | 99 (100) | AAX92028.1 | dUTPase_2 (PF08761) (3.7e−08) |
| 29 | 13852 | 14055 | 204 | 67 | 7.8 (9.80) | Hypothetical conserved protein | Staphylococcus φ42E (9e−30) | 98 (98) | YP_239920.1 | DUF1381 (PF07129) (7.4e−38) |
| 30 | 14052 | 14204 | 153 | 50 | 5.9 (4.33) | Transcriptional activator RinB | Staphylococcus φtp310-2 (1e−20) | 96 (98) | YP_001429932.1 | RinB (PF06116) (6.3e−35) |
| 31 | 14272 | 14472 | 201 | 66 | 7.8 (9.23) | Hypothetical conserved protein | Staphylococcus φ42E (2e−29) | 100 (100) | YP_239924.1 | DUF1514 (PF07438) (1.5e−48); signal peptide; transmembrane regions |
| 32 | 14524 | 16971 | 2,448 | 815 | 94.3 (4.97) | Hypothetical protein | S. aureus MRSA252 (0.0) | 99 (99) | YP_040926.1 | VirE (PF05272) (4.1e−127) |
| 33 | 17312 | 17602 | 291 | 96 | 11.2 (9.81) | Putative nuclease | Staphylococcus φ42E (4e−51) | 100 (100) | YP_239929.1 | VRR_NUC (PF08774) (1.1e−20) |
| 34 | 17583 | 18950 | 1,368 | 455 | 52.9 (6.66) | Putative helicase | Staphylococcus φ42E (0.0) | 100 (100) | YP_239930.1 | SNF2_N (PF00176) (3.7e−05); DEAD-like helicase superfamily (SM00487) (1.6e−13) |
| 35 | 18963 | 19400 | 438 | 145 | 17.0 (9.76) | Transcriptional regulator (RinA) | Staphylococcus prophage L54a (2e−78) | 100 (100) | YP_185256.1 | Phage_rinA; phage transcriptional regulator (IPR006523) (9.6e−16) |
| 36 | 19557 | 19871 | 315 | 104 | 12.5 (8.58) | HNH endonuclease family protein | Staphylococcus φ42E (5e−53) | 98 (100) | YP_239933.1 | HNH (PF01844) (1.6e−10) |
| 37 | 19980 | 20303 | 324 | 107 | 12.3 (8.98) | Terminase small subunit | Staphylococcus φ12 (3e−55) | 100 (100) | NP_803335.1 | Terminase_4 (PF05119) (9.4e−31) |
| 38 | 20284 | 21984 | 1,701 | 566 | 65.4 (5.64) | Terminase large subunit | S. aureus MSSA476 (0.0) | 100 (100) | YP_043059.1 | Terminase_1 (PF03354) (8.1e−93) |
| 39 | 21998 | 23227 | 1,230 | 409 | 47.3 (5.62) | Phage portal protein | Staphylococcus φ3A (0.0) | 100 (100) | YP_239936.1 | Phage portal (PF04860) (3.6e−116) |
| 40 | 23211 | 23984 | 774 | 257 | 29.1 (4.77) | Clp protease | Staphylococcus φSLT (9e−142) | 100 (100) | NP_075503.1 | Clp_protease (PF00574)(1.5e−51); transmembrane regions |
| 41 | 23996 | 25159 | 1,164 | 387 | 43.5 (4.95) | Major head protein | Staphylococcus φ12 (0) | 100 (100) | NP_803339 | Phage capsid (PF05065) (0.0012) |
| 42 | 25228 | 25506 | 279 | 92 | 10.8 (4.93) | Possible DNA packaging protein | Staphylococcus φSLT (0) | 100 (100) | NP_075505 | Put_DNA_pack: uncharacterized phage protein (2.7e−09) |
| 43 | 25518 | 25850 | 333 | 110 | 12.9 (5.77) | Hypothetical protein | Staphylococcus φ12 (3e−58) | 100 (100) | NP_803340.1 | |
| 44 | 25847 | 26248 | 402 | 133 | 15.1 (10.08) | Hypothetical protein | Staphylococcus φ12 (1e−68) | 99 (99) | NP_803341. | |
| 45 | 26249 | 26644 | 396 | 131 | 15.5 (8.76) | Hypothetical protein | Staphylococcus φSLT (1e−68) | 100 (100) | NP_075508 | |
| 46 | 26679 | 27320 | 642 | 213 | 23.4 (4.75) | Major tail protein | Staphylococcus φ12 (2e−120) | 100 (100) | NP_803343 | Phage_tail (PF04630) (2.2e−99) |
| 47 | 27412 | 27867 | 456 | 151 | 16.0 (4.39) | Tail protein | Staphylococcus φ12 (6e−81) | 99 (99) | NP_803344 | Big_2 (PF02368) (1.1e—07) |
| 48 | 27925 | 28275 | 351 | 116 | 13.6 (4.35) | Hypothetical protein | Staphylococcus p φSLT (4e−60) | 100 (100) | NP_075511 | |
| 49 | 28317 | 28475 | 159 | 52 | 6.1 (4.65) | Hypothetical protein | Staphylococcus φ42E (3e−21) | 100 (100) | YP_239870 | |
| 50 | 28489 | 34689 | 6,201 | 2,066 | 226.05 (10.23) | Tail tape measure protein | Staphylococcus φSLT | 99 (99) | YP_494090.1 | Lytic transglycosylase-like SLT (PF01464) (0.0013); Tape_meas_TP901 (1.4e−99); Peptidase_M23 (PF01551) (5.5e−36) |
| 51 | 34689 | 35513 | 825 | 274 | 31.2 (4.96) | Hypothetical protein | Staphylococcus φ12 (2e−158) | 99 (99) | NP_803347.1 | DUF1306 (PF06997) (1.4e−157) |
| 52 | 35522 | 37102 | 1,581 | 526 | 60.9 (5.66) | Conserved hypothetical protein | Staphylococcus φ47 (0.0) | 98 (99) | AAX91197.1 | DUF1142 (PF06605) (0) |
| 53 | 37102 | 37392 | 291 | 96 | 10.5 (5.04) | Unknown conserved protein | Staphylococcus φ42E (3e−49) | 100 (100) | YP_239876.1 | |
| 54 | 37408 | 39318 | 1,911 | 636 | 73.1 (5.80) | Minor structural protein | Staphylococcus φ12 (0.0) | 99 (100) | NP_803350.1 | |
| 55 | 39318 | 40784 | 1,467 | 488 | 54.1 (5.42) | Hypothetical protein | S. aureus MSSA476 (0.0) | 98 (99) | CAG42725.1 | |
| 56 | 40784 | 41173 | 390 | 129 | 14.7 (4.56) | Hypothetical protein | Staphylococcus φ12 (1e−65) | 100 (100) | NP_803352.1 | |
| 57 | 41166 | 41330 | 165 | 54 | 6.5 (4.06) | Hypothetical protein | Staphylococcus φ12 (1e−24) | 100 (100) | AAL82328.1 | Phage_XkdX: phage (TIGR01669) (7.5e−15) |
| 58 | 41376 | 41675 | 300 | 99 | 12.0 (7.63) | Hypothetical protein | Staphylococcus φSLT (9e−51) | 100 (100) | NP_075520.1 | Transmembrane regions |
| 59 | 41811 | 42113 | 303 | 100 | 11.1 (9.58) | Holin | Staphylococcus φ12 (3e−50) | 100 (100) | NP_803354.1 | Signal peptide; Phage_holin (PF04688) (1.1e−20) |
| 60 | 42124 | 43578 | 1,455 | 484 | 53.7 (9.62) | Amidase | Staphylococcus φtp310-1 (0.0) | 98 (99) | YP_001429893.1 | Amidase_3 (PF01520) (2.6e−09); CHAP (PF05257) (9.9e−43); SH3_5 (PF08460) (7.3e−08) |
| 61 | 43969 | 44319 | 351 | 116 | 13.3 (9.82) | Hypothetical protein | Staphylococcus p φ71 (2e−44) | 92 (93) | YP_240409.1 | Transmembrane regions |
| 62 | 44552 | 45007 | 456 | 151 | 17.1 (9.88) | Hypothetical protein | Staphylococcus p φ71 (4e−78) | 100 (100) | YP_240410.1 | Signal peptide; transmembrane regions |
ID, identifier; HTH, helix-turn-helix; PVL, Panton-Valentine leukocidin.
TABLE 2.
Features of bacteriophage phiIPLA88 ORFs, gene products, and functional assignments
| ORF | Position (nt) |
Length (nt) | No. of amino acids | Size (kDa [pI]) | Predictive function | Closest hit (e value) | % Amino acid identity (% similarity) | Accession no. | Predicted domain (protein ID) (e value)a | |
|---|---|---|---|---|---|---|---|---|---|---|
| From | To | |||||||||
| 1 | 1065 | 1 | 1,065 | 354 | 41.0 (9.63) | Integrase | S. aureus RF122 (0.0) | 99 (99) | YP_417222.1 | Phage integrase (PF00589) (2.2e−14) |
| 2 | 1639 | 1124 | 516 | 171 | 19.2 (8.98) | Hypothetical protein | S. aureus RF122 (6e−93) | 100 (100) | YP_417221.1 | Prokar_Lipoprotein(PS51257) (0.0) |
| 3 | 2143 | 1643 | 501 | 166 | 19.2 (9.58) | Putative excisionase | S. aureus RF122 (9e−89) | 100 (100) | YP_417220.1 | |
| 4 | 2834 | 2196 | 639 | 212 | 24.2 (5.67) | Mutated repressor | S. aureus phiNM (3e−107) | 98 (99) | YP_873953.1 | HTH (PF01381) (1.7e−14); Peptidase_S24 (PF00717) (0.00011) |
| 5 | 3025 | 3279 | 255 | 84 | 10 (9.03) | Cro | S. aureus phiNM (4e−37) | 97 (100) | YP_873954.1 | HTH (PF01381) (1.8e−13) |
| 6 | 4050 | 3460 | 591 | 196 | 22.3 (6.23) | Hypothetical protein | S. aureus RF122 (2e−107) | 100 (100) | YP_417216.1 | Transmembrane region |
| 7 | 4107 | 4856 | 750 | 249 | 28.5 (9.79) | Antirepressor | S. aureus RF122 (4e−144) | 100 (100) | YP_417215.1 | ANT (PF03374) (2.5e−102); AntA (PF08346) (3.8e−31) |
| 8 | 4872 | 5087 | 216 | 71 | 8.7 (9.70) | Hypothetical protein | S. aureus RF122 (4e−34) | 100 (100) | YP_417214.1 | DUF771 (PF05595) (3.3e−6) |
| 9 | 5102 | 5272 | 171 | 56 | 6.4 (8.60) | Hypothetical protein | S. aureus RF122 (5e−24) | 100 (100) | YP_417213.1 | DUF1270 (PF06900) (5.0e−34) |
| 10 | 5277 | 5591 | 315 | 104 | 12.4 (9.60) | Hypothetical protein | S. aureus RF122 (6e−53) | 100 (100) | YP_417212.1 | |
| 11 | 5656 | 5958 | 303 | 100 | 11.1 (4.48) | Hypothetical protein | S. aureus RF122 (8e−49) | 100 (100) | YP_417211.1 | |
| 12 | 5963 | 6223 | 261 | 86 | 10.1 (4.40) | Hypothetical protein | S. aureus φPVL108 (4e−46) | 94 (96) | YP_918906.1 | DUF1108 (PF06531)(6.2e−60) |
| 13 | 6233 | 6454 | 222 | 73 | 8.6 (5.42) | Hypothetical protein | S. aureus RF122 (4e−35) | 100 (100) | YP_417209.1 | |
| 14 | 6447 | 7067 | 621 | 206 | 23.5 (5.65) | Topoisomerase | S. aureus RF122 (6e−119) | 100 (100) | YP_417208.1 | DUF1071 (PF06378) (2.7e−12) |
| 15 | 7070 | 7495 | 426 | 141 | 15.8 (5.24) | ssDNA binding protein | S. aureus RF122 (9e−76) | 100 (100) | YP_417207.1 | SSB (PF00436) (1.6e−29) |
| 16 | 7509 | 8180 | 672 | 223 | 26 (6.60) | Hypothetical protein | S. aureus RF122 (8e−131) | 100 (100) | YP_417206.1 | DUF968 (PF06147) (0.0) |
| 17 | 8177 | 8932 | 756 | 251 | 29.4 (8.37) | DNA replication protein | S. aureus RF122 (1e−145) | 100 (100) | YP_417205.1 | DnaD and phage-associated region (IPR006343) (3.6e−10) |
| 18 | 8932 | 9288 | 357 | 118 | 13.7 (5.74) | Hypothetical protein | S. aureus RF122 (1e−64) | 100 (100) | YP_417204.1 | |
| 19 | 9285 | 10526 | 1,242 | 413 | 47.4 (5.14) | DNA helicase | S. aureus RF122 (0.0) | 100 (100) | YP_417203.1 | DnaB_C (PF03796) (7.2e−51) |
| 20 | 10523 | 10738 | 216 | 71 | 8.5 (5.13) | Hypothetical protein | S. aureus RF122 (1e−34) | 100 (100) | YP_417202.1 | |
| 21 | 10741 | 10962 | 222 | 73 | 8.5 (4.83) | Hypothetical protein | S. aureus RF122 (2e−35) | 100 (100) | YP_417201.1 | |
| 22 | 10973 | 11377 | 405 | 134 | 16.1 (9.79) | Hypothetical protein | S. aureus RF122 (1e−72) | 100 (100) | YP_417200.1 | DUF1064 (PF06356) (2.7e−86) |
| 23 | 11382 | 11567 | 186 | 61 | 7.2 (4.06) | Hypothetical protein | S. aureus φ29 (5e−25) | 93 (96) | YP_240593.1 | |
| 24 | 11568 | 11930 | 363 | 120 | 14.5 (10.42) | Hypothetical protein | S. aureus RF122 (2e−65) | 100 (100) | YP_417199.1 | PVL_ORF50 (PF07768)(1.5e-65) |
| 25 | 11930 | 12184 | 255 | 84 | 9.6 (4.99) | Hypothetical protein | S. aureus RF122 (8e−44) | 100 (100) | YP_417198.1 | |
| 26 | 12184 | 12432 | 249 | 82 | 9.8 (5.90) | Hypothetical protein | S. aureus RF122 (9e−42) | 100 (100) | YP_417197.1 | Phage_Orf51 (PF06194)(3.1e−59) |
| 27 | 12441 | 12677 | 237 | 78 | 9.6 (10.17) | Hypothetical protein | S. aureus RF122 (3e−39) | 100 (100) | YP_417196.1 | |
| 28 | 12737 | 12991 | 255 | 84 | 9.5 (3.95) | Hypothetical protein | S. aureus RF122 (3e−40) | 100 (100) | YP_417195.1 | DUF1024 (PF06260)(1.3e−51) |
| 29 | 12978 | 13139 | 162 | 53 | 6.1 (3.30) | Hypothetical protein | S. aureus phage 71 (1e−19) | 98 (98) | YP_240445.1 | |
| 30 | 13139 | 13675 | 537 | 178 | 20.6 (4.36) | dUTPase | S. aureus RF122 (6e−100) | 100 (100) | YP_417194.1 | dUTPase_2 (PF08761)(6.5e−6) |
| 31 | 13712 | 13957 | 246 | 81 | 9.5 (10.08) | Hypothetical protein | S. aureus RF122 (1e−35) | 100 (100) | YP_417193.1 | Transmembrane regions |
| 32 | 13954 | 14160 | 207 | 68 | 7.7 (7.31) | Hypothetical protein | S. aureus RF122 (6e−32) | 98 (100) | YP_417192.1 | DUF1381 (PF07129) (2.2e−43) |
| 33 | 14157 | 14330 | 174 | 57 | 6.5 (4.05) | Transcriptional activator | S. aureus RF122 (4e−25) | 100 (100) | YP_417191.1 | RinB (PF06116) (4.5e−38) |
| 34 | 14492 | 14911 | 420 | 139 | 16.1 (9.52) | Transcriptional activator | S. aureus RF122 (5e−72) | 100 (100) | YP_417190.1 | Phage transcriptional activator RinA (IPR006523) (2.1e−32) |
| 35 | 15099 | 15593 | 495 | 164 | 18.9 (6.24) | Terminase small subunit | S. aureus RF122 (3e−93) | 100 (100) | YP_417189.1 | Terminase_2 (PF03592) (1.3e−39) |
| 36 | 15596 | 16885 | 1,290 | 429 | 50.2 (9.33) | Terminase large subunit | S. aureus RF122 (0.0) | 100 (100) | YP_417188.1 | Terminase_3 (PF04466) (2.2e−11) |
| 37 | 16896 | 18437 | 1,542 | 513 | 59.6 (4.60) | Portal protein | S. aureus RF122 (0.0) | 100 (100) | YP_417187.1 | Phage_prot_Gp6 (PF05133) (2.8e−100) |
| 38 | 18319 | 18504 | 186 | 61 | 7.1 (11.05) | Hypothetical protein | Staphylococcus φ80α (2e−24) | 98 (98) | YP_001285357.1 | |
| 39 | 18549 | 19433 | 885 | 294 | 33.9 (9.38) | Head morphogenesis protein | S. aureus RF122 (1e−173) | 100 (100) | YP_417186.1 | Phage_Mu_F (PF04233) (5.1e−06) |
| 40 | 19506 | 19676 | 171 | 56 | 6.5 (9.70) | Hypothetical protein | S. aureus φ53 (5e−22) | 92 (94) | YP_239646.1 | |
| 41 | 19811 | 20425 | 615 | 204 | 23.6 (4.92) | Scaffold protein | S. aureus RF122 (4e−109) | 99 (100) | YP_417185.1 | |
| 42 | 20439 | 21413 | 975 | 324 | 36.6 (4.93) | Major head protein | S. aureus RF122 (3e−171) | 100 (100) | YP_417184.1 | Major capsid protein gp5superfamily (SSF56563)(2.8e-26) |
| 43 | 21435 | 21722 | 288 | 95 | 10.9 (5.21) | Hypothetical protein | S. aureus RF122 (9e−47) | 100 (100) | YP_417183.1 | |
| 44 | 21731 | 22063 | 333 | 110 | 12.8 (5.08) | DNA packaging protein | S. aureus RF122 (8e−56) | 100 (100) | YP_417182.1 | |
| 45 | 22362 | 22709 | 348 | 115 | 13.4 (5.60) | Head to tail joining protein | S. aureus RF122 (3e−62) | 100 (100) | YP_417180.1 | TP901-1_ORF40 (PF07772) (1.1e−38) |
| 46 | 22721 | 23104 | 384 | 127 | 14.7 (4.21) | Hypothetical protein | S. aureus RF122 (8e−70) | 100 (100) | YP_417179.1 | |
| 47 | 23122 | 23703 | 582 | 193 | 21.2 (4.55) | Major tail protein | S. aureus RF122 (4e−109) | 100 (100) | YP_417178.1 | Phage_tail_2 (PF06199)(4.1e−84) |
| 48 | 23765 | 24130 | 366 | 121 | 13.1 (4.30) | Hypothetical protein | S. aureus RF122 (4e−60) | 100 (100) | YP_417177.1 | |
| 49 | 24160 | 24504 | 345 | 114 | 13.5 (4.41) | Minor structural protein | φ11 (7e−60) | 100 (100) | NP_803294.1 | |
| 50 | 24521 | 27988 | 3,468 | 1,155 | 125.6 (10.37) | Tape measure protein | S. aureus φNM (0.0) | 98 (99) | YP_874001.1 | TMP (PF05017) (0.089); WD_REPEATS_1 (PS00678); transmembrane regions |
| 51 | 28001 | 28948 | 948 | 315 | 37 (7.28) | Minor tail protein | S. aureus RF122 (0.0) | 100 (100) | NP_803296.1 | WD_REPEATS_1 (PS00678) |
| 52 | 28957 | 30858 | 1,902 | 633 | 71.1 (6.96) | Putative anti-receptor protein | S. aureus RF122 (0.0) | 99 (100) | YP_417173.1 | Phage minor structural protein, N-terminal (IPR007119)(0.024); SGNH hydrolaseIPR13830 (1.1e−24) |
| 53 | 30873 | 32783 | 1,911 | 636 | 73.3 (6.64) | Minor structural protein | S. aureus RF122 (0.0) | 98 (99) | YP_417172.1 | |
| 54 | 32783 | 34606 | 1,824 | 607 | 66.5 (4.62) | Minor tail protein | S. aureus RF122 (0.0) | 99 (99) | YP_417171.1 | |
| 55 | 34606 | 34983 | 378 | 125 | 14.1 (4.30) | Hypothetical protein | S. aureus φ85 (2e−59) | 92 (95) | YP_239743.1 | |
| 56 | 34984 | 35166 | 183 | 60 | 7.2 (6.69) | Hypothetical protein | S. aureus φ29 (9e−29) | 98 (98) | YP_240554.1 | Phage uncharacterized protein XkdX (IPR010022) (3.3e-22) |
| 57 | 35207 | 35506 | 300 | 99 | 12.0 (10.23) | Hypothetical protein | S. aureus φNM (2e−35) | 89 (94) | YP_874008.1 | Transmembrane region |
| 58 | 35643 | 37547 | 1,905 | 634 | 72.5 (9.78) | Hydrolase | S. aureus RF122 (0.0) | 99 (99) | YP_417168.1 | CHAP (PF05257) (1.5e−30); LYZ2 (SM00047) (1e−51) |
| 59 | 37660 | 39573 | 1,914 | 637 | 71.8 (6.48) | Tail fiber protein | S. aureus RF122 (0.0) | 99 (99) | YP_417167.1 | Collagen alpha chain (PTHR10499) (0.00037) |
| 60 | 39624 | 40061 | 438 | 145 | 15.6 (4.52) | Holin | S. aureus RF122 (7e−79) | 100 (100) | YP_417166.1 | Transmembrane regions Phage_holin_1 (PF04531) (9.1e−50) |
| 61 | 40042 | 41487 | 1,446 | 481 | 53.7 (8.71) | Amidase | S. aureus RF122 (0.0) | 100 (100) | YP_417165.1 | Amidase_2 (PF01510) (2.5e−38); SH3b (SM00287) (1.4e−12); CHAP (PF05257) (4.7e−47) |
ID, identifier; HTH, helix-turn-helix; PVL, Panton-Valentine leukocidin; SSB, single-stranded DNA binding protein.
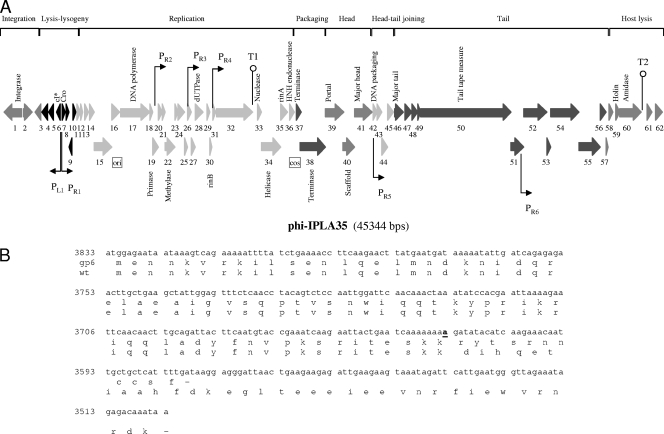
FIG. 3.
Physical and genetic map of the bacteriophage phiIPLA35. (A) The ORFs are sequentially numbered, indicated by arrows proportional to their lengths and pointing toward their direction of transcription (L = left, R = right). Some ORFs have been placed below for clarity. The functional modules are indicated on top of the scheme, and the names of several putatively or experimentally identified genes are shown below. Promoter (P) and terminator (T) sequences are also indicated. (B) DNA sequence of gp6 showing the insertion mutation (in bold and underlined) and alignment of the putative CI amino acid sequence of the mutant and the wild-type (wt) phages, respectively. Numbers to the left of the DNA sequence are the genome coordinates.
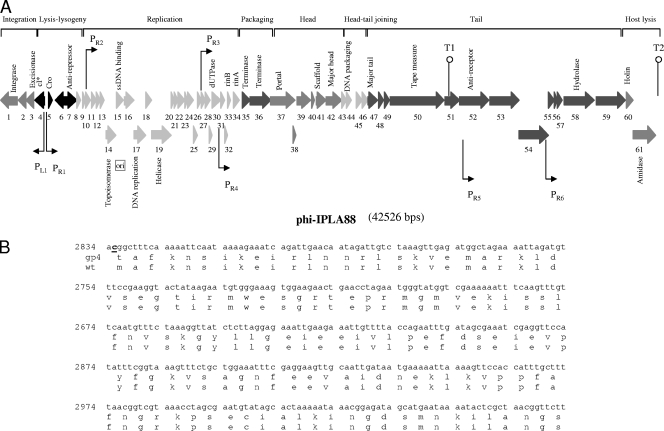
FIG. 4.
Physical and genetic map of the bacteriophage phiIPLA88. (A) The ORFs are sequentially numbered, indicated by arrows proportional to their lengths and pointing toward their directions of transcription. Some ORFs have been placed below for clarity. The functional modules are indicated on top of the scheme, and the names of several putatively or experimentally identified genes are shown below. Promoter (P) and terminator (T) sequences are also indicated. (B) Partial DNA sequence of gp4 showing the replacement mutation (in bold and underlined) and alignment of the putative CI amino acid sequence of the mutant and the wild-type (wt) phages, respectively. Numbers to the left of the DNA sequence are the genome coordinates.
The overall genome organization of phiIPLA35 and phiIPLA88 is described below. As shown in Fig. 3 and 4, both genomes are apparently organized into three major gene clusters, containing (i) the “lysogeny control” region, which is probably transcribed mostly leftwards based on the gene orientation; (ii) the “early” region, encoding products for the replication, recombination, and modification of the phage DNA; and (iii) the “late genes,” coding for structural and assembly proteins, DNA packaging proteins, and the lysis proteins.
The majority of ORFs presented an AUG start codon, while two (orf34 and orf38) initiated at GUG in phiIPLA35 and four (orf31, orf32, orf50, and orf57) in phiIPLA88. In addition, orf2 from phiIPLA35 and orf1 from phiIPLA88 have UUG as a start codon. The genomes are densely coded, having noncoding regions that comprise only 7.6% and 7.2% for phiIPLA35 and phiIPLA88, respectively. Furthermore, the phiIPLA35 and phiIPLA88 genomes encode 18 and 16 ORFs, respectively, in which the start codon overlaps with the stop codon of the preceding gene. Several putative promoters in phiIPLA35 and in phiIPLA88 were found by searching for the S. aureus σ70-dependent promoter consensus motif (Fig. 3 and 4). Regions forming stem-loop structures, probably representing factor-independent transcriptional terminators (Fig. 3 and 4), were also determined.
The amino acid sequences of the predicted ORFs were screened for similarities with sequences from the available databases for preliminary functional assignments (Tables 1 and 2). Significant matches were obtained for 26 ORFs from phiIPLA35 and for 32 ORFs from phiIPLA88, and biological functions were assigned. No tRNA genes were found. No virulence genes were clearly identified. However, a VirE motif, found in bacterial virulence protein E, was detected in phiIPLA35 gp32. VirE motifs had also been described in several phage DNA replication proteins in the databases and might have a particular role in DNA metabolism (51). Nevertheless, a putative role in promoting host pathogenicity cannot be excluded.
Comparative genomics of phiIPLA35 and phiIPLA88 phages.
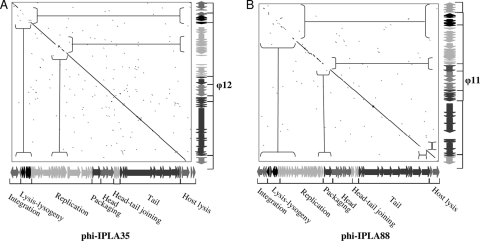
From a comparative perspective, BLAST results and subsequent proteome-based clustering using CoreGenes/CoreExtractor (29) quickly reveal that φ12, φ47, φ3A, φ42E, and φSLT are the closest relatives of phiIPLA35. The phiIPLA88 genome is practically identical to a prophage found in S. aureus RF122 and strongly related to φ11, φETA2, φ69, φ53, φNM, φ85, and φ187. Comparison of the phiIPLA35 sequence with the related φ12 sequence by the dot plot method (Fig. 5A) revealed colinearity throughout almost the entire genomic region. The extensive similarity of phiIPLA35 with these phages is underlined by the close relationship of some of them previously observed. The 3A, 42e, and 47 phages have been ascribed to class II, clade B (26), and display a similar B2 morphotype with a long, noncontractile tail and elongated head as described for phiIPLA35 (17). Detailed analysis between phiIPLA88 and φ11 by dot plot, as shown in Fig. 5B, revealed that the late gene cluster extending from the DNA packaging genes to the host lysis genes is more closely related than the early cluster comprising the lysis-lysogeny and replication clusters. However, no official taxonomic status can be given to phiIPLA35 and phiIPLA88 since both phages are artificially selected mutants (17).
FIG. 5.
Dot plot alignment of phiIPLA35 (A) and phiIPLA88 (B) genomes and related φ12 and φ11, respectively. The genome sequence is represented by the corresponding ORFs in each phage genome.
The deficient lysogeny module.
phiIPLA35 and phiIPLA88 are lytic derivatives selected after sodium pyrophosphate treatment of their parental temperate phages φA72 and φH5, respectively (17). Hence, the presence of a lysogeny module was expected. In both phages, gp1 was identified as the phage integrase (Tables 1 and 2), which would catalyze site-specific integration as it displays the C-terminal catalytic domain of DNA breaking-rejoining enzymes and of Cre integrases. The other module that is commonly present in temperate Siphoviridae is the lysis/lysogeny switch. The phiIPLA35 gp6 and phiIPLA88 gp4 proteins share extended similarity with repressors of the CI type. However, phiIPLA35 gp6 lacks 25 amino acids in the carboxy-terminal sequence (Fig. 3B), and the start codon was lost in phiIPLA88 gp4 (Fig. 4B). After the corresponding DNA regions of the parental temperate phages were sequenced and compared, two point mutations were mapped: (i) a 1-base insertion in phiIPLA35 at position 3614 and (ii) a 1-base replacement in phiIPLA88 that shifted the ATG codon to TTG. The presence (or absence) of a truncated CI repressor would explain why the lytic derivative phages are unable to lysogenize (17). It is worth mentioning that, in spite of the use of pyrophosphate that destabilizes the phage capsid and, consequently, selects for smaller genomes, no differences were observed between the restriction patterns of the temperate phages versus the lytic variants (data not shown), precluding the presence of relatively large DNA deletions. Therefore, it is likely that the lytic phages are spontaneous mutants, and the method used to obtain them would have been irrelevant. The second putative repressor-encoding gene (phiIPLA35 gp7 and phiIPLA88 gp5) specifies a polypeptide which resembles several phage-related transcription repressors and may represent a Cro analogue. Analysis of the phiIPLA88 orf7 reveals a strong sequence similarity to designated antirepressors from a number of very different phages and could be responsible for inactivation/bypass of the CI transcription repressor.
The DNA replication, recombination, and modification module.
Both phages have replication modules with genes involved in DNA metabolism and replication. Gene products predicted to be DNA polymerases are found in both phages, as well as primases, single-stranded DNA binding proteins, helicases, and topoisomerases (Tables 1 and 2). In phiIPLA35, orf22 encodes an N6-adenine methyltransferase, which probably specifically modifies newly synthesized viral DNA. phiIPLA35 gp28 and phiIPLA88 gp30 are predicted as dUTPases, the most conserved gene products among P335 phage species from Lactococcus lactis (27) and also present in many S. aureus phages. A region overlapping phiIPLA35 orf16 and phiIPLA88 orf15 also contains the putative oriC, as predicted from a cumulative GC skew analysis (Fig. 3 and 4). Within the phiIPLA35 replication module, gp36 is a zinc-dependent HNH homing endonuclease. It has been proposed that these endonuclease genes in phage genomes can be considered analogous to insertion or transposon elements in bacterial genomes (4, 43). phiIPLA35 orf33 encodes a nuclease that may be involved in host DNA degradation. In both phages, homologs to the transcriptional regulators rinA and rinB were detected (Tables 1 and 2). These genes have been shown to be required for the activation of the staphylococcal φ11 integrase gene expression (55).
The structural and lysis module.
The structural module can be divided into five submodules, ordered as packaging, head morphogenesis, head-tail connection, tail morphogenesis, and lysis (Fig. 3 and 4). Genes included specify proteins homologous to (or that bear domains typical of) small and large terminase subunits, portal proteins, tail tape measure, and head and tail structural proteins.
A putative translational frameshifting was observed in both phages in the two genes (orf48 and orf49) just upstream from the tail tape measure gene. We have found a canonical −1 slippery site (G GGA AAG) located 6 and 20 codons upstream from the termination codon of orf48 of phiIPLA35 and phiIPLA88, respectively. Putative pseudoknots downstream of each slippery sequence have also been detected (data not shown). This position is analogous to the phage lambda fusion protein gpG-T essential for tail assembly, even though it does not become part of the mature virion (32). In addition, in the case of tailed bacteriophages, the frameshifted products contained immunoglobulin-like domains which may be important to phage infection (14).
The lysis cassette would comprise the holin and lysin genes and was predicted in both phages (Fig. 3 and 4) and experimentally verified previously for recombinant phiIPLA88 gp61 (36). The lysins show typical domains of amidase and endopeptidase activities.
Proteomic analysis of virion particles.
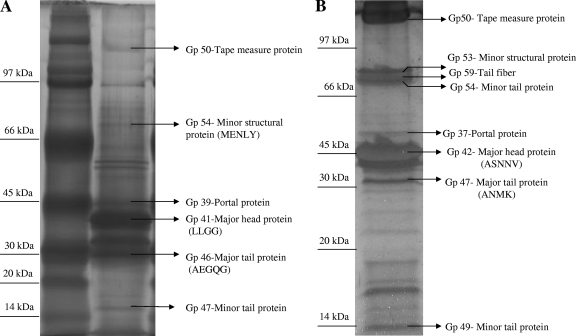
The bioinformatics analysis of the phiIPLA35 and phiIPLA88 genomes suggests that the regions from orf39 to orf57 in phiIPLA35 and from orf37 to orf59 in phiIPLA88 harbor the genes encoding the proteins that build up the mature virions. Combining both N-terminal sequencing and mass spectrometry analysis after trypsin digestion of Coomassie blue-stained and silver-stained gels, respectively, six proteins of phiIPLA35 and eight of phiIPLA88 were identified, and their structural roles predicted by genome analysis were confirmed (Fig. 6). The additional faint bands on the gel could not be identified or represented mixtures of degradation products from the structural proteins.
FIG. 6.
Silver-stained SDS-PAGE analysis of the phiIPLA35 (A) and phiIPLA88 (B) virion proteins. Proteins identified by mass spectrometry are indicated on the right. Unassigned bands contained mixes of two or more structural proteins. If available, the N-terminal sequence is shown in parentheses. The migration of the molecular mass markers is shown at the left.
Several examples of posttranslational processing were detected in both phages. N-terminal methionine processing takes place in each of the major tail proteins gp46 and gp47 (Fig. 6A and B). In phiIPLA88 virions, 14 amino acids were removed from the predicted major head protein gp42.
The N-terminal amino acid sequence obtained for the major head protein gp41 of phiIPLA35 is encoded upstream of the predicted ATG codon. This strongly suggests that gp41 might be synthesized as a fusion protein with gp40 and further processed. Indeed, closer inspection of the region between orf40 and orf41 reveals a potential −1 translational frameshifting element. Specifically, a slippery region, U UUA AAA (0 frame), and a pseudoknot that starts at position 23970 (3 nucleotides after the slippery sequence) and expands to position 24170 are observed at the 3′ end of orf40. The −1 frameshift would result in a 649-amino-acid protein combining both the scaffolding protein gp40 and the major head protein gp41. Bacteriophage frameshift occurrence is considered a strategy to compress as much information as possible into a very small genome or an alternative to obtain definite proportions of two proteins (16, 44). The fusion protein must be further processed to constitute the mature head protein detected in the virions. Interestingly, gp40 matches with proteases in its amino-terminal part (up to residue 186) and with scaffolding proteins in its carboxy-terminal part which, in addition, is predicted to be structured almost exclusively as a row of α-helices. It is tempting to speculate that the fusion protein could be autoprocessed by its own protease domain. The major capsid protein of Pseudomonas aeruginosa bacteriophage PAJU2 is thought to be synthesized as a protein fused to a prohead protease and is autocatalytically cleaved (53). Similar domain architecture is found in the capsid protein from Stenotrophomonas bacteriophage S1 (19). This protein matches three types of proteins in the databases: proteases, scaffold protein, and major head proteins. Matrix-assisted laser desorption ionization-time of flight analysis revealed that the major capsid protein is possibly autoprocessed by its protease domain, giving rise to the scaffold and the major capsid polypeptides.
Virion proteins with peptidoglycan hydrolytic activity.
Mature virions are often endowed with peptidoglycan hydrolases involved in host cell wall degradation prior to injecting their genetic material during infection (35, 40). The genomes of the two S. aureus phages were screened for the presence of putative peptidoglycan hydrolytic domains, different from those found within the lysis cassette. A lytic transglycosylase-like domain (nucleotides [nt] 1838 to 1964) and a peptidase domain (nt 1705 to 1806) were identified in the tail tape measure protein, gp50, of phiIPLA35. Recently, it has been demonstrated that some tail tape measure proteins have structural domains similar to peptidoglycan hydrolases, enabling the entry of phage DNA through the thick peptidoglycan layer of host bacteria such as Mycobacterium (8, 39). In phiIPLA88, gp58 could also be an infection-related hydrolase as it bears two domains for peptidoglycan hydrolysis (LYZ2 and CHAP).
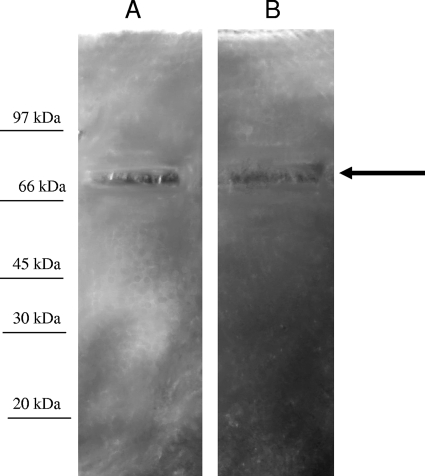
To confirm the presence of structural components with peptidoglycan hydrolytic activity in phiIPLA35 and phiIPLA88 virions, zymogram assays were performed with autoclaved S. aureus Sa9 cells. Upon electrophoretic separation of the virion proteins, the zymogram revealed a single 70-kDa band with muralytic activity in each phage (Fig. 7). The size is consistent with the virion protein gp58 (72.5 kDa) of phiIPLA88, which contains the predicted peptidoglycan hydrolytic domains. In contrast, for phiIPLA35 the size corresponding to this activity is smaller than the estimated size for gp50 (226 kDa). However, proteolytic processing of this protein might explain this result.
FIG. 7.
Zymogram gels of phiIPLA35 (A) and phiIPLA88 (B) phages. About 1011 particles were loaded on each lane for the zymogram analysis. Molecular mass markers are indicated at the left.
Conclusions.
In this study, we have presented a detailed genomic and molecular characterization of two S. aureus lytic derivative phages, phiIPLA35 and phiIPLA88, isolated from the dairy environment. The analysis revealed the mutations responsible for the loss of their ability to lysogenize. In addition, these genomes do not clearly encode known virulence factors, whereas new peptidoglycan lytic activities have been identified. All these elements suggest that these phages could be useful as natural antimicrobials against S. aureus in dairy products.
Acknowledgments
This research study was supported by grants AGL2006-03659/ALI from the Ministry of Education of Spain and PC06-009 from FICYT (Regional Government of Asturias). P.G. is a fellow of the Spanish Ministry of Education Ramón y Cajal Research Programme. J.M.O. is a fellow of the Spanish Ministry of Education (I3P Programme-CSIC).
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Bae, T., T. Baba, K. Hiramatsu, and O. Schneewind. 2006. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 62:1035-1047. [DOI] [PubMed] [Google Scholar]
- 2.Casjens, S., W. M. Huang, M. Hayden, and R. Parr. 1987. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J. Mol. Biol. 194:411-422. [DOI] [PubMed] [Google Scholar]
- 3.Casjens, S. R., and E. B. Gilcrease. 2009. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 502:91-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceyssens, P. J., K. Hertveldt, H.-W. Ackermann, J.-P. Noben, M. Demeke, G. Volckaert, and R. Lavigne. 2008. The intron-containing genome of the lytic Pseudomonas phage LUZ24 resembles the temperate phage PaP3. Virology 377:233-238. [DOI] [PubMed] [Google Scholar]
- 5.Chiang, Y. C., W. W. Liao, C. M. Fan, W. Y. Pai, C. S. Chiou, and H. Y. Tsen. 2008. PCR detection of staphylococcal enterotoxins (SEs) N, O, P, Q, R, U, and survey of SE types in Staphylococcus aureus isolates from food-poisoning cases in Taiwan. Int. J. Food Microbiol. 121:66-73. [DOI] [PubMed] [Google Scholar]
- 6.Clark, J. R., and J. B. March. 2006. Bacteriophages and biotechnology: vaccines, gene therapy and antibacterials. Trends Biotechnol. 24:212-218. [DOI] [PubMed] [Google Scholar]
- 7.Courchesne, N. M., A. Parisien, and C. Q. Lan. 2009. Production and application of bacteriophage and bacteriophage-encoded lysins. Recent Pat. Biotechnol. 3:37-45. [DOI] [PubMed] [Google Scholar]
- 8.Dusthackeer, A., V. N. Hassan, and V. Kumar. 2008. Tape measure protein having MT3 motif facilitates phage entry into stationary phase cells of Mycobacterium tuberculosis. Comp. Biol. Chem. 32:367-369. [DOI] [PubMed] [Google Scholar]
- 9.El-Sharoud, W. M., and G. Spano. 2008. Diversity and enterotoxigenicity of Staphylococcus spp. associated with domiati cheese. J. Food Prot. 71:2567-2571. [DOI] [PubMed] [Google Scholar]
- 10.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 11.Fischetti, V. A., D. Nelson, and R. Schuch. 2006. Reinventing phage therapy: are the parts greater than the sum? Nat. Biotechnol. 24:1508-1511. [DOI] [PubMed] [Google Scholar]
- 12.Fischetti, V. A. 2008. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11:393-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Fraser, J. S., Z. Yu, K. L. Maxwell, and A. R. Davidson. 2006. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359:496-507. [DOI] [PubMed] [Google Scholar]
- 15.García, P., V. Ladero, and J. E. Suárez. 2003. Analysis of the morphogenetic cluster and genome of the temperate Lactobacillus casei bacteriophage A2. Arch. Virol. 148:1051-1070. [DOI] [PubMed] [Google Scholar]
- 16.García, P., I. Rodríguez, and J. E. Suárez. 2004. A −1 ribosomal frameshift in the transcript that encodes the major head protein of bacteriophage A2 mediates biosynthesis of a second essential component of the capsid. J. Bacteriol. 186:1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García, P., C. Madera, B. Martínez, and A. Rodríguez. 2007. Biocontrol of Staphylococcus aureus in curd manufacturing processes using bacteriophages. Int. Dairy J. 17:1232-1239. [Google Scholar]
- 18.Garcia, P., B. Martinez, J. M. Obeso, and A. Rodríguez. 2008. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 47:479-485. [DOI] [PubMed] [Google Scholar]
- 19.García, P., C. Monjardín, R. Martín, C. Madera, N. Soberón, E. García, A. Meana, and J. E. Suárez. 2008. Isolation of new Stenotrophomonas bacteriophages and genomic characterization of the temperate phage S1. Appl. Environ. Microbiol. 74:7552-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García, P., C. Madera, B. Martínez, A. Rodríguez, and J. E. Suárez. 2009. Prevalence of bacteriophages infecting Staphylococcus aureus in dairy samples and their potential as biocontrol agents. J. Dairy Sci. 92:3019-3026. [DOI] [PubMed] [Google Scholar]
- 21.Goerke, C., C. Wirtz, U. Flückiger, and C. Wolz. 2006. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 61:1673-1685. [DOI] [PubMed] [Google Scholar]
- 22.Górski, A., M. Targońska, J. Borysowski, and B. Weber-Dąbrowska. 2009. The potential of phage therapy in bacterial infections of the eye. Ophthalmologica 223:162-165. [DOI] [PubMed] [Google Scholar]
- 23.Hagens, S., and M. J. Loessner. 2007. Application of bacteriophages for detection and control of foodborne pathogens. Appl. Microbiol. Biotechnol. 76:513-519. [DOI] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Herrero, M., C. G. de los Reyes-Gavilán, J. L. Caso, and J. E. Suárez. 1994. Characterization of φ393-A2, a bacteriophage that infects Lactobacillus casei. Microbiology 140:2585-2590. [Google Scholar]
- 26.Kwan, T., J. Liu, M. DuBow, P. Gros, and J. Pelletier. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. USA 102:5174-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lavigne, R., D. Seto, P. Mahadevan, H. W. Ackermann, and A. M. Kropinski. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406-414. [DOI] [PubMed] [Google Scholar]
- 30.Le Loir, Y., F. Baron, and M. Gautier. 2003. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2:63-76. [PubMed] [Google Scholar]
- 31.Lepeuple, A. S., E. Van Gemert, and M. P. Chapot-Chartier. 1998. Analysis of the bacteriolytic enzymes of the autolytic Lactococcus lactis subsp. cremoris strain AM2 by renaturing polyacrylamide gel electrophoresis: identification of a prophage-encoded enzyme. Appl. Environ. Microbiol. 64:4142-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levin, M. E., R. W. Hendrix, and S. R. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage lambda tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J., M. Dehbi, G. Moeck, F. Arhin, P. Bauda, D. Bergeron, M. Callejo, V. Ferretti, N. Ha, T. Kwan, J. McCarty, R. Srikumar, D. Williams, J. J. Wu, P. Gros, J. Pelletier, and M. DuBow. 2004. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22:185-191. [DOI] [PubMed] [Google Scholar]
- 34.Matsuzaki, S., M. Yasuda, H. Nishikawa, M. Kuroda, T. Ujihara, T. Shuin, Y. Shen, Z. Jin, S. Fujimoto, M. D. Nasimuzzaman, H. Wakiguchi, S. Sugihara, T. Sugiura, S. Koda, A. Muraoka, and S. Imai. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J. Infect. Dis. 187:613-624. [DOI] [PubMed] [Google Scholar]
- 35.Moak, M., and I. J. Molineux. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169-1183. [DOI] [PubMed] [Google Scholar]
- 36.Obeso, J. M., B. Martínez, A. Rodríguez, and P. García. 2008. Lytic activity of the recombinant staphylococcal bacteriophage ΦH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 128:212-218. [DOI] [PubMed] [Google Scholar]
- 37.O'Flaherty, S., A. Coffey, R. Edwards, W. Meaney, G. F. Fitzgerald, and R. P. Ross. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira, V., C. Lopes, A. Castro, J. Silva, P. Gibbs, and P. Teixeira. 2009. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 26:278-282. [DOI] [PubMed] [Google Scholar]
- 39.Piuri, M., and G. F. Hatfull. 2006. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62:1569-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rashel, M., J. Uchiyama, I. Takemura, H. Hoshiba, T. Ujihara, H. Takatsuji, K. Honke, and S. Matsuzaki. 2008. Tail-associated structural protein gp61 of Staphylococcus aureus phage phi MR11 has bifunctional lytic activity. FEMS Microbiol. Lett. 284:9-16. [DOI] [PubMed] [Google Scholar]
- 41.Reeder, J., and R. Giegerich. 2004. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics 5:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhoads, D. D., R. D. Wolcott, M. A. Kuskowski, B. M. Wolcott, L. S. Ward, and A. Sulakvelidze. 2009. Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J. Wound Care 18:237-243. [DOI] [PubMed] [Google Scholar]
- 43.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318:245-266. [DOI] [PubMed] [Google Scholar]
- 44.Rodríguez, I., P. García, and J. E. Suárez. 2005. A second case of −1 ribosomal frameshifting affecting a major virion protein of the Lactobacillus bacteriophage A2. J. Bacteriol. 187:8201-8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schmid, D., R. Fretz, P. Winter, M. Mann, G. Höger, A. Stöger, W. Ruppitsch, J. Ladstätter, N. Mayer, A. de Martin, and F. Allerberger. 2009. Outbreak of staphylococcal food intoxication after consumption of pasteurized milk products, June 2007, Austria. Wien. Klin. Wochenschr. 121:125-131. [DOI] [PubMed] [Google Scholar]
- 47.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 48.Spiess, E., and R. Lurz. 1988. Electron microscopic analysis of nucleic acids and nucleic acid-protein complexes. Methods Microbiol. 20:293-323. [Google Scholar]
- 49.Stenholm, A. R., I. Dalsgaard, and M. Middelboe. 2008. Isolation and characterization of bacteriophages infecting the fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 74:4070-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sulakvelidze, A., and E. Kutter. 2005. Bacteriophage therapy in humans, p. 381-436. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and application. CRC Press, Boca Raton, FL.
- 51.Summer, E. J., J. J. Gill, C. Upton, C. F. Gonzalez, and R. Young. 2007. Role of phages in the pathogenesis of Burkholderia, or where are the toxin genes in Burkholderia phages? Curr. Opin. Microbiol. 10:410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tormo, M. A., M. D. Ferrer, E. Maiques, C. Úbeda, L. Selva, I. Lasa, J. J. Calvete, R. P. Novick, and J. R. Penadés. 2008. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J. Bacteriol. 190:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchiyama, J., M. Rashel, T. Matsumoto, Y. Sumiyama, H. Wakiguchi, and S. Matsuzaki. 2009. Characteristics of a novel Pseudomonas aeruginosa bacteriophage, PAJU2, which is genetically related to bacteriophage D3. Virus Res. 139:131-134. [DOI] [PubMed] [Google Scholar]
- 54.Reference deleted.
- 55.Ye, Z. H., and C. Y. Lee. 1993. Cloning, sequencing, and genetic characterization of regulatory genes, rinA and rinB, required for the activation of staphylococcal phage φ11 int expression. J. Bacteriol. 175:1095-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zouharova, M., and D. Rysanek. 2008. Multiplex PCR and RPLA Identification of Staphylococcus aureus enterotoxigenic strains from bulk tank milk. Zoonoses Public Health 55:313-319. [DOI] [PubMed] [Google Scholar]