Abstract
Arthrobacter sp. strain IF1 is able to grow on 4-fluorophenol (4-FP) as a sole source of carbon and energy. To clone the 4-FP degradation genes, DNA libraries were constructed and screened with a probe obtained by PCR using primers designed on the basis of conserved regions of aromatic two-component monooxygenases. Sequencing of positive clones yielded two gene clusters, each harboring a gene encoding a monooxygenase with high sequence similarity to the oxygenase component of 4-nitrophenol and 4-chlorophenol monooxygenase systems. Both these monooxygenase genes were differentially expressed during growth on 4-FP, as revealed by Northern blotting and reverse transcription-PCR. One cluster also contained a gene for a flavin reductase. The monooxygenase and reductase were purified from Escherichia coli cells expressing the corresponding genes, and together they catalyzed NADH-dependent hydroxylation and dehalogenation of 4-halophenols. The results indicate that strain IF1 transforms 4-FP to hydroquinone by a two-component monooxygenase system of which one component provides reduced flavin adenine dinucleotide at the expense of NADH and the other catalyzes para-hydroxylation of 4-FP and other 4-substituted phenols.
Halogenated phenols are used as building blocks in the synthesis of pharmaceuticals, agrochemicals, and performance materials. They also occur frequently as pollutants in water and soil and may cause serious environmental problems. Some soil microorganisms have evolved biodegradation pathways that allow growth on these compounds (9, 18, 26, 47). Most studies of such pathways have been conducted with organisms that degrade and dehalogenate chlorinated phenols. Since the 1990s, the industrial use of fluorinated compounds has been growing (20, 45).
Pathways for the biodegradation of fluorinated compounds and the enzymes catalyzing defluorination have scarcely been examined, although some routes are known (34). The cleavage of the carbon-fluorine bond is especially interesting in view of its kinetic stability and high bond energy. Defluorination of fluoroaromatics can occur prior to ring cleavage, e.g., via an oxygenase that defluorinates fluorobenzoate (11, 36) or fluorobenzene (8). In other cases, defluorination occurs after ring cleavage via the formation of fluorinated muconolactones (42), which can be produced from 4-fluorobenzoate (19) and fluorobenzene (8) via 4-fluorocatechol. Bacterial and fungal phenol hydroxylases can convert fluorophenols to fluorocatechols or fluoropyrogallols, which are metabolized to fluoromuconic acids by ring cleavage dioxygenases (5, 6). Defluorination of 4-fluorophenol (4-FP) prior to ring cleavage in a strain of Arthrobacter has recently been described by us, but the enzymatic basis of defluorination was not solved (13).
For chlorinated phenols, two main metabolic routes have been described. Pathways in which the chlorophenol is oxidized to a substituted catechol, in some cases with partial dehalogenation, followed by ortho-cleavage of the aromatic ring and post-ring cleavage dehalogenation, occur in bacteria that degrade mono- and dichlorophenols (18, 21, 47, 48). On the other hand, routes in which the substituted phenol is converted via hydroquinone (or a substituted hydroquinone) to maleylacetate are also known, mainly in organisms that grow on polyhalophenols (28, 30, 31, 38, 44, 50). The further aerobic metabolism of hydroquinone may proceed via direct ring fission (7, 32) or via hydroxylation to hydroxyhydroquinone (1,2,4-trihydroxybenzene) (12), which can undergo ring fission by an intradiol dioxygenase (25, 29, 33, 35). Genes for the latter hydroquinone degradation route have been cloned from Cupriavidus necator (formerly Ralstonia eutropha) strain JMP134 and Ralstonia pickettii DTP0602, both of which grow on 2,4,6-trichlorophenol (20, 30, 31), and from a strain of Sphingobium chlorophenolicum that can grow on pentachlorophenol (9, 37).
In the present paper, we report the characterization of two 4-FP catabolic gene clusters from Arthrobacter sp. strain IF1 (13) and characterize the expression and function of the two-component flavin monooxygenase genes that are involved in the initial steps of 4-FP degradation. The organism was isolated on the basis of its ability to utilize 4-FP as a carbon source for growth (13).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Arthrobacter sp. strain IF1 (13) was grown in Luria-Bertani (LB) medium or in a synthetic medium (13) at 30°C. Escherichia coli BL21(DE3) (Stratagene) was grown in LB medium, and when necessary, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 100 μg/ml of ampicillin were added.
Isolation and analysis of DNA.
Small plasmid DNA was isolated as described by Sambrook et al. (41). Genomic DNA was isolated from strain IF1 as previously described (14), and plasmid DNA was isolated with a large-construct isolation kit from Qiagen.
The following degenerate primer set was used for the initial amplification and cloning in pGEM-T Easy (Promega) of segments of the 4-FP monooxygenase genes: primer 1 (5′-AACGTIGCNACNCAYCC-3′; for sequence NVATHP) and primer 2 [5′-CGCCANGGGATRAANACRT-3′; reverse complement for sequence (N/D)VFIPWS]. PCR was performed with these primers in 50-μl reaction mixtures containing 4 μl of a deoxynucleoside triphosphate mixture, 0.5 μl ExTaq DNA polymerase (Takara Bio, Shiga, Japan), and 50 ng of strain IF1 genomic DNA. PCR schedules were as follows: 94°C for 8 min; 34 cycles consisting of 94°C for 30 s, a gradient from 45°C to 55°C for 40 s, and 72°C for 50 s; and a final incubation at 72°C for 3 min.
Cloning and sequencing of monooxygenase genes.
Genomic DNA was separately digested by ApaI and BamHI, subjected to gel electrophoresis, and transferred to a Hybond-N+ nylon membrane (Amersham Pharmacia Biotech, Hercules, CA) by blotting. Putative monooxygenase sequences were detected with a digoxigenin hybridization system (Roche Diagnostics, Mannheim, Germany) as described previously (24), using a probe obtained by labeling of PCR products that were produced by amplification of genomic DNA with the degenerate primers mentioned above. A 5-kb band was detected with genomic DNA that was treated with ApaI, and fragments of this size were cloned into pBluescript II KS(+) (Stratagene) to produce library A. Fragments of 9 kb were detected with BamHI-restricted DNA and were cloned into pHSG397 (Takara) to produce library B. For screening, the libraries were transformed into E. coli cells, and transformants were inoculated into several Falcon tubes containing 2 ml of LB medium and chloramphenicol (for pHSG397) or ampicillin [for pBluescript II KS(+)]. After overnight growth at 30°C, DNA was isolated and screened for the presence of the 4-FP monooxygenase gene by PCR with the primers used earlier for preparation of the probe. Positive cultures were plated, and colonies were screened again by PCR. DNA was isolated from the positive clones, subcloned into pUC19 (Takara), and used for sequencing.
Dideoxy sequencing was done using an ABI Prism BigDye ready reaction kit and an ABI sequencer, model 3700. Sequences were analyzed as described previously (24).
Analysis of mRNA.
Cellular mRNA levels were determined by Northern blotting and reverse transcription-PCR (RT-PCR). Cells were grown in LB medium, collected at an optical density at 600 nm (OD600) of 0.7, washed with saline-phosphate buffer (pH 7.0), and resuspended in a mineral medium (MM) (13) to an OD600 of 3.5. The cells were distributed to test tubes and incubated with a carbon source (10 mM glucose, 1.0% glycerol, 10 mM succinate, 1 mM 4-FP, or 1 mM 4-nitrophenol) for 6 h at 30°C with shaking. Then cells were collected and disrupted (23), and total RNA was extracted using an Isogen RNA extraction kit (Nippon Gene, Tokyo, Japan).
For Northern hybridization, RNA was transferred to a membrane by capillary blotting, followed by hybridization with labeled probes prepared from PCR products of the fpdA1 and fpdA2 open reading frames (ORFs) with the primer pairs that were also used for making the expression constructs. The primer sequences were as follows: fpdA1/A2-5′w/NdeI, CATATGAGGACAGGAAAAGAGTACCT; fpdA2-3′w/BamHI, GGATCCTTAGGCCGGTGCGGTGACT; fpdA1-3′w/BamHI, GGATCCTTAGGCCGGCGTGGTGACTG (where underlined sequences indicate restriction sites).
Reverse transcription was performed using RNA treated with RNase-free DNase (Toyobo, Osaka, Japan). A primer specific for and complementary to the 3′ end of a monooxygenase gene was extended with ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) at 50°C for 30 min, according to the manufacturer's instructions. The following two primer sets were used for subsequent PCR analyses: primer set Apa (forward, 5′-GCGGCACTGGGAAGAAGTCACCT-3′; reverse, 5′-CGTGGTGACTGCCGTGTCGCTCTG-3′), which amplified a 1,189-bp region from fpdA1, and primer set Bam (forward, 5′-GCTTCCGTGAGACGTGTCGCGCA-3′; reverse, 5′-TGCGGTGACTGCCGAGTCATTCGT-3′), which amplified a 599-bp region from fpdA2. Amplification was performed with 100-fold-diluted cDNA samples and ExTaq DNA polymerase (Takara Bio, Shiga, Japan) with the following schedule: 94°C for 5 min; 25 cycles consisting of 94°C for 20 s, 67°C for 20 s, and 72°C for 30 s; and a final incubation at 72°C for 5 min. DNase-treated RNA samples that were not subjected to reverse transcriptase treatment were used as controls in PCRs to verify the absence of contaminating genomic DNA. PCR products were analyzed on a 1.0% agarose gel.
Expression of fpd genes in E. coli.
The nucleotide sequences of fpdA1, fpdA2, and fpdB were amplified with PCR primers (sequences mentioned above) and cloned into pET17b (Novagen) as translational fusions in the NdeI restriction site of the vector. E. coli BL21(DE3) was used for expression.
Purification of 4-FP monooxygenase (FpdA2).
The 4-FP monooxygenase (FpdA2) was purified from E. coli BL21(DE3)(pETfpdA2). Cells were grown in LB medium containing ampicillin until the OD600 reached 0.5. IPTG was then added (0.5 mM), and the culture was incubated overnight at 20 to 22°C with shaking. Cells were harvested by centrifugation, washed twice with TEMG buffer (50 mM Tris·SO4 [pH 7.5], 0.5 mM EDTA, 1 mM β-mercaptoethanol, 5% glycerol), resuspended in the same buffer, and disrupted by sonication. After centrifugation (at 40,000 × g for 60 min), the extract was loaded onto a DEAE Sepharose column (bed volume, 60 ml) preequilibrated with TEMG buffer. FpdA2 was eluted with a linear gradient of 0 to 0.5 M (NH4)2SO4 in TEMG buffer, concentrated by ultrafiltration (Amicon YM-30 membrane), and separated on a hydroxyapatite column (50 ml) using 10 to 400 mM potassium phosphate (pH 7.0) containing 1 mM β-mercaptoethanol and 5% glycerol. FpdA2 was concentrated by ultrafiltration and stored at −20°C.
Purification of flavin reductase (FpdB).
Flavin reductase was purified from E. coli BL21(DE3)(pETfpdB), cultivated, induced, and lysed as described above for FpdA2. The cell extract was fractionated on a DEAE Sepharose column, after which FpdB protein was concentrated and dialyzed against 1.5 M (NH4)2SO4 in TEMG buffer, which caused precipitation. The protein pellet was dissolved in 4 ml TEMG buffer (pH 7.5) and was fractionated on a Superdex 200 column (bed volume, 320 ml) using TEMG buffer containing 0.15 M NaCl. FpdB was concentrated by ultrafiltration and stored at −20°C.
Enzyme assays.
4-FP monooxygenase was measured at 25°C in incubations containing 50 mM phosphate buffer (pH 7.0), a suitable amount of monooxygenase, 1 mM ascorbic acid, 10 μM flavin adenine dinucleotide (FAD), 3 μg of reductase (FpdB), 180 U ml−1 catalase (Fluka), 2.5 mM NADH, and 400 to 600 μM substrate. Reactions were started by adding NADH. Samples of 25 μl were taken with intervals of 5 to 25 min and were quenched by addition of high-performance liquid chromatography (HPLC) eluent (see below). The samples were centrifuged, and the supernatants were analyzed by HPLC. One unit of enzyme activity corresponds to 1 μmol of 4-FP converted per min.
Flavin reductase activity was determined by monitoring the oxidation of NADH at 340 nm (ɛ340 = 6.22 mM−1 cm−1). Reaction mixtures contained 50 mM phosphate buffer (pH 7.5), 300 μM NADH, and 100 μM FAD or flavin mononucleotide (FMN). The reaction was initiated by addition of enzyme, and initial rates were used for calculating kinetic parameters.
Analytical methods.
Isocratic HPLC of 20-μl samples was carried out using a LiChrospher 100 RP8 reversed-phase column (250 mm by 4.6 mm; particle size, 5 μm) in connection with Jasco PU-980 pumps, a Jasco MD-910 diode array detector, and a Jasco UV-2075 detector. The mobile phase (1 ml min−1) was 70:30 (vol/vol) acetic acid-methanol containing 0.02 M ammonium acetate, pH 4.5.
For gas chromatographic (GC) analysis, samples (300 μl) were extracted with an equal volume of ethyl acetate containing mesitylene as the internal standard, followed by analysis on a Hewlett-Packard 6890 GC equipped with a flame ionization detector and an HP-5 column (Agilent 19091J-413). Helium (1 ml min−1) was the carrier gas, and the temperature schedule was 5 min at 50°C, followed by an increase of 15°C per min to 250°C.
Fluoride, chloride, bromide, and nitrite were measured in 50-μl samples using a Dionex DX 120 ion chromatograph (Dionex, Sunnyvale, CA) equipped with an Alltech A-2 anion column (100 by 4.6 mm; particle size, 7 μm) and an Alltech guard column (50 by 4 mm). The eluent was a mixture of NaHCO3 and Na2CO3 in deionized water with a flow rate of 1.2 ml min−1.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the DNA Data Bank of Japan under accession no. AB530680 and AB530681.
RESULTS
Cloning of 4-FP monooxygenase genes.
In order to obtain the 4-FP degradation genes from strain IF1, we first used a PCR approach. Using degenerate primers, designed on the basis of the alignment of six published gene sequences for two-component aromatic monooxygenases (15, 22, 27, 30, 40), a 731-bp product was obtained. Its nucleotide sequence revealed similarity with 4-nitrophenol (39) and 4-chlorophenol monooxygenase genes (35). Using a large-construct isolation kit, two plasmids were detected in strain IF1. Southern hybridization with a probe derived from the PCR-amplified monooxygenase gene fragment indicated that the corresponding genes are not located on these plasmids (data not shown) but on genomic DNA (see Fig. S1 in the supplemental material). Hybridization analysis of genomic DNA restricted with ApaI, BamHI, EcoRI, HindIII, or PstI gave two positive signals in each case, whereas four bands were obtained when the DNA was restricted with SalI. The fact that the probe contained only one SalI restriction site indicates that strain IF1 has two highly similar or identical copies of the 4-FP monooxygenase gene, with different flanking regions.
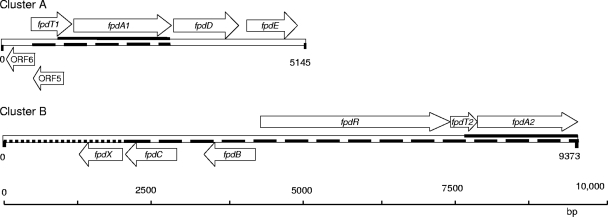
Based on the hybridization results, two DNA libraries were constructed and screened for the presence of the 4-FP monooxygenase by PCR, which yielded a positive clone from each library. The sequence of the insert of an ApaI clone consisted of 5,145 bp (cluster A). For a BamHI clone, an insert of 9,373 bp was found (cluster B). BLAST sequence similarity searches with the deduced amino acid sequences of the ORFs identified a number of homologs, allowing annotation (Fig. 1; see also Tables S1 and S2 in the supplemental material). Both clusters have a large segment that is similar to a p-nitrophenol catabolism gene cluster (GenBank accession no. EF052871) of Arthrobacter sp. strain JS443 (39). The putative genes involved in 4-FP degradation were designated fpd.
FIG. 1.
Organization of the ORFs in the fpd gene regions of Arthrobacter sp. strain IF1. Open arrows indicate the size and direction of each ORF. ORFs that are expected to be involved in 4-FP metabolism are labeled with fpd. Heavy solid lines delineate regions of high similarity between cluster A and cluster B; heavy dashed lines delineate regions of high similarity between the clusters indicated and the 4-nitrophenol gene region (39); heavy dotted line delineates regions of lower similarity between cluster B and the 4-nitrophenol gene region.
Each cluster contained a putative monooxygenase gene, and these were designated fpdA1 and fpdA2 for cluster A and B, respectively. They are 93% identical at the DNA sequence level and 98.9% identical at the deduced amino acid sequence level. The closest homologs of which the function is established are the hydroxylase proteins of the two-component 4-nitrophenol monooxygenases from Arthrobacter sp. strain JS443 (99% amino acid sequence identity) (39) and Rhodococcus opacus SAO101 (72% identity) (27) and the hydroxylase component of 4-chlorophenol monooxygenase from Arthrobacter chlorophenolicus A6 (96% identity) (35). These monooxygenase systems consist of a reductase that reduces FAD at the expense of NADH and a hydroxylase that uses reduced FAD (FADH2) and O2 to hydroxylate the substrate; they are classified as class D flavoprotein monooxygenases (two-component flavin-diffusible monooxygenases [TC-FDM]) (46).
A putative reductase gene, termed fpdB, was detected only in cluster B. It is located upstream of fpdA2 on the opposite strand, and the encoded protein has high similarity to the reductase component of similar monooxygenases from Arthrobacter sp. strain JS443 (39) (92% amino acid sequence identity), R. opacus SAO101 (27) (47% identity), and A. chlorophenolicus A6 (86% identity). Analysis of FpdB with the Pfam database showed the presence in the N terminus of a flavin reductase-like domain (Pfam01613), characteristic for proteins that provide FADH2 to the hydroxylase component. The C-terminal segment of FpdB aligns weakly with the N-terminal segment of GntR-type transcriptional regulators (Pfam00392), indicating the presence of a C-terminal regulator domain.
Some other genes presumably involved in haloaromatic metabolism were detected (Fig. 1). In cluster A, the fpdD and fpdE ORFs encode proteins with sequence similarity to maleylacetate reductases that are involved in the degradation of p-nitrophenol (27, 39) and to α/β-hydrolase fold family enzymes, respectively. The translated sequences from ORF5 and ORF6 in cluster A showed the highest similarity to proteins involved in conjugational plasmid transfer (Pfam02534.12). In cluster B, ORF fpdC encodes a putative protein with high similarity to hydroxyquinol dioxygenases, e.g., the dioxygenase involved in 4-chlorophenol degradation (35), and ORF fpdX encodes a putative periplasmic binding protein. An ORF designated fpdR is present in front of the fpdA2 ORF and may encode a transcriptional regulator, based on the presence of a nucleotide-binding domain and a helix-turn-helix (HTH) motif. It is similar to putative regulator genes encoded in p-nitrophenol and 4-chlorophenol degradation gene clusters (39, 35). Finally, ORF fpdT2, with unknown function, occurs at a similar position in the p-nitrophenol gene cluster (39).
Expression and induction of fpdA1 and fpdA2.
The expression of the fpdA genes was tested with strain IF1 cultures exposed to aromatic compounds and with controls. The results of Northern hybridization revealed that fpdA1 and fpdA2 were not expressed in cells incubated with glycerol, glucose, or succinate (see Fig. S2 in the supplemental material), whereas RNA corresponding to the fpdA1 and fpdA2 genes was clearly observed in cells exposed to 4-FP. With 4-nitrophenol as an inducer, the signal was weaker than with 4-FP-grown cells, but induction was still visible for fpdA2.
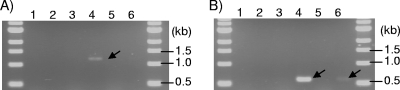
RT-PCR was used to further analyze the expression of the monooxygenase genes (Fig. 2). When the 3′-specific primer for fpdA1 was used with the 5′-specific primer for fpdA2, or when the 3′-specific primer for fpdA2 was used with the 5′-specific primer for fpdA1, no amplification was obtained, indicating that the primers were specific for their target DNAs. RT-PCR analysis showed no expression of the fpd genes when glucose was used as the carbon source, but with 4-FP, transcription of both fpdA1and fpdA2 was observed (Fig. 2). The sizes of the amplified fragments (1,189 and 599 bp) were in agreement with the primer positions.
FIG. 2.
RT-PCR analysis of the expression of the monooxygenase genes. PCRs were carried out with primer sets specific for fpdA1 (A) or fpdA2 (B). The carbon sources used for induction were 10 mM glucose (lanes 1 and 2), 1 mM 4-FP (lanes 3 and 4), and 1 mM 4-nitrophenol (lanes 5 and 6). Lanes 1, 3, and 5 were loaded with samples for which the PCR templates were DNase treated but not subjected to the reverse transcriptase reaction (negative control).
The results of Northern blot analysis and RT-PCR indicate that the expression of fpdA1and fpdA2 is stimulated by the presence of 4-FP and that fpdA2 is more strongly expressed than fpdA1.
Properties of 4-FP monooxygenase (FpdA2) and flavin reductase (FpdB).
To confirm the activities of the proteins encoded by the putative monooxygenase genes, fpdA2 and fpdB were expressed in E. coli BL21(DE3), yielding proteins with molecular masses of approximately 62 and 30 kDa. FpdA2 was purified by a protocol that involved two chromatographic steps (Table 1). Solutions of purified FpdA2 were colorless and showed no absorption in the region of 320 to 500 nm, suggesting that FpdA2 does not contain a flavin cofactor. The reductase component (FpdB) was also purified by column chromatography, after which only one band was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (see Fig. S3 in the supplemental material). Gel filtration chromatography indicated that FpdB behaved as an octamer. The FpdB protein used NADH to reduce either FAD or FMN (Table 2) but did not use NADPH or riboflavin as a substrate.
TABLE 1.
Purification of FpdA2 from E. coli BL21(DE3)(pET17b)
| Step | Total vol (ml) | Total protein (mg) | Total enzyme activitya (U) | Sp act (U mg−1) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|---|
| Cell extract | 38 | 1,350 | 9.5 | 0.007 | 100 | 1 |
| DEAE Sepharose | 15 | 250 | 7.5 | 0.030 | 79 | 4.3 |
| Hydroxyapatite | 7 | 38 | 6.0 | 0.160 | 63 | 23 |
Reaction mixtures contained 400 μM 4-FP, a suitable amount of FpdA2, and the components given in Materials and Methods.
TABLE 2.
Kinetic parameters of FpdBa
| Fixed substrateb | Varied substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| FAD | NADH | 13 ± 2 | 33 ± 4 | 2.5 × 106 |
| FMN | NADH | 122 ± 14 | 12 ± 2 | 1.0 × 105 |
| NADH | FMN | 10 ± 1 | 63 ± 3 | 6.6 × 106 |
| NADH | FAD | 6 ± 1 | 23 ± 2 | 3.6 × 106 |
Values refer to the varied substrate and are means for triplicate experiments with standard deviations.
FAD and FMN concentrations were fixed at 100 μM, and the NADH concentration was fixed at 300 μM.
In the presence of reductase, the activity of the purified hydroxylase was 160 nmol/min·mg of protein. Thus, the enzyme would have to be present in strain IF1 at a level of at least 5 to 10% of the total cellular protein in order to allow for the observed growth rate (μ = 0.1 h−1), assuming a yield of about 50 mg of cells per mmol of fluorophenol consumed (13).
Conversion of phenols by FpdA2 and FpdB.
The purified FpdA2 in combination with FpdB transformed various substituted phenols, with the release of the respective anions (Table 3). No transformation was observed in the absence of NADH or FpdB. The highest transformation rate was observed with 4-bromophenol, whereas 4-chlorophenol, 4-FP, and 4-nitrophenol were converted at lower rates. The disappearance of the para-substituted phenols was accompanied by the release of more than 90% of the substituent as halide or nitrite anions and the formation of hydroquinone (Table 3). Slow conversion of hydroquinone was also observed, both in incubations where it was formed from a 4-substituted phenol and in incubations where it was added from the start, explaining why the amount of hydroquinone detected was smaller than the amount of phenol converted. Some formation of trihydroxybenzene was seen by HPLC when hydroquinone was added (Table 3), but the instability of trihydroxybenzene, leading to brown products, made it impossible to establish mass balances, and rates were too low to conclude that FpdA2 is responsible for hydroquinone metabolism. 4-Nitrocatechol was not formed from 4-nitrophenol or degraded by purified FpdA2 and FpdB.
TABLE 3.
Conversion of 4-substituted phenols by purified FpdA2 and FpdBa
| Substrate | Rate (μmol/min mg−1) | Concn (μM)b |
||
|---|---|---|---|---|
| Substrate degraded | Anion formed | Hydroquinone | ||
| 4-FP | 46 | 344 | 312 | 145 |
| 4-Chlorophenol | 39 | 292 | 279 | 92 |
| 4-Bromophenol | 72 | 541 | 492 | 98 |
| 4-Nitrophenol | 40 | 303 | 280 | 150 |
| Hydroquinone | 64 | 130 | 10c | |
Reaction mixtures contained 600 μM substrate, 0.15 mg ml−1 of 4-FP monooxygenase, and the components described in Materials and Methods. Aromatics were analyzed by HPLC or GC at different times, and anions were analyzed by ion chromatography.
After 20 min of incubation.
Hydroxyhydroquinone detected by GC.
DISCUSSION
The 4-FP degradation genes from Arthrobacter sp. strain IF1 were detected on two different but related gene clusters. Cluster A harbors fpdA1DE, which comprise a FADH2-dependent monooxygenase, a putative maleylacetate reductase, and a hydrolase gene, while in cluster B fpdA2 encodes a similar monooxygenase, fpdB encodes a flavin reductase, and fpdC encodes a putative hydroxyquinol dioxygenase (Fig. 1). The sequences suggest that the 4-FP monooxygenases belong to the class D flavoprotein monooxygenases, for which a 4-hydroxyphenylacetate 3-hydroxylase can be regarded as a prototype (15, 16, 40). Several enzymes of this class have been reported to act on aromatic substrates such as 4-hydroxyphenylacetate (16, 40) or substituted phenols (22, 27, 30, 35, 44). Expression of the fpdA genes, especially fpdA2, is induced during growth on 4-FP, as indicated by Northern hybridization and RT-PCR, in agreement with a role in 4-FP metabolism. When 4-nitrophenol was the inducer, only the fpdA2 gene was well expressed, indicating that FpdA1 is not primarily involved in 4-nitrophenol metabolism in strain IF1. A recent proteomics study showed that the homologous 4-chlorophenol hydroxylase is induced in 4-chlorophenol- and 4-nitrophenol-grown cells of Arthrobacter chlorophenolicus (45a).
Comparison of the regions flanking the fpdA genes led to the identification of a segment of about 290 nucleotides that is present upstream of the monooxygenase genes in both clusters and that probably was copied together with the monooxygenase gene in a duplication event. Since the sequence present in cluster A is more complete (larger) than the sequence found in cluster B, it is likely that fpdA2 of cluster B was copied from (a predecessor of) cluster A during the assembly of the clusters. Thus, cluster A may have served as the source of DNA segments for a new cluster that is involved in 4-FP transformation. The deduced amino acid sequences and gene organization in cluster B further indicate a close evolutionary relationship with a cluster involved in hydroxyquinol degradation in A. chlorophenolicus A6 (35). An even more similar gene cluster was isolated by Perry and Zylstra from the 4-nitrophenol degrader Arthrobacter sp. strain JS443 (39). Strain IF1 also grows on 4-nitrophenol (13), and 4-nitrophenol induced the expression of the fpdA2 gene.
Like the majority of the genes encoding class D flavoprotein monooxygenases (members of the TC-FDM family), fpdA2 has a reductase gene (fpdB) in its proximity. This reductase has two distinct domains: the flavin reductase domain, which is common to this family of enzymes, and a C-terminal region with an HTH motif, which has not been described for these proteins (20, 27, 30, 43), and which is also present in the C-terminal region of the reductases encoded by the 4-chlorophenol and 4-nitrophenol gene clusters (35, 39). Based on sequence similarity to a putative regulator from Thermotoga maritima (29% identity), the 3-dimensional structure of which was recently solved, it can be predicted that the C-terminal region of FpdB forms a domain containing three α-helices and a small β-sheet of two strands: a two-stranded winged HTH, which is one of the two major structural HTH classes (2). HTH domains incorporated in enzymes may have a role in substrate recognition, localization, or feedback regulation of metabolic pathways (1, 2).
The FpdA2 and FpdB proteins, purified from E. coli transformants, were capable of catalyzing hydroxylation reactions with 4-substituted phenols and released the corresponding anions. The kinetic properties of FpdB (Table 2) are comparable to those of other flavin reductases. The kcat is lower than that of the reductase of a trichlorophenol monooxygenase (TcpX) (4), but the kinetic parameters are similar to those of the reductase of a chlorophenol 4-monooxygenase system (TftD) (17).
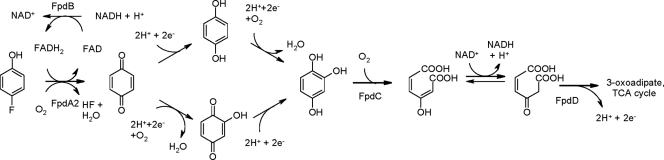
On the basis of genetic and biochemical information, we propose a pathway for 4-FP degradation by Arthrobacter sp. strain IF1 that starts with monooxygenation at the para position, with the release of fluoride (Fig. 3). We and others observed the formation of hydroquinone during the transformation of 4-FP (13), 4-chlorophenol (3), or 4-nitrophenol (25, 39). However, if one NADH molecule is consumed per monooxygenase catalytic cycle with anion release, the aromatic product should be benzoquinone. Reduction of benzoquinone to hydroquinone is possible, and a distinct enzyme for this conversion has been proposed for the trichlorophenol (4), pentachlorophenol (10), and 4-nitrophenol (51) degradation pathways. Strain IF1 may have such a protein, but apparently it is not required for the formation of hydroquinone in vitro, since we also detected this product with purified enzymes. Chemical reduction of a quinone by reduced nicotinamide cofactors has been suggested for 2,6-dichlorobenzoquinone (10, 17) and 6-chlorohydroxybenzoquinone (4, 49), and we observed NADH oxidation with benzoquinone (data not shown). Hydroquinone was slowly converted both by whole cells (13) and by mixtures of FpdA2 and FpdB, with some formation of hydroxyhydroquinone, but at this stage we are not certain about the physiological significance. For the trichlorophenol (4, 49) and 4-nitrophenol (39) pathways, a hydrolysis reaction catalyzed by the initial monooxygenase was proposed, but we consider it impossible to convert benzoquinone to hydroxybenzoquinone in this way, because the stoichiometry does not fit.
FIG. 3.
Proposed catabolic pathway for 4-FP degradation. See the text for details.
The product undergoing ring fission in strain IF1 most likely is hydroxyquinol (trihydroxybenzene), formed either by hydroxylation of hydroquinone or by reduction of hydroxybenzoquinone (Fig. 3). The presence of a putative intradiol cleavage type hydroxyquinol dioxygenase gene (fpdC) supports the possibility that hydroxyquinol is an intermediate in the degradation pathways for para-substituted phenols. The fpdC-encoded dioxygenase would produce maleylacetate, which likely can be converted into β-ketoadipate by the fpdD-encoded maleylacetate reductase (Fig. 3). Together with the work of others (35, 39), our results show that the initial steps of 4-nitrophenol, 4-chlorophenol, and 4-FP metabolism are highly similar.
Supplementary Material
Acknowledgments
This work was supported by a postdoctoral fellowship for foreign researchers (short-term) from JSPS (Japan Society for the Promotion of Science) and also in part by the European Community Human Potential Programme under contract HPRTN-CT-2002-00213 (BIOSAP).
Footnotes
Published ahead of print on 16 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anantharaman, V., E. V. Koonin, and L. Aravind. 2001. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 307:1271-1292. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., V. Anantharaman, S. Balaji, M. M. Babu, and L. M. Iyer. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29:231-262. [DOI] [PubMed] [Google Scholar]
- 3.Bae, H. S., J. M. Lee, and S. T. Lee. 1996. Biodegradation of 4-chlorophenol via a hydroquinone pathway by Arthrobacter ureafaciens CPR706. FEMS Microbiol. Lett. 145:125-129. [DOI] [PubMed] [Google Scholar]
- 4.Belchik, S. M., and L. Xun. 2008. Functions of flavin reductase and quinone reductase in 2,4,6-trichlorophenol degradation by Cupriavidus necator JMP134. J. Bacteriol. 190:1615-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boersma, M. G., T. Y. Dinarieva, W. J. Middelhoven, W. J. H. van Berkel, J. Doran, J. Vervoort, and I. M. C. M. Rietjens. 1998. 19F nuclear magnetic resonance as a tool to investigate microbial degradation of fluorophenols to fluorocatechols and fluoromuconates. Appl. Environ. Microbiol. 64:1256-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bondar, V. S., M. G. Boersma, E. Golovlev, J. Vervoort, W. J. H. van Berkel, Z. I. Finkelstein, I. P. Solyanikova, L. A. Golovleva, and I. M. C. M. Rietjens. 1998. 19F NMR study on the biodegradation of fluorophenols by various Rhodococcus species. Biodegradation 9:475-486. [DOI] [PubMed] [Google Scholar]
- 7.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho, M. F., M. I. Ferreira, I. S. Moreira, P. M. Castro, and D. B. Janssen. 2006. Degradation of fluorobenzene by Rhizobiales strain F11 via ortho cleavage of 4-fluorocatechol and catechol. Appl. Environ. Microbiol. 72:7413-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, R. L., C. M. Jung, and J. L. Strap. 2007. The recent evolution of pentachlorophenol (PCP)-4-monooxygenase (PcpB) and associated pathways for bacterial degradation of PCP. Biodegradation 18:525-539. [DOI] [PubMed] [Google Scholar]
- 10.Dai, M., J. B. Rogers, J. R. Warner, and S. D. Copley. 2003. A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD). J. Bacteriol. 185:302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engesser, K. H., and P. Schulte. 1989. Degradation of 2-bromo-, 2-chloro- and 2-fluorobenzoate by Pseudomonas putida CLB 250. FEMS Microbiol. Lett. 51:143-147. [DOI] [PubMed] [Google Scholar]
- 12.Eppink, M. H. M., E. Cammaart, D. van Wassenaar, W. J. Middelhoven, and W. J. H. van Berkel. 2000. Purification and properties of hydroquinone hydroxylase, a FAD-dependent monooxygenase involved in the catabolism of 4-hydroxybenzoate in Candida parapsilosis CBS604. Eur. J. Biochem. 267:6832-6840. [DOI] [PubMed] [Google Scholar]
- 13.Ferreira, M. I. M., J. R. Marchesi, and D. B. Janssen. 2008. Degradation of 4-fluorophenol by Arthrobacter sp. strain IF1. Appl. Microbiol. Biotechnol. 78:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabor, E. M., E. J. de Vries, and D. B. Janssen. 2004. Construction, characterization, and use of small-insert gene banks of DNA isolated from soil and enrichment cultures for the recovery of novel amidases. Environ. Microbiol. 6:948-958. [DOI] [PubMed] [Google Scholar]
- 15.Galán, B., E. Diaz, M. A. Prieto, and J. L. Garcia. 2000. Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin:NAD(P)H reductase subfamily. J. Bacteriol. 182:627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibello, A., M. Suarez, J. L. Allende, and M. Martin. 1997. Molecular cloning and analysis of the genes encoding the 4-hydroxyphenylacetate hydroxylase from Klebsiella pneumoniae. Arch. Microbiol. 167:160-166. [PubMed] [Google Scholar]
- 17.Gisi, M. R., and L. Y. Xun. 2003. Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:flavin adenine dinucleotide oxidoreductase (TftC) of Burkholderia cepacia AC1100. J. Bacteriol. 185:2786-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häggblom, M. M. 1992. Microbial breakdown of halogenated aromatic pesticides and related compounds. FEMS Microbiol. Rev. 9:29-71. [DOI] [PubMed] [Google Scholar]
- 19.Harper, D. B., and E. R. Blakley. 1971. The metabolism of p-fluorobenzoic acid by a Pseudomonas sp. Can. J. Microbiol. 17:1015-1023. [DOI] [PubMed] [Google Scholar]
- 20.Hatta, T., O. Nakano, N. Imai, N. Takizawa, and H. Kiyohara. 1999. Cloning and sequence analysis of hydroxyquinol 1,2-dioxygenase gene in 2,4,6-trichlorophenol-degrading Ralstonia pickettii DTP0602 and characterization of its product. J. Biosci. Bioeng. 87:267-272. [DOI] [PubMed] [Google Scholar]
- 21.Hollender, J., J. Hopp, and W. Dott. 1997. Degradation of 4-chlorophenol via the meta cleavage pathway by Comamonas testosteroni JH5. Appl. Environ. Microbiol. 63:4567-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hübner, A., C. E. Danganan, L. Xun, A. M. Chakrabarty, and W. Hendrickson. 1998. Genes for 2,4,5-trichlorophenoxyacetic acid metabolism in Burkholderia cepacia AC1100: characterization of the tftC and tftD genes and locations of the tft operons on multiple replicons. Appl. Environ. Microbiol. 64:2086-2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iida, T., Y. Mukouzaka, K. Nakamura, I. Yamaguchi, and T. Kudo. 2002. Isolation and characterization of dibenzofuran-degrading actinomycetes: analysis of multiple extradiol dioxygenase genes in dibenzofuran-degrading Rhodococcus species. Biosci. Biotechnol. Biochem. 66:1462-1472. [DOI] [PubMed] [Google Scholar]
- 24.Iida, T., K. Nakamura, A. Izumi, Y. Mukouzaka, and T. Kudo. 2006. Isolation and characterization of a gene cluster for dibenzofuran degradation in a new dibenzofuran-utilizing bacterium, Paenibacillus sp. strain YK5. Arch. Microbiol. 184:305-315. [DOI] [PubMed] [Google Scholar]
- 25.Jain, R. K., J. H. Dreisbach, and J. C. Spain. 1994. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl. Environ. Microbiol. 60:3030-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Key, B., R. Howell, and C. S. Criddle. 1997. Fluorinated organics in the biosphere. Environ. Sci. Technol. 31:2445-2454. [Google Scholar]
- 27.Kitagawa, W., N. Kimura, and Y. Kamagata. 2004. A novel p-nitrophenol degradation gene cluster from a gram-positive bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 186:4894-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiyohara, H., T. Hatta, Y. Ogawa, T. Kakuda, H. Yokoyama, and N. Takizawa. 1992. Isolation of Pseudomonas pickettii strains that degrade 2,4,6-trichlorophenol and their dechlorination of chlorophenols. Appl. Environ. Microbiol. 58:1276-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latus, M., H. Seitz, J. Eberspacher, and F. Lingens. 1995. Purification and characterization of hydroxyquinol 1,2-dioxygenase from Azotobacter sp. strain GP1. Appl. Environ. Microbiol. 61:2453-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louie, T. M., C. M. Webster, and L. Xun. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J. Bacteriol. 184:3492-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matus, V., M. A. Sanchez, M. Martinez, and B. Gonzalez. 2003. Efficient degradation of 2,4,6-trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 69:7108-7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyauchi, K., Y. Adachi, Y. Nagata, and M. Takagi. 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of γ-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moonen, M. J., S. A. Synowsky, W. A. van den Berg, A. H. Westphal, A. J. Heck, R. H. van den Heuvel, M. W. Fraaije, and W. J. van Berkel. 2008. Hydroquinone dioxygenase from Pseudomonas fluorescens ACB: a novel member of the family of nonheme-iron(II)-dependent dioxygenases. J. Bacteriol. 190:5199-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Natarajan, R., R. Azerad, B. Badet, and E. Copin. 2005. Microbial cleavage of C-F bond. J. Fluorine Chem. 126:424-435. [Google Scholar]
- 35.Nordin, K., M. Unell, and J. K. Jansson. 2005. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6. Appl. Environ. Microbiol. 71:6538-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oltmanns, R. H., R. Muller, M. K. Otto, and F. Lingens. 1989. Evidence for a new pathway in the bacterial degradation of 4-fluorobenzoate. Appl. Environ. Microbiol. 55:2499-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orser, C. S., C. C. Lange, L. Xun, T. C. Zahrt, and B. J. Schneider. 1993. Cloning, sequence analysis, and expression of the Flavobacterium pentachlorophenol-4-monooxygenase gene in Escherichia coli. J. Bacteriol. 175:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padilla, L., V. Matus, P. Zenteno, and B. Gonzalez. 2000. Degradation of 2,4,6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J. Basic Microbiol. 40:243-249. [DOI] [PubMed] [Google Scholar]
- 39.Perry, L. L., and G. J. Zylstra. 2007. Cloning of a gene cluster involved in the catabolism of p-nitrophenol by Arthrobacter sp. strain JS443 and characterization of the p-nitrophenol monooxygenase. J. Bacteriol. 189:7563-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prieto, M. A., and J. L. Garcia. 1994. Molecular characterization of 4-hydroxyphenylacetate 3-hydroxylase of Escherichia coli. A two-protein component enzyme. J. Biol. Chem. 269:22823-22829. [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Schlömann, M., P. Fischer, E. Schmidt, and H.-J. Knackmuss. 1990. Enzymatic formation, stability, and spontaneous reactions of 4-fluoromuconolactone, a metabolite of the bacterial degradation of 4-fluorobenzoate. J. Bacteriol. 172:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeo, M., T. Yasukawa, Y. Abe, S. Niihara, Y. Maeda, and S. Negoro. 2003. Cloning and characterization of a 4-nitrophenol hydroxylase gene cluster from Rhodococcus sp. PN1. J. Biosci. Bioeng. 95:139-145. [PubMed] [Google Scholar]
- 44.Takizawa, N., H. Yokoyama, K. Yanagihara, T. Hatta, and H. Kiyohara. 1995. A locus of Pseudomonas pickettii DTP0602, had, that encodes 2,4,6-trichlorophenol-4-dechlorinase with hydroxylase activity, and hydroxylation of various chlorophenols by the enzyme. J. Ferment. Bioeng. 80:318-326. [Google Scholar]
- 45.Thayer, A. M. 2006. Fabulous fluorine. Chem. Engin. News 23:15-24. [Google Scholar]
- 45a.Unell, M., P. E. Abraham, M. Shah, B. Zhang, C. Rückert, N. C. VerBerkmoes, and J. K. Jansson. 2009. Impact of phenolic substrate and growth temperature on the Arthrobacter chlorophenolicus proteome. J. Proteome Res. 8:1953-1964. [DOI] [PubMed] [Google Scholar]
- 46.van Berkel, W. J. H., N. M. Kamerbeek, and M. W. Fraaije. 2006. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124:670-689. [DOI] [PubMed] [Google Scholar]
- 47.van der Meer, J. R. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie van Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 48.Wieser, M., J. Eberspaicher, B. Vogler, and F. Lingens. 1994. Metabolism of 4-chlorophenol by Azotobacter sp. GPI: structure of the m-cleavage product of 4-chlorocatechol. FEMS Microbiol. Lett. 116:73-78. [DOI] [PubMed] [Google Scholar]
- 49.Xun, L., and C. M. Webster. 2004. A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J. Biol. Chem. 279:6696-6700. [DOI] [PubMed] [Google Scholar]
- 50.Xun, L., E. Topp, and C. S. Orser. 1992. Confirmation of oxidative dehalogenation of pentachlorophenol by a Flavobacterium pentachlorophenol hydroxylase. J. Bacteriol. 174:5745-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, J. J., H. Liu, Y. Xiao, X. E. Zhang, and N. Y. Zhou. 2009. Identification and characterization of catabolic para-nitrophenol 4-monooxygenase and para-benzoquinone reductase from Pseudomonas sp. strain WBC-3. J. Bacteriol. 191:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.