Abstract
We analyzed the temporal and spatial diversity of the microbiota in a low-usage and a high-usage hospital tap. We identified a tap-specific colonization pattern, with potential human pathogens being overrepresented in the low-usage tap. We propose that founder effects and local adaptation caused the tap-specific colonization patterns. Our conclusion is that tap-specific colonization represents a potential challenge for water safety.
Humans are exposed to and consume large amounts of tap water in their everyday life, with the tap water microbiota representing a potent reservoir for pathogens (8). Despite the potential impact, our knowledge about the ecological diversification processes of the tap water microbiota is limited (4, 11).
The aim of the present work was to determine the temporal and spatial distribution patterns of the planktonic tap water microbiota. We compared the summer and winter microbiota from two hospital taps supplied from the same water source. We analyzed 16S rRNA gene clone libraries by using a novel alignment-independent approach for operational taxonomic unit (OTU) designation (6), while established OTU diversity and richness estimators were used for the ecological interpretations.
Tap water samples (1 liter) from a high-usage kitchen and a low-usage toilet cold-water tap in Akershus University Hospital, Lørenskog, Norway, were collected in January and July 2006. The total DNA was isolated and the 16S rRNA gene PCR amplified and sequenced. Based on the sequences, we estimated the species richness and diversity, we calculated the distances between the communities, and trees were constructed to reflect the relatedness of the microbiota in the samples analyzed. Details about these analytical approaches are given in the materials and methods section in the supplemental material.
Our initial analysis of species composition was done using the RDPII hierarchical classifier. We found that the majority of pathogen-related bacteria in our data set belonged to the class Gammaproteobacteria. The genera encompassed Legionella, Pseudomonas, and Vibrio (Table 1). We found a significant overrepresentation of pathogen-related bacteria in the toilet tap (P = 0.04), while there were no significant differences between summer and winter samples. Legionella showed the highest relative abundance for the pathogen-related bacteria. With respect to the total diversity, we found that Proteobacteria dominated the tap water microbiota (representing 86% of the taxa) (see Table S1 in the supplemental material). There was, however, a large portion (56%) of the taxa that could not be assigned to the genus level using this classifier.
TABLE 1.
Cloned sequences related to human pathogensa
| Sampling place | Sampling time | Pathogen | NCBI accession no. | Identity (%) |
|---|---|---|---|---|
| Toilet | Summer | Escherichia coli | EF418614 | 99 |
| Toilet | Summer | Escherichia sp. | EF074307 | 99 |
| Toilet | Summer | Legionella sp. | AY924155 | 95 |
| Toilet | Summer | Legionella sp. | AY924153 | 95 |
| Toilet | Summer | Legionella sp. | AY924153 | 96 |
| Toilet | Winter | Legionella sp. | AY924061 | 96 |
| Toilet | Winter | Legionella sp. | AY924158 | 97 |
| Toilet | Winter | Legionella sp. | AY924158 | 97 |
| Kitchen | Winter | Legionella sp. | AY923996 | 97 |
| Toilet | Summer | Pseudomonas fluorescens | EF413073 | 98 |
| Toilet | Summer | Pseudomonas fluorescens | EF413073 | 98 |
| Kitchen | Summer | Pseudomonas fluorescens | DQ207731 | 99 |
| Toilet | Winter | Vibrio sp. | DQ408388 | 98 |
| Toilet | Winter | Vibrio sp. | AB274760 | 98 |
| Kitchen | Winter | Vibrio sp. | DQ408388 | 98 |
| Kitchen | Winter | Vibrio lentus | AY292936 | 99 |
| Kitchen | Winter | Vibrio sp. | AM183765 | 97 |
| Toilet | Winter | Stenotrophomonas maltophilia | AY837730 | 99 |
| Kitchen | Winter | Stenotrophomonas maltophilia | DQ424870 | 98 |
| Toilet | Winter | Streptococcus suis | AF284578 | 98 |
| Toilet | Winter | Streptococcus suis | AF284578 | 98 |
The relatedness between the cloned sequences and potential pathogens was determined by BLAST searches of the NCBI database, carried out using default settings.
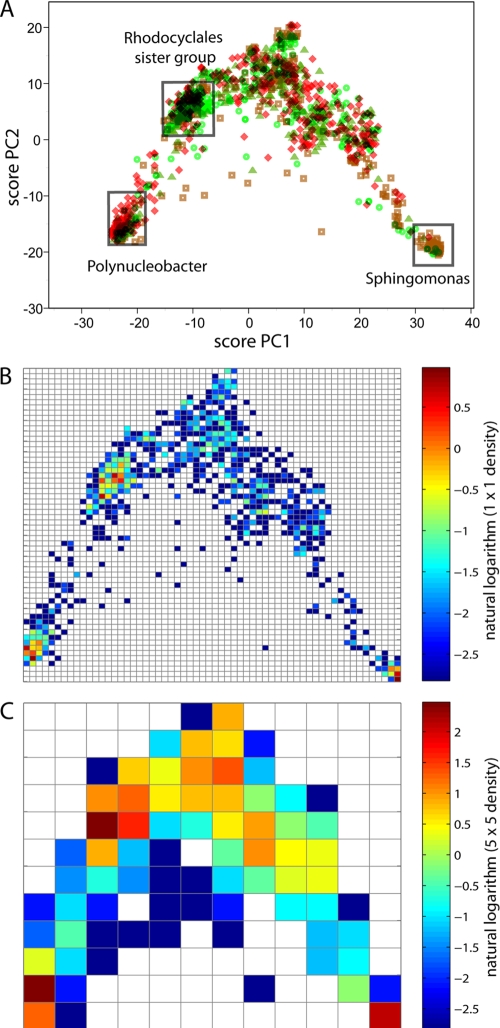
To obtain a better resolution of the uncharacterized microbiota, we analyzed the data using a clustering approach that is not dependent on a predefined bacterial group (see the materials and methods section in the supplemental material for details). These analyses showed that there were three relatively tightly clustered groups in our data set (Fig. 1A). The largest group (n = 590) was only distantly related to characterized betaproteobacteria within the order Rhodocyclales. We also identified another large betaproteocaterial group (n = 320) related to Polynucleobacter. Finally, a tight group (n = 145) related to the alphaproteobacterium Sphingomonas was identified.
FIG. 1.
Tap water microbiota diversity, determined by use of a principal component analysis coordinate system. (A) Each bacterium is classified by coordinates, with the following color code: brown squares, kitchen summer; red diamonds, toilet summer; green triangles, kitchen winter; and green circles, toilet winter. (B and C) Each square represents a 1 × 1 (B) or 5 × 5 (C) OTU. PC1, first principal component; PC2, second principal component.
The tap-specific distributions of the bacterial groups were investigated using density distribution analyses. A dominant population related to Polynucleobacter was identified for the toilet summer samples, while for the winter samples there was a dominance of the Rhodocyclales-related bacteria. The kitchen summer samples revealed a dominance of Sphingomonas. The corresponding winter samples did not reveal distinct high-density bacterial populations (see Table S2 in the supplemental material).
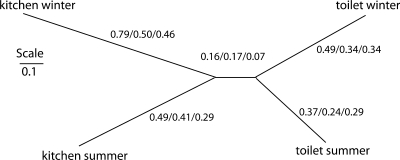
Hierarchical clustering for the 1 × 1 OTU density distribution confirmed the relatively low overlap for the microbiota in the samples analyzed (Fig. 2). We found that the microbiota clustered according to tap and not season.
FIG. 2.
Hierarchical clustering for the density distribution of the tap water microbiota. The density of 1 × 1 OTUs was used as a pseudospecies for hierarchical clustering. The tree for the Cord distance matrix is presented, while the distances calculated using the three distance matrices Cord, Brad Curtis, and Sneath Sokal, respectively, are shown for each branch.
We have described the species diversity and richness of the microbiota in Table S3 in the supplemental material. For the low taxonomic level, these analyses showed that the diversity and species richness were greater for the winter samples than for the summer samples. Comparing the two taps, the diversity and richness were greater in the kitchen tap than in the toilet tap. In particular, the winter sample from the kitchen showed great richness and diversity. The high taxonomic level, however, did not reveal the same clear differences as did the low level, and the distributions were more even. Rarefaction analyses for the low taxonomic level confirmed the richness and diversity estimates (see Fig. S1 in the supplemental material).
Our final analyses sought to fit the species rank distributions to common rank abundance curves. Generally, the rank abundance curves were best fitted to log series or truncated log normal distributions (see Table S4 in the supplemental material). The log series distribution could be fit to all of the samples except the kitchen summer samples at the low taxonomic level, while the truncated log normal distribution could not be fit to the kitchen samples at the high taxonomic level. Interestingly, however, the kitchen winter sample was best fit to a geometric curve at both the high and the low taxonomic level.
Diversifying, adaptive biofilm barriers have been documented for tap water bacteria (7), and it is known that planktonic bacteria can interact with biofilms in an adaptive manner (3). On the other hand, tap usage leads to water flowthrough and replacement of the global with the local water population by stochastic founder effects (1).Therefore, we propose that parts of the local diversity observed can be explained by local adaptation (10) and parts by founder effects (9).
Most prokaryote diversity measures assume log normal or log series OTU dominance density distributions (5). The kitchen winter sample, however, showed deviations from these patterns by being correlated to geometric distributions (in addition to the log series and truncated log normal distributions for the high taxonomic level). This sample also showed a much greater species richness than the other samples. A possible explanation is that the species richness of the tap water microbiota can be linked to usage and that the kitchen tap is driven toward a founder microbiota by high usage.
Since our work indicates an overrepresentation of Legionella in the low-usage tap, it would be of high interest to determine whether the processes for local Legionella colonization can be related to tap usage. Understanding the ecological forces affecting Legionella and other pathogens are of great importance for human health. At the Akerhus University Hospital, this was exemplified by a Pseudomonas aeruginosa outbreak in an intensive care unit, where the outbreak could be traced back to a single tap (2).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences generated were deposited in GenBank with the accession numbers GQ163803 to GQ165464.
Supplementary Material
Acknowledgments
We give special thanks to Dung Tran for providing technical assistance. We also thank Tone Tønjum for her help with the water samples and Geir Bukholm for helpful discussions.
We thank Helse Sørøst, EU project no. FP6-2006-036272, and Hedmark Sparebank for their financial support.
Footnotes
Published ahead of print on 16 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alonso, D., R. S. Etienne, and A. J. McKane. 2006. The merits of neutral theory. Trends Ecol. Evol. 21:451-457. [DOI] [PubMed] [Google Scholar]
- 2.Bukholm, G., T. Tannaes, A. B. Kjelsberg, and N. Smith-Erichsen. 2002. An outbreak of multidrug-resistant Pseudomonas aeruginosa associated with increased risk of patient death in an intensive care unit. Infect. Control Hosp. Epidemiol. 23:441-446. [DOI] [PubMed] [Google Scholar]
- 3.Jefferson, K. K. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236:163-173. [DOI] [PubMed] [Google Scholar]
- 4.Keinanen-Toivola, M. M., R. P. Revetta, and J. W. Santo Domingo. 2006. Identification of active bacterial communities in a model drinking water biofilm system using 16S rRNA-based clone libraries. FEMS Microbiol. Lett. 257:182-188. [DOI] [PubMed] [Google Scholar]
- 5.Prosser, J. I., B. J. Bohannan, T. P. Curtis, R. J. Ellis, M. K. Firestone, R. P. Freckleton, J. L. Green, L. E. Green, K. Killham, J. J. Lennon, A. M. Osborn, M. Solan, C. J. van der Gast, and J. P. Young. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384-392. [DOI] [PubMed] [Google Scholar]
- 6.Rudi, K., M. Zimonja, B. Kvenshagen, J. Rugtveit, T. Midtvedt, and M. Eggesbo. 2007. Alignment-independent comparisons of human gastrointestinal tract microbial communities in a multidimensional 16S rRNA gene evolutionary space. Appl. Environ. Microbiol. 73:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simões, L. C., M. Simões, and M. J. Vieira. 2007. Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl. Environ. Microbiol. 73:6192-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szewzyk, U., R. Szewzyk, W. Manz, and K. H. Schleifer. 2000. Microbiological safety of drinking water. Annu. Rev. Microbiol. 54:81-127. [DOI] [PubMed] [Google Scholar]
- 9.Volkov, I., J. R. Banavar, S. P. Hubbell, and A. Maritan. 2003. Neutral theory and relative species abundance in ecology. Nature 424:1035-1037. [DOI] [PubMed] [Google Scholar]
- 10.Waxman, D., and S. Gavrilets. 2005. 20 questions on adaptive dynamics. J. Evol. Biol. 18:1139-1154. [DOI] [PubMed] [Google Scholar]
- 11.Williams, M. M., J. W. Domingo, M. C. Meckes, C. A. Kelty, and H. S. Rochon. 2004. Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J. Appl. Microbiol. 96:954-964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.