Abstract
Although the level of diversity of root-associated fungi can be quite high, the effect of plant distribution and soil environment on root-associated fungal communities at fine spatial scales has received little attention. Here, we examine how soil environment and plant distribution affect the occurrence, diversity, and community structure of root-associated fungi at local patch scales within a mature forest. We used terminal restriction fragment length polymorphism and sequence analysis to detect 63 fungal species representing 28 different genera colonizing tree root tips. At least 32 species matched previously identified mycorrhizal fungi, with the remaining fungi including both saprotrophic and parasitic species. Root fungal communities were significantly different between June and September, suggesting a rapid temporal change in root fungal communities. Plant distribution affected root fungal communities, with some root fungi positively correlated with tree diameter and herbaceous-plant coverage. Some aspects of the soil environment were correlated with root fungal community structure, with the abundance of some root fungi positively correlated with soil pH and moisture content in June and with soil phosphorous (P) in September. Fungal distribution and community structure may be governed by plant-soil interactions at fine spatial scales within a mature forest. Soil P may play a role in structuring root fungal communities at certain times of the year.
In temperate forests, most trees form relationships with ectomycorrhizal (ECM) fungi, and the diversity of this fungal group alone can approach 100 species within a forest stand (17, 20, 60). The ECM mutualism may be necessary for the success of some native plant species, as approximately 90% of roots of some tree species are colonized by ECM fungi (65). Nevertheless, we still know surprisingly little about what controls the community structure and distribution of root-associated fungi in forest systems (44, 46). The occurrence of root-associated fungi may broadly reflect soil environmental conditions and the presence of preferred plant hosts (28, 61), but how these factors interact to influence the diversity, distribution, and community structure of these fungi within forest habitat patches at a local scale is uncertain.
The distribution of root-associated fungi may be primarily a species response to local soil environmental conditions. For example, both the quality (i.e., nutrient content) and the quantity of soil organic matter are known to influence the diversity of ECM communities (18, 20, 32). ECM fungi also vary in drought tolerance (14, 36), resistance to fire (61, 65), and tolerance to soil acidity (19) and temperature (56). Changes in soil chemistry, especially as they relate to pH and the availability of nitrogen (N) and phosphorous (P), might favor selection of fungi most capable of tolerating environmental extremes (2, 28, 29).
Plant distribution and identity may, however, play the strongest role in structuring the below-ground diversity of root-associated fungi. Many ECM fungi can colonize a wide range of plant species, and plant species can be host to a large number of ECM fungi (63), especially those in the families Russulaceae and Thelephoraceae (34, 35, 62). Moreover, some ECM fungi are also specific to certain tree species (e.g., Suillus and Rhizopogon species are specific to species in the family Pinaceae [38, 39]). At the local scale, fungal distribution and richness might be influenced by differences in root growth and architecture (30, 42), by the distance to the bole of the tree (11, 42, 49), or by the presence of neighboring trees (29, 64). Temporal changes in ECM communities could be associated with seasonal changes in plant physiology and phenology (3, 8, 17).
An often overlooked factor influencing root-associated fungi of tree roots is the occurrence of herbaceous plant species within forest stands. Many species of parasitic, achlorophyllous angiosperms obtain carbon (C) from ECM fungi that colonize tree roots (43), and some autotrophic plants could also obtain C from ECM fungi during certain times of the year (58). Herbaceous plants also influence the cycling of nutrients, including N, P, and K (potassium) (31, 50), within forests, which could affect the distribution of root-associated fungi. Herbaceous plants can also produce secondary compounds that inhibit colonization of tree roots (68).
In this study, we examine the effect of soil environment and plant distribution on root-associated fungi of tree roots in a mature beech-maple forest at two points in the growing season. We predict that plant distribution, both the distribution of host trees and that of herbaceous plants, influences fungi associated with tree roots in terms of both community structure and diversity. Molecular typing protocols, including a site-specific database of fungal sequences and fingerprints, were used to identify fungi on tree roots (i.e., beech or maple trees) to the species level.
MATERIALS AND METHODS
Site description and soil sampling.
Stebbins Gulch is a 360-ha mature, mixed-mesophytic forest located within the Holden Arboretum in northeastern Ohio (82°28′N and 40°57′W). Approximately 80 ha is old-growth beech-maple forest, within which we established our study site. American beech (Fagus grandifolia, ∼75% of canopy coverage) and sugar maple (Acer saccharum, ∼15% of canopy coverage) are the dominant overstory species. White ash (Fraxinus americana, ∼7% of canopy coverage) and basswood (Tilia americana, ∼3% of canopy coverage) are also important overstory species. Understory woody species include A. saccharum, F. grandifolia, and Lindera benzoin, while abundant herbaceous plants include Allium tricoccum, Arisaema triphyllum, Claytonia virginica, Dicentra canadensis, and Uvularia grandiflora. The total precipitation averages around 116 cm per year, with an average of 287 cm of snowfall per season. The site is characterized by gently sloping ground (2 to 6% slope) and moderately drained silt loam soils.
Soil and root sampling was conducted during the 2006 growing season. Sixty cores were collected between June 5 and June 16 and between September 11 and September 22 (n = 120) to a depth of 5 cm by use of a 10-cm-diameter metal soil corer. Preliminary sampling found that 90% of all tree root tips are found within the top 5 cm of soil. Samples were placed in bags and kept on ice until processing. Soil cores were collected along three 100-m-long parallel transects. Twenty cores, spaced 5 m apart, were collected along each transect (three transects by 20 cores by two samplings = 120 cores). Transects were placed at least 50 m apart and run perpendicular to the natural contour. Cores collected between June and September at the same point along a transect were no more than 25 cm apart. We monitored the presence of beech and maple near each sample point along transects and found that greater than 93% of soil cores contained at least beech saplings within 5 m of the point (>2.0-cm diameter at breast height [dbh]).
Soil was sieved (2 mm) to separate soil from root tissue and then divided into several fractions: one fraction was used for pH and water content analysis, and a second fraction was placed in a −70°C freezer for analysis of soil C, N, and P. Soil separation from root tissue was completed within 4 h of sampling. Roots were placed into a series of nested sieves (smallest sieve, 250 μm) and washed of soil, and viable roots were removed under a ×2 magnification lens and placed in cold physiological saline (8.5 g NaCl liter−1) (9). Only woody tree roots were retained for molecular analysis. Herbaceous roots recovered from the samples were discarded and not included in our analysis. Tree roots were further examined under a dissecting scope, and nonviable (senescent) roots and large-diameter roots (>1 mm) were discarded. All remaining viable root tissue, including ECM root tips and root tips not visually colonized by ectomycorrhizae, were placed in centrifuge tubes and stored at −70°C until DNA extraction. Since we did not separate tree roots based on plant species (i.e., beech or maple), our analysis reflects root-associated fungal communities colonizing tree roots within a discrete volume of forest soil. Abundant ectomycorrhizae were subsampled for individual DNA extraction and analysis prior to −70°C storage. Abundant ectomycorrhizae were morphotyped, photographed, and placed in individual microcentrifuge tubes for separate DNA extraction and molecular analysis. These morphotyped tips were used to construct a terminal restriction fragment length polymorphism (TRFLP) database for molecular identification of fungi on tree roots.
In addition, we conducted sporocarp surveys of Stebbins Gulch and Bole Woods (a beech-maple forest also located at the Holden Arboretum) in the spring, summer, and fall of 2006, 2007, and 2008. Surveys were conducted every 2 weeks throughout the spring and summer and every week throughout the fall. Sporocarps were photographed, cataloged, and used for DNA sequencing and TRFLP analysis to expand our molecular database of fungal sequences and TRFLP patterns obtained from root tips.
Analysis of soil environment and vegetation.
Field fresh soil was used to measure soil pH (1:1 H2O) and gravimetric water content and is expressed here as follows: (g water g fresh weight soil−1) × 100% (37). Soil for C and N was oven dried and pulverized in a Precellys homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) and analyzed with an ECS 4010 CHNSO elemental analyzer (Costech Analytical, Valencia, CA). Labile soil inorganic P (Pi) (readily available) and organic P (Po) (easily mineralizable) were extracted from pulverized, oven-dried soil by adding 0.5 M NaHCO3 (pH 8.5) and shaking at 100 rpm on an orbital shaker (Lab-Line, Melrose Park, IL) for 30 min (51). Pi was determined colorimetrically using a modified ascorbic acid method (40) directly on the NaHCO3 extracts, while Po was determined by the increase in Pi after NaHCO3 extract digestion with 1.8 N H2SO4 and (NH4)2S2O2 (26).
We measured the diameter at breast height of each tree of >2 cm in diameter located within 5 m of where each soil sample was taken, as well as the distance of each tree from the sampling location. Trees were placed in four size classes (2 to 10 cm, 10 to 30 cm, 30 to 60 cm, and >60 cm). At each sampling location, we also established a 25- by 25-cm quadrat and recorded the presence and estimated percent coverage of all herbaceous plants. In our forest, the herbaceous community consists primarily of a spring ephemeral community, and we therefore used percent coverage of the spring community to examine the effects on fungal species in both early-summer (June) and late-summer (September) samples.
DNA extraction and purification.
DNA from all viable roots in each soil core was extracted using a bead-beating protocol. Viable roots (up to 200 mg fresh weight) were placed in a 1.5-ml bead-beating tube containing 500 mg of 400 μM glass beads (VWR, West Chester, PA) and 750 μl 2% CTAB (cetyltrimethyl-ammonium bromide). Samples were then beaten for 90 s in a Precellys homogenizer at 6,500 rpm and incubated at 65°C for 1 h. Approximately 500 ml of the supernatant was removed and DNA purified by phenol-chloroform extraction and precipitation with 20% polyethylene glycol 8000 in 2.5 M NaCl (9, 11). All extracted DNA was suspended in PCR-grade water and stored at −20°C. DNA extraction of subsampled ECM roots and sporocarps was performed using the same procedure as described above except that subsampled roots and sporocarps were manually ground with a micropestle prior to incubation at 65°C for 1 h.
Fungal TRFLP and sequence database.
The TRFLP and sequence database was created from three sources of environmental DNA: (i) subsampled and morphotyped ECM root tips, (ii) sporocarps periodically collected during the 2006 to 2008 growing seasons, and (iii) Stebbins forest soil collected at the site. Subsampled and morphotyped root tips and sporocarps were extracted as described above, and DNA was used as the template for PCR using primers NSI1 and NLB4, which target the internal transcribed spacer regions (ITS1, 5.8S rRNA, and ITS2) of the rRNA gene (9, 11, 48). These primers have been tested extensively over 5 years of research on various topics and have successfully amplified ECM fungi from more than 2,000 single-root-tip DNA extracts, including ascomycetes, such as Cenoccocum sp. and Tuber sp. (48). PCR was carried out with 50-μl reaction volumes using 1 μl of purified DNA (approximately 100 ng), 0.2 μm of primers NSI1 and NLB4, 2.0 mM MgCl, 0.2 mM deoxynucleoside triphosphate, 0.15 mg ml−1 bovine serum albumin, and 2.0 U Taq DNA polymerase (Promega, Madison, WI) on a PTC 100 thermal cycler (MJ Research, Boston, MA). An initial denaturation step of 5 min at 94°C was followed by amplification for 35 cycles under the following conditions: 30 s at 94°C, 60 s at 50°C, and 90 s at 72°C. A final 5-min extension at 72°C completed the protocol. The PCR product was purified using a PCR purification kit (Promega, Madison, WI) following the manufacturer's instructions and used as the template for sequencing that was completed through the Cornell Bioresource Center using an Applied BioSystems 3730xl DNA analyzer. Generated sequences were compared to EMBL/GenBank/DDBJ database entries using the FASTA program (European Bioinformatics Institute) and confirmed through UNITE (http://unite.ut.ee) to determine the putative identities of ECM fungi. Some root samples failed to sequence adequately using this procedure. Consequently, DNA of problematic samples was cloned using a pGEM-T Easy vector system (Promega, Madison, WI), following the manufacturer's instructions. Ten randomly selected colonies were incubated overnight at 37°C in LB medium, and plasmids were harvested using a Wizard Plus SV Miniprep DNA purification system (Promega, Madison, WI). Harvested plasmids were used as the template for sequencing in these cases. To further expand our database, DNA was also extracted from forest soil using a MoBio DNA extraction system and bead beating. Soil DNA was used for PCR, and the PCR product was cloned as described above. Two soil samples were processed in this matter, and 50 clones (25 clones per core; soil cores collected along transects) were harvested and sequenced as described above (see Fig. S1 in the supplemental material for sampling scheme details).
Environmental DNA (i.e., morphotyped root tips and sporocarps) and plasmid DNA (i.e., soil clones) were also used for TRFLP analysis. Labeled primers 58A2F (6-carboxyfluorescein) and NLB4 (4,7,2′,4′,5′,7′-hexachloro-6-carboxyfluorescein) were used to amplify ITS2, located between the 5.8S and the 28S rRNA genes (9, 11, 48; see reference 48 for details on primer testing and design). We focused on this region because it appeared to provide the best discrimination among species based on TRFs generated from morphotyped tips and sporocarps and predicted from sequence information. In a previous study, we also found that the ITS2 primers amplified species of the ECM genus Cenococcum more consistently than did ITS1 primers (9). PCR was performed using the conditions noted above, and endonucleases AluI and HaeIII (Promega, Madison WI) were used for TRFLP analyses (9-11). TRFLP analyses were completed through the Cornell Bioresource Center using an Applied BioSystems 3730xl DNA analyzer. TRFLP results were analyzed with PeakScanner software version 1 (Applied Biosystems). Because the AluI digest resulted in NLB4 TRFs that were nearly identical in size for most of the recovered fungi, our procedure provided us with three distinct restriction fragments to use for the discrimination of fungi within our database and for the identification of fungi within the complex root fungal communities. At the time of community analysis, the site-specific database consisted of 266 typed and identified fungi.
Detection and analysis of root fungal communities through TRFLP analysis.
DNA extracted from all viable roots separated from each soil core was used for amplification of all of the root fungal communities within that core. DNA was amplified with ITS2 primers and used for TRFLP analysis with restriction enzymes AluI and HaeIII, as noted above. For each core, we generated three TRFLP profiles and used these profiles to identify fungal species using the program Fragsort (57) to facilitate identification of fungi in TRFLP profiles. The three TRFLP profiles for each core were used as input for Fragsort, which then used our TRFLP database to identify fungal species in each core by matching TRFs in the community sample to the species in our database. For a species type to be considered present on roots, the appropriate TRF must appear in all three community profiles (see Fig. S1 in the supplemental material). Only TRFLP peaks with >50 fluorescence units (scale of 5,000) were included in our analysis (i.e., major TRFs). We found that the TRFs of separate isolates of the same fungal species type could vary by as much as 2 bp (see Table S1 in the supplemental material). Consequently, we considered TRFs in community profiles a match if they were within 1 to 2 bp of the TRFs within the fungal database. Using this technique, we were able to identify more than 50% of all TRFs in a TRFLP profile, comprising more than 75% of the total area (i.e., all major peaks were identified). Since the database we used for fungal identification was constructed largely from ECM root tips and sporocarps of basidiomycetes, our approach may underrepresent ascomycetes present on root samples. Our analysis, therefore, is limited to those fungi that we could positively identify with our database.
Statistical analyses.
Differences in soil chemistry between transects and sampling locations and across sampling dates were analyzed using the Mann-Whitney rank sum test and SigmaStat 3.5 (Systat Software Inc., CA). Differences in fungal richness and Shannon diversity (H′) were determined through the use of Student's t test. Richness represents the fungal species positively identified on root tips from our soil cores by using the TRFLP database. H′ was calculated for identified fungi, where the average TRF peak area was used as a measure of proportional abundance in that core (9, 10, 11). H′ was calculated using procedures available through PC-ORD 4 (MjM Software, OR). To assess the relationship between the community structure of the detected fungi and the environmental conditions and to determine whether detected fungal species were correlated with soil chemistry and plant data, we used nonmetric multidimensional scaling (NMS) procedures available through PC-ORD 4 (MjM Software, OR). The Sørenson distance with a random starting configuration was used for these analyses. A maximum of 400 iterations were used for 50 runs, with data for the Monte Carlo test randomized. We also calculated Pearson correlation coefficients to further elucidate correlations between fungal community patterns and soil chemistry and plant data. Although we were able to determine species identity using our site-specific database, 50% of detected fungal species occurred in less than 5% of the soil cores. Consequently, we lumped fungal species together and performed NMS on fungal genera to reduce the variance within the data set. To determine whether differences between transects and sampling times existed, we used multiresponse permutation procedures (MRPP) through PC-ORD 4. For both MRPP and NMS, the proportional abundance of all detected fungal species was used as an indicator of abundance within each sample (9, 10, 12), and all proportional abundance data were transformed before analysis. Although peak area may not represent a true quantification of fungal species, in previous work we found that the TRFLP peak area was an accurate approximation of the relative number of ECM root tips colonized by a fungal species in a soil core (11). Because MRPP analysis indicated that the June and September communities were significantly different, we performed NMS on all samples together and also separately on samples collected during June and September.
RESULTS
Analysis of soil environment and vegetation.
We found that soil chemistry changed significantly from June to September (Table 1), with soil C, N, and C/N all increasing and labile Pi and Po decreasing. Gravimetric soil moisture contents ranged from 23 to 70% (mean ± standard deviation, 35% ± 1%) but were not significantly different between the two sampling dates. Soil pH levels ranged from 3.5 to 5.6, with a mean of 3.98 ± 0.05 in June, and did not exhibit an effect of sampling date. Beech trees of 10- to 30-cm dbh were encountered near 25% of soil sampling locations, while 45% of the sampling locations were near beech trees of >60-cm dbh. Beech trees between 30- and 60-cm dbh were less frequently encountered near soil sampling locations, with only 15% of cores having trees of that size class nearby. The frequency of maples in the largest size classes was low (maples between 30- and 60-cm and >60-cm dbh were encountered near less than 5% of soil cores), but maples in the smallest size class were frequently encountered near soil sampling locations (near 25% of cores). Herbaceous community coverage averaged 15.1% ± 2.2%, and herbaceous coverage ranged from 0 to 60%. A. tricoccum and D. canadensis had the highest average percent coverage of the herbaceous species encountered (8.3% ± 1.7% and 6.2% ± 1.2%, respectively).
TABLE 1.
Results of soil chemical analysis for Stebbins Gulch in June and September of 2006a
| Soil parameter | Result from: |
P value | |
|---|---|---|---|
| June | September | ||
| pH | 3.98 (0.05) | 3.95 (0.05) | 0.723 |
| N (mg·g−1) | 0.428 (0.027) | 0.509 (0.027) | 0.011 |
| C (mg·g−1) | 6.54 (0.57) | 8.35 (0.57) | 0.002 |
| C/N | 14.86 (0.25) | 15.93 (0.25) | 0.007 |
| Labile Pi (mg·kg−1) | 60.86 (3.40) | 39.76 (3.40) | <0.001 |
| Labile Po (mg·kg−1) | 129.73 (4.64) | 61.80 (4.64) | <0.001 |
Means are shown, with standard errors of the means in parentheses. The Mann-Whitney rank sum test was used for this analysis (n = 60 for all tests). Significant differences between seasons are shown in bold.
Fungal community diversity and structure.
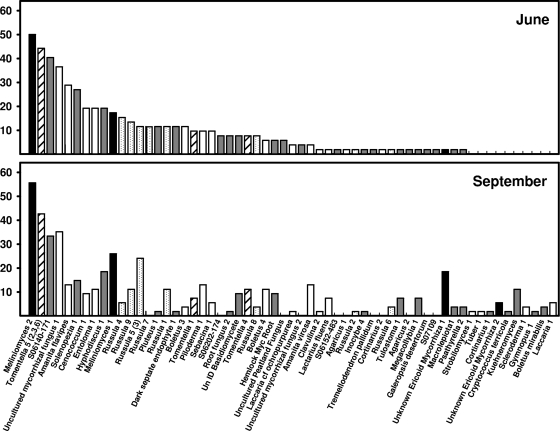
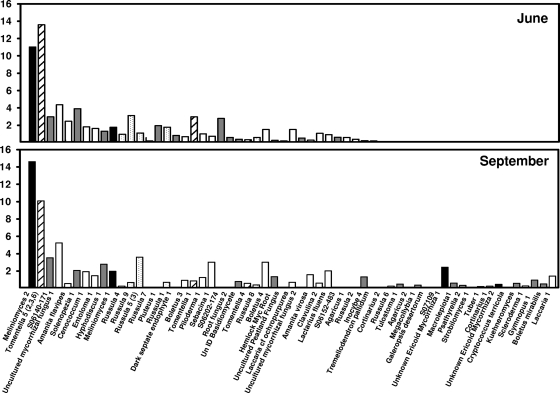
Overall fungal richness ranged from 1 to 12 species per soil core, and H′ ranged from 0 to 2.1 per core when combined across sampling dates. Richness averaged 5.2 ± 0.4 and 5.0 ± 0.3 species per core and H′ averaged 1.19 ± 0.08 and 1.14 ± 0.07 in June and September, respectively, but differences were not significant. Fungal richness was negatively correlated with labile Pi (−0.195; P = 0.04), but no other significant correlations between richness and environmental variables were found (Table 2). In total, we detected 63 different species of fungi colonizing the tree roots in the soil cores, representing 28 different genera (Fig. 1 and 2). We were able to confirm that at least 32 of these species had high similarity to known mycorrhizal fungi, while the remaining fungi showed high similarity to either ericoid mycorrhizae or saprotrophic fungi (Fig. 1 and 2; also see Table S1 in the supplemental material). The most frequently encountered ECM fungi were from the genera Russula and Tomentella, which were encountered in 49% and 43% of soil cores and were represented by eight and six species, respectively (Fig. 1 and 2). Fungi with similarity to Meliniomyces and Rhizoscyphus (Rhizoscyphus ericae aggregate [33]), genera known to form ericoid mycorrhizae, were the fungi most frequently and abundantly encountered on roots from our soil cores (because we could not distinguish between these closely related genera with our method, they are lumped together and referred to herein as Meliniomyces). Frequency and abundance patterns suggested that fungal communities were different in the June and September samplings (Fig. 1 and 2). Although many fungal species remained both frequent and abundant throughout the study, the abundance and frequency patterns of other species varied between June and September. For example, Cenococcum sp. was detected in approximately 19% of soil cores in June but only 9% of cores in September and represented only 2% of the community proportional abundance during both times. In contrast, Boletus species 4 increased in frequency from approximately 6% to 11% from June to September, and the proportional abundance increased from 1.5 to 3%. Species within the same genera also behaved differently over time. Russula species 4 declined in frequency from June to September (15 to 6%), while other Russula species remained constant or increased in frequency. The overall fungal communities differed significantly between June and September based on MRPP analysis (agreement statistic [A] = 0.09; P < 0.001).
TABLE 2.
Relationships between fungal richness, diversity, and environmental variables, determined using Pearson correlationa
| Parameter | Pearson correlation coefficient |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| H′ | Soil pH | Soil N | Soil C | C/N | Pi | Po | % Moisture | Total herbaceous-plant cover | |
| Fungal richness | 0.909 | −0.0359 | −0.0445 | −0.0553 | −0.0986 | −0.195 | 0.106 | −0.0371 | −0.0656 |
| H′ | −0.0169 | −0.0171 | −0.0319 | −0.093 | −0.105 | 0.138 | 0.0149 | −0.0574 | |
Variables with positive values tend to increase together; where correlations are negative, one variable tends to increase while the other decreases. Only Pi was significantly correlated (P = 0.04), with fungal richness (shown in bold). Correlations were also performed with the distance of the core to beech and maple of various size classes and with the percent cover of specific herbaceous species. No significant correlations were found for these environmental variables (data not shown).
FIG. 1.
Frequencies of root-colonizing fungi encountered within a mature beech-maple forest. The frequency is expressed as the number of cores in which the species type was observed (out of 60). Black bars represent ericoidlike mycorrhizal fungi, gray bars represent fungi of unknown habit, and white bars represent ECM fungi. The genus Tomentella is denoted by hatched bars and the genus Russula by stippled bars. UnID, unidentified.
FIG. 2.
Relative levels of abundance of root-colonizing fungi encountered within a mature beech-maple forest. The bars represent the relative levels of abundance (percent) of fungal types averaged across all cores (n = 60), as determined by TRFLP analysis. Peak area is used as a proxy measure of abundance in this study. Black bars represent ericoidlike mycorrhizal fungi, gray bars represent fungi of unknown habit, and white bars represent ECM fungi. The genus Tomentella is denoted by hatched bars and the genus Russula by stippled bars. UnID, unidentified.
Correlations between fungal communities and environmental parameters.
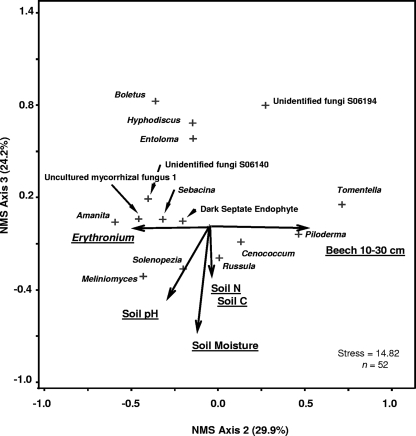
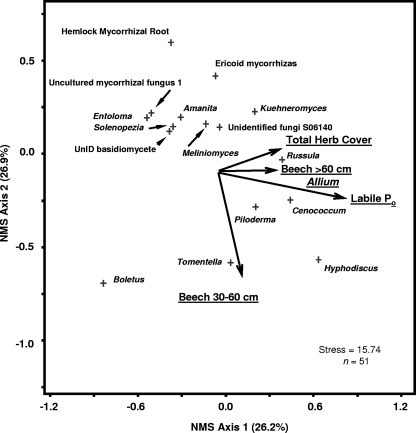
NMS analysis of all collected samples (June and September combined) determined that the first dimension of the ordination was significantly correlated with the coverage of herbaceous plants, while the second and third dimensions of the ordination were significantly correlated with the soil environment, with negative correlations between community structure and soil moisture and soil N and positive correlations with soil Po (Table 3). The cumulative variance for all three dimensions of the ordination was r2 = 0.489. The first dimension of the June fungal ordination was not significantly correlated with environmental metrics (Fig. 3; Table 3). The second dimension was significantly positively correlated with the distance of the core from beech trees between 10- and 30-cm dbh and significantly negatively correlated with coverage by Erythronium. The third dimension of the June ordination was significantly negatively correlated with soil pH, moisture content, and soil C and N (Fig. 3; Table 3). Root fungal communities in June were also influenced by the transect location (MRPP analysis, P < 0.001). Samples along transect 1 had fungal communities more similar to each other than to samples from transects 2 and 3. Analysis of variance performed on ranks suggested that the genus Amanita, an uncultured mycorrhizal fungus (UMF1), and the genus Meliniomyces were significantly more abundant on transect 1, while the genus Russula was significantly more abundant on transect 2 (data not shown). Unlike June ordinations, the first dimension of the September ordination was significantly correlated with Po, the total percent cover of herbaceous plants, and specifically the percent cover of A. tricoccum, as well as the distance to beech trees of >60-cm dbh (Fig. 4; Table 3). The second dimension of the September ordination was strongly negatively correlated with the distance to beech trees between 30- and 60-cm dbh, while the third dimension was positively correlated with soil C, C/N ratio, and the smallest beech diameter class (Fig. 4; Table 3). Both the June and September ordinations suggest a separation between many ECM genera and nonmycorrhizal genera. The genera Cenococcum, Piloderma, Russula, and Tomentella tended to cluster together and were more likely than other ECM fungal genera and nonmycorrhizal genera to be positively correlated with many of the environmental metrics. The genus Russula in particular seemed positively correlated with herbaceous plants during the September sampling (Fig. 4).
TABLE 3.
Relationships between soil environmental and vegetation variables and NMS dimensions, determined using Pearson correlationa
| Parameter | Pearson correlation coefficient |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| All samples |
June samples |
September samples |
|||||||
| Dim1 | Dim2 | Dim3 | Dim1 | Dim2 | Dim3 | Dim1 | Dim2 | Dim3 | |
| Soil pH | 0.09 | 0.05 | 0.07 | 0.08 | −0.23 | −0.32 | −0.02 | −0.02 | −0.17 |
| Soil N | 0.16 | −0.20 | 0.02 | 0.02 | −0.01 | −0.26 | 0.06 | 0.11 | 0.24 |
| Soil C | 0.17 | −0.19 | 0.02 | 0.08 | −0.01 | −0.25 | 0.09 | 0.07 | 0.25 |
| C/N | 0.18 | −0.12 | −0.04 | 0.24 | 0.11 | −0.09 | 0.20 | 0.18 | 0.27 |
| Labile Pi | 0.14 | 0.14 | 0.16 | 0.12 | 0.01 | −0.19 | 0.02 | 0.15 | 0.10 |
| Labile Po | 0.14 | 0.30 | 0.44 | −0.12 | 0.03 | 0.06 | 0.43 | −0.18 | −0.03 |
| % Moisture | 0.18 | −0.24 | −0.02 | 0.03 | −0.12 | −0.39 | 0.09 | 0.11 | 0.20 |
| Fagus 10- to 30-cm dbh | 0.16 | −0.06 | 0.04 | 0.15 | 0.35 | −0.05 | −0.10 | 0.16 | 0.27 |
| Fagus 30- to 60-cm dbh | −0.12 | 0.11 | 0.08 | −0.01 | 0.08 | −0.08 | 0.19 | −0.36 | −0.12 |
| Fagus >60-cm dbh | −0.11 | −0.09 | 0.11 | −0.10 | 0.08 | 0.13 | 0.25 | 0.01 | 0.12 |
| Acer 10- to 30-cm dbh | −0.01 | 0.02 | 0.03 | −0.16 | 0.06 | 0.04 | −0.24 | 0.11 | 0.04 |
| Acer 30- to 60-cm dbh | 0.12 | −0.05 | 0.11 | 0.02 | 0.05 | −0.06 | −0.23 | 0.16 | 0.20 |
| Acer >60-cm dbh | 0.06 | 0.01 | −0.03 | −0.12 | −0.15 | 0.01 | −0.04 | −0.02 | 0.18 |
| Allium | 0.23 | −0.07 | 0.09 | −0.10 | 0.20 | 0.13 | 0.30 | −0.01 | −0.08 |
| Dicentra | −0.10 | −0.15 | 0.05 | −0.03 | −0.01 | −0.10 | 0.14 | 0.24 | −0.16 |
| Erythronium | 0.11 | −0.04 | −0.05 | −0.02 | −0.28 | −0.03 | −0.08 | 0.15 | 0.14 |
| Maianthemum | 0.01 | −0.02 | −0.04 | 0.09 | 0.09 | −0.03 | −0.02 | 0.22 | −0.05 |
| Total herbaceous-plant cover | −0.23 | −0.14 | 0.09 | −0.09 | 0.13 | 0.04 | 0.30 | 0.15 | −0.14 |
Significance of correlations was determined using the critical values for correlation coefficients (71). For all samples (n = 120), P was <0.05 for an r of >0.20; for June and September samples (n = 60), P was <0.05 for an r of >0.25 (two-tailed test). Significant correlations are shown in bold. Dim1, first dimension; Dim2, second dimension; Dim3, third dimension.
FIG. 3.
NMS ordination based on relative abundance of identified root fungal genera, with joint plots of the most important environmental variables for early-summer (June) sampling. The joint-plot vector lengths indicate the strength and direction of the strongest correlations. The proportion of variance explained by axes 2 and 3 is shown. The cumulative variance was r2 = 0.780. See Table 3 for correlations with all environmental variables. The vectors for soil C and N are in the same approximate positions, so only one vector is shown for illustration.
FIG. 4.
NMS ordination based on relative abundance of identified root fungal genera, with joint plots of the most important environmental variables for late-summer (September) sampling. The joint-plot vector lengths indicate the strength and direction of the strongest correlations. The proportion of variance explained by axes 1 and 2 is shown. The cumulative variance was r2 = 0.813. See Table 3 for correlations with all environmental variables. The vectors for beech trees of >60-cm dbh and Allium are in the same approximate positions, so only one vector is shown for illustration. UnID, unidentified.
The ordinations suggested that positive correlations existed between some ECM fungal genera. For example, the genera Russula and Piloderma always appeared near each other in ordination space (Fig. 3 and 4). Pearson correlations for fungal genera found some significant positive and negative correlations among ECM genera (Table 4). Russula was significantly positively correlated with Piloderma and significantly negatively correlated with Tomentella. Tomentella was significantly negatively correlated with Amanita, Meliniomyces, and Russula. Cenococcum also shared ordination space with both Russula and Piloderma; however, Pearson correlations for all data found positive associations between Cenococcum and only Hyphodiscus and a dark septate endophyte (DSE19). A separate analysis of the June data determined significant correlations between Russula and Cenococcum (r = 0.44; P = 0.001) and between Russula and Piloderma (r = 0.35; P = 0.011), but these relationships were not apparent for September samples (data not shown), suggesting that these fungi may have similar environmental preferences but do not necessarily associate with one another.
TABLE 4.
Relationships among root fungal genera, determined using Pearson correlationa
| Organism | Pearson correlation coefficient |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aman. | Bole. | Ceno. | Hyph. | Hyme. | Pilo. | Russ. | Tome. | Ericoid | DSE19 | Kueh. | UMF1 | |
| Amanita | −0.06 | −0.03 | −0.13 | 0.17 | −0.07 | −0.03 | −0.21** | 0.04 | −0.03 | 0.11 | 0.36 | |
| Boletus | −0.05 | −0.02 | −0.20** | −0.05 | −0.12 | −0.12 | −0.08 | −0.04 | −0.06 | 0.05 | ||
| Cenococcum | 0.23** | −0.09 | −0.05 | 0.05 | −0.11 | −0.06 | 0.21** | −0.04 | −0.07 | |||
| Hyphodiscus | −0.20** | −0.01 | 0.12 | −0.05 | −0.09 | −0.03 | 0.07 | −0.15 | ||||
| Hymenocyphus | −0.16 | −0.14 | −0.21** | 0.18* | −0.05 | 0.02 | 0.04 | |||||
| Piloderma | 0.18* | −0.02 | −0.05 | −0.05 | 0.27*** | −0.07 | ||||||
| Russula | −0.20** | −0.03 | 0.09 | 0.01 | 0.01 | |||||||
| Tomentella | −0.10 | −0.11 | −0.09 | −0.25*** | ||||||||
| Ericoid | −0.06 | 0.14 | −0.02 | |||||||||
| DSE19 | −0.04 | −0.05 | ||||||||||
| Kuehneromyces | 0.01 | |||||||||||
| UMF1 | ||||||||||||
Significant correlations are indicated in bold (*, P < 0.06; **, P < 0.05; ***, P < 0.01). DSE19 has high similarity to dark septate fungi, and UMF1 has high similarity to an uncultured mycorrhizal fungus. Analysis was run on all samples (June and September) (n = 107 cores). Aman., Amanita; Bole., Boletus; Ceno., Cenococcum; Hyph., Hyphodiscus; Hyme., Hymenocyphus; Pilo., Piloderma; Russ., Russula; Tome., Tomentella; Kueh., Kuehneromyces.
DISCUSSION
Effect of vegetation on root-associated fungi.
We found evidence to support our prediction that the distributions of both host trees and nonhost herbaceous plants would have an effect on the distribution and community structure of root-associated fungi. Our analysis indicates that plant distribution was, in fact, strongly correlated with root-associated fungi in our forest and that both trees and herbaceous plants could affect tree root fungal communities. Although the distance from the soil core to beech trees of certain diameter classes did appear to affect the community structure of root-associated fungi, fungal richness and diversity were unaffected. This is in agreement with a previous study, where ECM morphotype diversity was lower in soil collected near isolated trees than in soil from trees growing in groups, but diversity was not affected by tree diameter (64). In a loblolly pine plantation, the distance of the soil core to the tree bole did not affect ECM diversity or richness; however, the distribution of some fungal taxa was affected (11). Changes in community structure, without alteration of species richness or diversity, may reflect different colonization strategies and resource requirements of ECM fungi.
In our study, the presence and abundance of the genera Cenococcum, Piloderma, and Tomentella appeared strongly correlated with distance to beech trees in both June and September, whereas Boletus was strongly correlated with distance only in June. These changes in community structure could reflect differences in below-ground root density and resource availability nearest the tree bole or differences in tree age. The distance to adjoining trees can affect sporocarp production, especially in plantations (42, 49), and root system age, class, and development can affect ECM fungal colonization (30). Fungi may possess different strategies, with root age and resource availability affecting the ability of some fungi to successfully colonize roots (45, 60). Differences in root density and turnover among trees of different sizes might influence fungal colonization, but tree size might be related to differences in plant physiology that could also affect root fungal communities. Larger and older canopy trees may have different carbon acquisition and whole-tree carbon allocation strategies (54), and tree size and canopy structure could affect stem flow following storm events, thereby altering soil physiochemical conditions (e.g., soil N and moisture) closest to the tree (23). Examining the different effects of root system structure and tree size on root-associated fungi will require further study.
We observed a strong correlation between the community structure of root fungi and the coverage of spring ephemeral plants. Surprisingly, the strongest correlation between herbaceous plants and root fungi occurred during the September sampling, several months after the senescence of these herbaceous plants. The genus Russula appeared most strongly correlated with the coverage of herbaceous plants, especially Allium, during the September sampling, but the cause of these correlations is uncertain. Russula has been reported as an associate of mycoheterotrophic plants in previous studies (5, 69, 70); however, mycoheterotrophic plants are not present along the transects, and these herbaceous communities are generally dominated by Allium. The genus Russula is capable of producing extracellular enzymes that can degrade organic matter in litter and soil (1). Therefore, it is possible that the strong correlation between the genus Russula and areas that contained actively growing herbaceous plants 4 months earlier is a response to the presence of a litter/resource pulse coming from the senescence and fall of herbaceous litter in some forest patches. Herbaceous plants can contribute as much as 16% of total forest litter fall on an annual basis (31), and this could create temporary and spatially patchy nutrient hot spots within soil that some ECM fungi exploit. The exact relationship between herbaceous plants and fungi associated with tree roots requires further study.
Effect of soil environment on root-associated fungi.
In this mature forest, we found that root-associated fungi responded to soil environmental conditions and that these relationships changed between the June and September samplings. Soil chemistry changes between June and September may reflect decompositional losses and seasonal turnover of litter that would result in increases in soil C and N contents in the top organic layer. In June, fungal communities were significantly correlated with soil pH, soil moisture, and soil C and N at fine spatial scales, while in September, fungal communities were significantly correlated with labile Po, soil C, and C/N ratio. Soil water availability can affect mycorrhizae on beech (36, 59), and this could be an important factor controlling fungi during early summer. A substantial body of evidence suggests that soil N can significantly alter and affect the distribution and community structure of ECM fungi (3, 11, 24, 47, 53). Although C and N were significantly correlated with fungal community structure in June and C was significantly correlated with root fungi in September, the correlations were not strong. We restricted our analysis to the top 5 cm of soil, containing >90% of all root tips (data not shown). Thus, we were unable to analyze differences in fungal communities based on vertical separation in soil or between mineral and organic soil. Niche differentiation with respect to organic matter content and vertical distribution in soil has been shown previously (7, 20, 32, 41, 62), and it is possible that different fungal communities may exist on roots near the soil surface and 5 cm below. By combining all roots within a 5-cm core into one analysis, we could have obscured some environmental relationships and failed to detect a stronger relationship between C and N and root fungi.
September root-associated fungal communities were significantly correlated with labile Po, and the richness of root-associated fungal communities was also negatively correlated with concentrations of labile Pi, indicating that fungal diversity declines as soil Pi levels increase. Plant P deficiency can increase carbon allocation to roots (27) and has also been found to increase the production of ECM extraradical hyphae (25, 67). In a study of an old-growth stand of Douglas fir, some ECM morphotypes were significantly correlated with soil extractable P (32). Soil P is often not considered to be limiting in forest systems or an important factor affecting ECM communities (60); however, forests that experience low soil pH (pH of <5.5) and high exchangeable aluminum concentrations can experience a reduced availability of P (66), and this could affect root and ECM fungal communities. Fungi that were positively correlated with Po in late summer included the genera Russula, Cenococcum, and Piloderma, whereas the genera Amanita, Entoloma, and Boletus were negatively correlated with Po. One of the most abundant genera in this study, Tomentella, did not appear to be positively or negatively correlated with Po. Although labile Pi was rather high in our study and labile Po is available to plants only after mineralization to Pi (6), different ECM species may have different abilities to capture Pi (15, 21) and many ECM fungi also vary in their abilities to produce extracellular enzymes that liberate nutrients, such as N and P, from organic matter and detritus (7, 8, 16, 60). Differences in the abilities to produce such enzymes can explain the differential ability of ECM fungi to mineralize Po (4, 52) and the shifts in communities observed along N deposition gradients toward taxa adapted for acidic, P-limited conditions (47). Changes in ECM communities within our forest may reflect differences in the functional abilities of ECM species to acquire P from the different fractions present in soil.
We analyzed fungal communities and their relationship to environmental conditions for both sampling events together and also performed these analyses separately by sampling time (June or September). Since the life span of ECM roots can be as short as a few months (22, 55) and chemical conditions in soil can change over the same time frame, it seemed most appropriate to analyze the relationship between soil chemistry and ECM communities for the June and September samplings separately. Analyzing all sampling events together may be more effective for questions that address the effects of plants or major seasonal changes (e.g., winter-to-summer changes) on ECM communities. Whether sampling events are analyzed together or separately will depend upon the questions being investigated.
Fungal community structure and coexistence.
Root-associated fungal communities in our study were significantly different between the June and September samplings and were dominated by the ECM genera Russula and Tomentella. The families Russulaceae and Thelephoraceae commonly dominate roots in many ECM studies (34). Previous studies have observed seasonal changes in ECM communities (3, 8), including changes over periods as short as a month (17). The functional life span of ECM root tips is estimated to be few months in length (22, 55), which suggests that ECM communities can also change over the course of one growing season. Although some ECM fungi were present in an oak forest throughout the year, others were detected only during certain portions of the year (e.g., winter, in the case of Clavulina) (17). A similar pattern was observed in a beech forest, with Clavulina cristata, Laccaria amethystina, and a Russula species more abundant and active in winter than in summer (8). Our results confirm these previous studies suggesting changes in ECM fungal communities between early and late summer. Seasonal changes in fungal communities may be driven, in part, by differences in fungal ecology, environmental tolerance, or resource availability (8, 17, 35). We also observed a large number of ericoid-like fungi on our root samples, as well as dominance of the communities in terms of frequency and abundance by Meliniomyces, a genus that forms ericoid mycorrhizae. This may not be surprising, as an increasing number of studies have found ericoid mycorrhizal fungi colonizing the roots of tree species in the family Pinaceae and other nonericoid plant species (13, 33). Whether tree roots act as alternative or primary hosts for these fungi in the absence of ericaceous plants is uncertain.
We found some evidence that root-associated fungi may be capable of coexistence at fine spatial scales. Several previous studies have observed high species richness and diversity of ECM in small soil cores, suggesting that even at a very local scale (<10 cm), root systems may contain several different ECM species existing in close juxtaposition (11, 24, 64). The factors that promote this coexistence are uncertain, but differences in resource utilization between fungi, as evidenced by differences in hyphal morphology, may be one answer. Some fungi were strongly correlated with one another in our ordination, with some indication that Russula and Piloderma, and to a lesser extent Russula and Cenococcum, are more likely than other fungi to cooccur in soil cores. Tomentella is frequently described as a multistage, highly competitive ECM genus in mature forests (61, 65), and it was negatively correlated with all major groups in our study, with significant negative correlations between it and Russula, Amanita, and Meliniomyces. These data support the perception that the genus Tomentella is capable of competitive exclusion of other fungi. It should be noted, however, that by conducting our analysis at the genus level, we could mask differences among species, since not all species within a genus may share environmental preferences. A larger sampling effort, and analysis of individual species, would allow the environmental preferences of different species to be analyzed more rigorously.
Supplementary Material
Acknowledgments
This work was supported by funding from the Holden Arboretum Trust, The Reinberger Foundation, The Kelvin and Eleanor Smith Foundation, and the Corning Institute for Education and Research.
Footnotes
Published ahead of print on 23 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agerer, R. 2001. Exploration types of ectomycorrhizae: a proposal to classify ectomycorrhizal mycelial systems according to their patterns of differentiation and putative ecological importance. Mycorrhiza 11:107-114. [Google Scholar]
- 2.Agerer, R., A. F. S. Taylor, and R. Treu. 1998. Effect of acid irrigation and liming on the production of fruit bodies by ecto-mycorrhizal fungi. Plant Soil 199:83-89. [Google Scholar]
- 3.Avis, P. G., G. M. Mueller, and J. Lussenhop. 2008. Ectomycorrhizal fungal communities in two North American oak forests respond to nitrogen addition. New Phytol. 179:472-483. [DOI] [PubMed] [Google Scholar]
- 4.Bending, G. D., and D. J. Read. 1995. The structure and function of the vegetative mycelium of ectomycorrhizal plants. V. Foraging behaviour and translocation of nutrients from exploited organic matter. New Phytol. 130:401-409. [Google Scholar]
- 5.Bidartondo, M. I., and T. D. Bruns. 2001. Extreme specificity in epiparasitic Monotropoideae: widespread phylogenetic and geographic structure. Mol. Ecol. 10:2285-2295. [DOI] [PubMed] [Google Scholar]
- 6.Bowman, R. A., and C. V. Cole. 1978. An exploratory method for fractionations of organic phosphorus from grassland soils. Soil Sci. 125:95-101. [Google Scholar]
- 7.Buée, M., P. E. Courty, D. Mignot, and J. Garbaye. 2007. Soil niche effect on species diversity and catabolic activities in an ectomycorrhizal fungal community. Soil Biol. Biochem. 39:1947-1955. [Google Scholar]
- 8.Buée, M., D. Vairelles, and J. Garbaye. 2005. Year-round monitoring of diversity and potential metabolic activity of the ectomycorrhizal community in a beech (Fagus silvatica) forest subjected to two thinning regimes. Mycorrhiza 15:235-245. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D. J., K. J. Martin, P. T. Rygiewicz, and M. A. Topa. 2005. Ectomycorrhizal fungi identification in single and pooled root samples: terminal restriction fragment length polymorphism (TRFLP) and morphotyping compared. Soil Biol. Biochem. 37:1683-1694. [Google Scholar]
- 10.Burke, D. J., A. M. Kretzer, P. T. Rygiewicz, and M. A. Topa. 2006. Soil bacterial diversity in a loblolly pine plantation: influence of ectomycorrhizas and fertilization. FEMS Microbiol. Ecol. 57:409-419. [DOI] [PubMed] [Google Scholar]
- 11.Burke, D. J., K. J. Martin, P. T. Rygiewicz, and M. A. Topa. 2006. Relative abundance of ectomycorrhizas in a managed loblolly pine (Pinus taeda L.) genetics plantation as determined through terminal restriction fragment length polymorphism (TRFLP) profiles. Can. J. Bot. 84:924-932. [Google Scholar]
- 12.Burke, D. J., S. M. Dunham, and A. M. Kretzer. 2008. Molecular analysis of bacterial communities associated with the roots of Douglas fir (Pseudotsuga menziesii) colonized by different ectomycorrhizal fungi. FEMS Microbiol. Ecol. 65:299-309. [DOI] [PubMed] [Google Scholar]
- 13.Chambers, S. M., N. J. A. Curlevski, and J. W. G. Cairney. 2008. Ericoid mycorrhizal fungi are common root inhabitants of non-Ericaceae plants in a south-eastern Australian sclerophyll forest. FEMS Microbiol. Ecol. 65:263-270. [DOI] [PubMed] [Google Scholar]
- 14.Coleman, M. D., C. S. Bledsoe, and W. Lopushinsky. 1989. Pure culture response of ectomycorrhizal fungi to imposed water stress. Can. J. Bot. 67:29-39. [Google Scholar]
- 15.Colpaert, J. V., K. K. van Tichelen, J. A. van Assche, and A. van Laere. 1999. Short-term phosphorus uptake rates in mycorrhizal and non-mycorrhizal roots of intact Pinus sylvestris seedlings. New Phytol. 143:589-597. [DOI] [PubMed] [Google Scholar]
- 16.Courty, P. E., N. Bréda, and J. Garbaye. 2007. Relation between oak tree phenology and the secretion of organic matter degrading enzymes by Lactarius quietus ectomycorrhizas before and during bud break. Soil Biol. Biochem. 39:1655-1663. [Google Scholar]
- 17.Courty, P. E., A. Franc, J.-C. Pierrat, and J. Garbaye. 2008. Temporal changes in the ectomycorrhizal community in two soil horizons of a temperate oak forest. Appl. Environ. Microbiol. 74:5792-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullings, K. W., M. H. New, S. Makhija, and V. T. Parker. 2003. Effects of litter addition on ectomycorrhizal associates of a lodgepole pine (Pinus contorta) stand in Yellowstone National Park. Appl. Environ. Microbiol. 69:3772-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danielson, R. M., and S. Visser. 1989. Effects of forest soil acidification on ectomycorrhizal and vesicular-arbuscular mycorrhizal development. New Phytol. 112:41-47. [Google Scholar]
- 20.Dickie, I. A., X. Bing, and R. T. Koide. 2002. Vertical niche differentiation of ectomycorrhizal hyphae in soil as shown by TRFLP analysis. New Phytol. 156:527-535. [DOI] [PubMed] [Google Scholar]
- 21.Dighton, J., P. A. Mason, and J. M. Poskitt. 1990. Field use of 32P to measure phosphate uptake by birch mycorrhizas. New Phytol. 116:635-661. [Google Scholar]
- 22.Downes, G. M., I. J. Alexander, and J. W. G. Cairney. 1992. A study of spruce (Picea stichensis (Bong) Carr.) ectomycorrhizas. I. Morphological and cellular changes in mycorrhizas formed by Tylospora fibrillose (Burt) Donk and Paxillus involutus (Batsch ex Fr.) Fr. New Phytol. 122:141-152. [DOI] [PubMed] [Google Scholar]
- 23.Eaton, J. S., G. E. Likens, and F. H. Bormann. 1973. Throughfall and stemflow chemistry in a northern hardwood forest. J. Ecol. 61:495-508. [Google Scholar]
- 24.Edwards, I. P., J. L. Cripliver, A. R. Gillespie, and R. F. Turco. 2004. Long-term optimal fertilization changes the community structure of basidiomycetes associated with loblolly pine on a nitrogen poor soil. New Phytol. 162:755-770. [DOI] [PubMed] [Google Scholar]
- 25.Ekblad, A., H. Wallander, R. Carlsson, and D. K. Huss. 1995. Fungal biomass in roots and extramatrical mycelium in relation to macronutrients and plant biomass of ectomycorrhizal Pinus sylvestris and Alnus incana. New Phytol. 131:443-451. [DOI] [PubMed] [Google Scholar]
- 26.Environmental Protection Agency (EPA). 1971. Methods of chemical analysis of water and wastes. EPA, Cincinnati, OH.
- 27.Ericsson, T. 1995. Growth and shoot:root ratio of seedlings in relation to nutrient availability. Plant Soil 169:205-214. [Google Scholar]
- 28.Erland, S., and A. F. S. Taylor. 2002. Diversity of ecto-mycorrhizal fungal communities in relation to the abiotic environment, pp 163-200. In M. G. A. van der Heijden and I. Sanders (ed.). Mycorrhizal ecology. Ecological studies 157. Springer, Heidelberg, Germany.
- 29.Gehring, C. A., T. C. Theimer, T. G. Whitham, and P. Keim. 1998. Ectomycorrhizal fungal community structure of pinyon pines growing in two environmental extremes. Ecology 79:1562-1572. [Google Scholar]
- 30.Gibson, F., and J. W. Deacon. 1988. Experimental study of establishment of ectomycorrhizas in different regions of birch root systems. Trans. Br. Mycol. Soc. 91:239-251. [Google Scholar]
- 31.Gilliam, F. S. 2007. The ecological significance of the herbaceous layer in temperate forest ecosystems. Bioscience 57:845-858. [Google Scholar]
- 32.Goodman, D. M., and J. A. Trofymow. 1998. Distribution of ectomycorrhizas in micro-habitats in mature and old-growth stands of Douglas-fir on southeastern Vancouver Island. Soil Biol. Biochem. 30:2127-2138. [Google Scholar]
- 33.Hambleton, S., and L. Sigler. 2005. Meliniomyces, a new anamorph genus for root-associated fungi with phylogenetic affinities to Rhizoscyphus ericae (☰ Hymenoscyphus ericae), Leotiomycetes. Studies Mycol. 53:1-27. [Google Scholar]
- 34.Horton, T. R., and T. D. Bruns. 2001. The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol. Ecol. 10:1855-1871. [DOI] [PubMed] [Google Scholar]
- 35.Izzo, A., J. A. Agbowo, and T. D. Bruns. 2005. Detection of plot-level changes in ectomycorrhizal communities across years in an old-growth mixed-conifer forest. New Phytol. 166:619-630. [DOI] [PubMed] [Google Scholar]
- 36.Jany, J.-L., F. Martin, and J. Garbaye. 2003. Respiration activity of ectomycorrhizas from Cenococcum geophilum and Lactarius sp. in relation to soil water potential in five beech forests. Plant Soil 255:487-494. [Google Scholar]
- 37.Jarrell, W. M., D. E. Armstrong, D. F. Grigal, E. F. Kelley, H. C. Monger, and D. A. Wedin. 1999. Soil water and temperature status, pp 55-73. In G. Robertson, D. C. Coleman, C. S. Bledsoe, and P. Sollins (ed.), Standard soil methods for long-term ecological research. Oxford University Press, New York, NY.
- 38.Kretzer, A. M., Y. Li, T. Szaro, and T. D. Bruns. 1996. Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: phylogenetic and taxonomic implications. Mycologia 88:776-785. [Google Scholar]
- 39.Kretzer, A. M., and T. D. Bruns. 1999. Use of atp6 in fungal phylogenetics: an example from the Boletales. Mol. Phylogenet. Evol. 13:483-492. [DOI] [PubMed] [Google Scholar]
- 40.Kuo, S. 1996. Phosphorus, p. 869-919. In D. L. Sparks (ed.), Methods of soils analysis, part 3, chemical methods. ASA and SSSA, Madison, WI.
- 41.Landeweert, R., C. Veenman, T. W. Kuyper, H. Fritze, K. Wernars, and E. Smit. 2003. Quantification of ectomycorrhizal mycelium in soil by real-time PCR compared to conventional quantification techniques. FEMS Microbiol. Ecol. 45:283-292. [DOI] [PubMed] [Google Scholar]
- 42.Last, F. T., P. A. Mason, J. Wilson, and J. W. Deacon. 1983. Fine roots and sheathing mycorrhizas: their formation, function and dynamics. Plant Soil 71:9-21. [Google Scholar]
- 43.Leake, J. R. 1994. The biology of myco-heterotrophic (“saprophytic”) plants. New Phytol. 127:171-216. [DOI] [PubMed] [Google Scholar]
- 44.Leake, J. R. 2001. Is diversity of ectomycorrhizal fungi important for ecosystem function? New Phytol. 152:1-3. [DOI] [PubMed] [Google Scholar]
- 45.Lilleskov, E. A., and T. D. Bruns. 2003. Root colonization dynamics of two ectomycorrhizal fungi of contrasting life history strategies are mediated by addition of organic nutrient patches. New Phytol. 159:141-151. [DOI] [PubMed] [Google Scholar]
- 46.Lilleskov, E. A., T. D. Bruns, T. R. Horton, D. L. Taylor, and P. Grogan. 2004. Detection of forest stand-level spatial structure in ectomycorrhizal fungal communities. FEMS Microbiol. Ecol. 49:319-332. [DOI] [PubMed] [Google Scholar]
- 47.Lilleskov, E. A., T. J. Fahey, T. R. Horton, and G. M. Lovett. 2002. Belowground ectomycorrhizal fungal community change over a nitrogen deposition gradient in Alaska. Ecology 83:104-115. [Google Scholar]
- 48.Martin, K. J., and P. T. Rygiewicz. 2005. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mason, P. A., F. T. Last, J. Pelham, and K. Ingleby. 1982. Ecology of some fungi associated with an ageing stand of birches (Betula pendula and B. pubescens). For. Ecol. Manage. 4:19-39. [Google Scholar]
- 50.Muller, R. N., and F. H. Bormann. 1976. Role of Erythronium americanum Ker. in energy flow and nutrient dynamics of a northern hardwood forest ecosystem. Science 193:1126-1128. [DOI] [PubMed] [Google Scholar]
- 51.Olsen, S. R., C. V. Cole, F. S. Watanabe, and L. A. Dean. 1954. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA circular no. 939. U.S. Government Print Office, Washington, DC.
- 52.Pacheco, S., J. Cambraia, and M. C. M. Kasuya. 1991. Effect of different levels of phosphorus on acid phosphatase activity and mineral composition of some ectomycorrhizal fungi. Rev. Microbiol. 22:345-348. [Google Scholar]
- 53.Peter, M., F. Ayer, and D. Egli. 2001. Nitrogen addition in a Norway spruce stand altered macromycete sporocarp production and below-ground ectomycorrhizal species composition. New Phytol. 149:311-325. [DOI] [PubMed] [Google Scholar]
- 54.Ryan, M., and R. Waring. 1992. Maintenance respiration and stand development in a subalpine lodgepole pine forest. Ecology 73:2100-2108. [Google Scholar]
- 55.Rygiewicz, P. T., M. G. Johnson, L. M. Ganio, D. T. Tingey, and M. J. Storm. 1997. Lifetime and temporal occurrence of ectomycorrhizae on ponderosa pine (Pinus ponderosa Laws.) seedlings grown under varied atmospheric CO2 and nitrogen levels. Plant Soil 189:275-287. [Google Scholar]
- 56.Samson, J., and J. A. Fortin. 1986. Ectomycorrhizal fungi of Larix laricina and the interspecific and intraspecific variation in response to temperature. Can. J. Bot. 64:3020-3028. [Google Scholar]
- 57.Sciarini, S. M., and F. Michel. 2002. tRFLP fragment sorter 2.5b3. http://www.oardc.ohio-state.edu/trflpfragsort/index.php.
- 58.Selosse, M.-A., and M. Roy. 2009. Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci. 14:64-70. [DOI] [PubMed] [Google Scholar]
- 59.Shi, L., M. Guttenberger, I. Kottke, and R. Hampp. 2002. The effect of drought on mycorrhizas of beech (Fagus sylvatica L.): changes in community structure, and the content of carbohydrates and nitrogen storage bodies of the fungi. Mycorrhiza 12:303-311. [DOI] [PubMed] [Google Scholar]
- 60.Smith, S. E., and D. J. Read. 2008. Mycorrhizal symbiosis, p. 787. Academic Press, Inc., London, United Kingdom.
- 61.Taylor, D. L., and T. D. Bruns. 1999. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: minimal overlap between the mature forest and resistant propagule communities. Mol. Ecol. 8:1837-1850. [DOI] [PubMed] [Google Scholar]
- 62.Tedersoo, L., T. Suvi, T. Jairus, and U. Kõljalg. 2008. Forest microsite effects on community composition of ectomycorrhizal fungi on seedlings of Picea abies and Betula pendula. Environ. Microbiol. 10:1189-1201. [DOI] [PubMed] [Google Scholar]
- 63.Trappe, J. M. 1977. Selection of fungi for ectomycorrhizal inoculation in nurseries. Annu. Rev. Phytopathol. 15:203-222. [Google Scholar]
- 64.Valentine, L. L., T. L. Fiedler, A. N. Hart, C. A. Petersen, H. K. Berninghausen, and D. Southworth. 2004. Diversity of ectomycorrhizas associated with Quercus garryana in southern Oregon. Can. J. Bot. 82:123-135. [Google Scholar]
- 65.Visser, S. 1995. Ectomycorrhizal fungal succession in jack pine stands following wildfire. New Phytol. 129:389-401. [Google Scholar]
- 66.Walker, L., and J. Syers. 1976. The fate of phosphorus during pedogenesis. Geoderma 15:1-19. [Google Scholar]
- 67.Wallander, H., and J.-E. Nylund. 1992. Effects of excess nitrogen and phosphorous starvation on the extraradical mycelium of Pinus sylvestris L. ectomycorrhiza. New Phytol. 120:495-503. [Google Scholar]
- 68.Wolfe, B. E., V. L. Rodgers, K. A. Stinson, and A. Pringle. 2008. The invasive plant Alliaria petiolata (garlic mustard) inhibits ectomycorrhizal fungi in its introduced range. J. Ecol. 96:777-783. [Google Scholar]
- 69.Yang, S., and D. H. Pfister. 2006. Monotropa uniflora plants of eastern Massachusetts form mycorrhizae with a diversity of russulacean fungi. Mycologia 98:535-540. [DOI] [PubMed] [Google Scholar]
- 70.Young, B., H. Massicotte, L. Tackaberry, Q. Baldwin, and K. Egger. 2002. Monotropa uniflora: morphological and molecular assessment of mycorrhizae retrieved from sites in the sub-boreal spruce biogeoclimatic zone in central British Columbia. Mycorrhiza 12:75-82. [DOI] [PubMed] [Google Scholar]
- 71.Zar, J. H. 1998. Biostatistical analysis, p. 929. Prentice Hall, Upper Saddle River, NJ.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.