Abstract
Since the development of green fluorescent protein (GFP) and other fluorescent proteins (FPs) with distinct colors, genetically-encoded probes and biosensors have been widely applied to visualize the molecular localization and activities in live cells. In particular, biosensors based on fluorescence resonance energy transfer (FRET) have significantly advanced our understanding of the dynamic molecular hierarchy at subcellular levels. These biosensors have also been extensively applied in recent years to study how cells perceive the mechanical environment and transmit it into intracellular molecular signals (i.e. mechanotransduction). In this review, we will first provide a brief introduction of the recent development of FPs. Different FRET biosensors based on FPs will then be described. The last part of the review will be dedicated to the introduction of examples applying FRET biosensors to visualize mechanotransduction in live cells. In summary, the integration of FRET technology and the different cutting-edge mechanical stimulation systems can provide powerful tools to allow the elucidation of the mechanisms regulating mechanobiology at cellular and molecular levels in normal and pathophysiological conditions.
I. Introduction
The development of genetic technology has allowed the sequencing of human genome and those of other species. However, genome sequences lack the spatial and temporal information for the target molecules. It becomes clear that imaging technologies should play a central role in this aspect to obtain the spatiotemporal characteristics of different molecules, particularly in cells and organisms. The development of molecular probes has become increasingly powerful in molecular imaging, along with the developmental progress in microscopic technologies. The development of fluorescent-antibodies in 1940s provided the first breakthrough in visualizing the cellular proteins. The imaging of deoxyribonucleic acids (DNAs) and ribonucleic acids (RNAs) also became available in 1980s with the development of in situ hybridization techniques. However, these techniques require the cells to be killed, which may cause the alteration of the information. During the 1980s, cell-permeable fluorescent dyes were developed to monitor the intracellular signals in live cells including pH and Ca2+.81 However, the kinds of molecules which can be monitored are limited. Furthermore, it is very difficult to control the localization of these small molecule dyes and hence to monitor the localized molecular signals at subcellular compartments continuously in live cells. Hence, there was a great need to develop a general and convenient approach which can allow the labeling and monitoring of different molecules in live cells. The cloning and expression of green fluorescent protein (GFP) timely served this need.
While GFP has allowed a variety of revolutionary discoveries, the limitation of this GFP-tagging approach is that only the position of the molecules can be traced. To monitor the molecular activities or interactions in live cells, fluorescence resonance energy transfer (FRET) becomes particularly appealing. In this article, we will focus on the introduction of FRET technologies, in particular of those based on GFP and its derivatives. We will then discuss the application of these FRET biosensors in visualizing signaling cascades in mechanobiology, i.e. how cells perceive mechanical cues and transmit them into biochemical/molecular signals.
II. Fluorescent proteins (FPs) and fluorescence resonance energy transfer (FRET)
FRET technology, in particular in combination with FPs, has been increasingly applied to detect intracellular molecular interactions and activities in live cells with high spatiotemporal resolutions. In this section, the development of different kinds of FPs will be introduced. The FRET principle and FP-based FRET biosensors will then be described.
II.1 Green fluorescent protein (GFP) and FPs with different colors
GFP is an accompanying protein for a bioluminescent protein aequorin in Aequorea jellyfish. When jellyfish are provoked, calcium can be released to bind aequorin and cause blue fluorescence emission. This blue emission can stimulate GFP to result in bright green fluorescence. Shimomura first purified GFP in 1960s and noticed that GFP appears slightly greenish under sunlight while it emits bright green light under UV light.77 About 30 years later, Prasher successfully sequenced the gene encoding GFP when he was in the Woods Hole Marine Biological Laboratory.68 The idea is to fuse the gene of GFP with that of a target molecule so that the resultant recombinant protein can emit fluorescence while mimicking the traffic of the target molecule. As such, one can, in principle, visualize and track the dynamic localization of any interesting molecules in different kinds of live cells (Fig. 1). Indeed, Chalfie and others demonstrated that the expression of GFP gene alone is sufficient to produce fluorescence in organisms other than jellyfish,8,27 verifying that GFP can mature and generate fluorescence independent of aequorin or other co-factors and hence can serve as a fluorescent marker for various molecules.
Fig. 1.

Upper panel: a schematic drawing of a recombinant gene containing cDNA of a target molecule fused to that of GFP. Lower panel: the protein production of the recombinant gene in the upper panel allows the observation of the recombinant protein with excitation and emission wavelengths of GFP at 488 and 510 nm, respectively.
GFP forms a ‘can’ structure consisting of 11 β-barrel strands. Therefore, the chromophore of GFP is very well protected and hence stable at physiological environment of living organisms. The wild-type GFP, however, has two different populations: the major one is excitable at 395 nm with an emission at 508 nm and the minor one excitable at 475 nm with an emission at 503 nm.22 Roger Tsien first revealed that the three amino acids, Ser65-Tyr66-Gly67, will undergo conformational changes, cyclization and dehydration between Ser65 and Gly67, and oxidation of Tyr66 to form a mature chromophore.82 About 85% of the chromophores are in neutral form whereas 15% of them are anions.82 The neutral chromophores are responsible for the major population with 395 nm excitation wavelength whereas the anionic ones are excitable at 475 nm.
A mutation of Ser65 to the bulky Thr has prompted a neighboring Glu222 to remain neutral such that the polar groups surrounding the chromophore can promote its anionic form. This mutation has led to the development of enhanced GFP (EGFP) and Emerald, which mature efficiently at 37 °C and are the most popular GFP versions used in cell biology.11,62,82 Several FPs with different colors were further developed based on GFP. By introducing a Y66H mutation in the chromophore, a blue shifted fluorescent protein (FP), BFP, can be generated together with other mutations beneficial for quantum yield and photostability.22,23,62 BFP is typically inferior in its brightness and photostability. Recent development of BFP derivatives, Azurite and EBFP2, may however offer significantly improved characteristics in these aspects.3,42 A Y66W mutation in the chromophore, on the other hand, can produce a cyan FP (CFP) and its enhanced version ECFP.22,23 A variety of CFP derivatives have since been developed in the hope of improving the brightness. Among them, mTFP1 and Cerulean are of particular interest to cell biologists.2,69 Although both CFP derivatives are significantly improved in their general fluorescence characteristics comparing to ECFP, Cerulean appears to be less photostable.75 By engineering an aromatic ring proximal to the anionic chromophore, the mutations of T203Y and S65T led to the development of yellow FP (YFP).59,82 Early versions of YFP suffered from low photostability.75 Recently improved versions, namely Citrine and Venus, provide excellent photostability and brightness.21,52 A new YFP developed via a quantitative evolutionary strategy, YPet, offers outstanding brightness and can serve as a superior acceptor for a CFP derivative during fluorescence resonance energy transfer (FRET) experiments.57,61 The last example of FP that we want to introduce is the T209I mutation which can prevent the ionization of the chromophore. This mutation of T209I on the wild-type GFP resulted in Sapphire and Turbo Sappire (T-Sapphire).13,22 Both FPs can be excited by UV light and undergo a large Stokes shift to emit green fluorescence.82,98
However, mutagenesis efforts on GFP and its derivatives failed to generate FPs with fluorescence emission ranging from orange to far red. A bright FP with red emission was discovered from Discosoma, namely DsRed.41 Despite its outstanding brightness and photostability, DsRed unfortunately tends to form tetramers.5 This tetramerization tendency of DsRed has significantly limited the general application of DsRed for live cell imaging. After numerous attempts, a real breakthrough came when Campbell and Tsien developed the monomeric RFP (mRFP1) with 33 mutations on the parental DsRed.6 Tsien’s laboratory has further applied directed evolution and iterative somatic hyper-mutation on mRFP1 to develop other monomeric FPs with their emission extending from deep yellow to far red, including mHoneydew, mBanana, mOrange, mTangerine, mStrawberry, mCherry, mRaspberry, mPlum and mGrape.73,83,88 Several other FPs with orange or red color were also developed independently from other species, including monomeric mKO,31 monomeric TagRFP,43 and monomeric mKate.76 Together with the FPs derived from GFP, these FPs with distinct colors allow the simultaneous visualization of multiple signaling molecules in a single live cell (Fig. 2). Most recently, an infrared FP has also been developed in Tsien’s lab from a bacteriophytochrome of Deinococcus radiodurans.78 This is a revolutionary step in FP development since the wavelength of the infrared FP can allow the deep penetration of tissues, thus having paved the way for the whole-animal optical imaging. This infrared FP can also serve as a FRET acceptor for red FPs to allow more choices in FRET pairs.
Fig. 2.

The top panel shows the purified FPs under white light and the bottom panel demonstrates the fluorescence of these FPs. Image courtesy of R. Y. Tsien.83
2.2 FP-based FRET biosensors
FRET occurs when two fluorophores are in proximity, with the emission spectrum of the donor overlapping the excitation spectrum of the acceptor.9,10 The efficiency of FRET is mainly dependent on three factors: (1) the overlap area between the emission spectrum of the donor and the excitation spectrum of the acceptor (Fig. 3A), with a larger area allowing a more efficient energy transfer; (2) the distance between the donor and acceptor. The FRET efficiency is inversely proportional to the 6th power of the distance between donor and acceptor. Hence, a slight modification of this distance can significantly affect the FRET signals. Typically, sufficient FRET between FPs occurs only when this distance is within 10 nm (Fig. 3B); and (3) the relative orientation between the donor and acceptor dipoles (Fig. 3B). Indeed, FRET is particularly sensitive to the relative orientation between donor and acceptor since a dramatic effect on FRET can be observed by changing the orientation while maintaining the distance of chromophores.20,54 Because FRET can provide such a high resolution in detecting distance/orientation, it has been widely applied to design biosensors for the measurement of signals with high precision.67
Fig. 3.

(A) The excitation and emission spectra of a typical FRET pair: CFP as the donor and YFP as the acceptor. The broken lines represent the excitation spectra and the solid lines represent the emission spectra of CFP and YFP. The spectra curves of CFP and YFP are color-coded with cyan and yellow, respectively. The shaded red area represents the overlap between the CFP emission and the YFP excitation. (B) The cartoon shows that the FRET efficiency between a typical FRET pair, ECFP as the donor and EYFP as the acceptor, is mainly dependent on the distance and the relative orientation between the donor and acceptor.
Different kinds of biosensors have been developed utilizing FRET technology. For example, nucleotide-based biosensors have been applied to detect GTPases and ATPases.24,91 Hybridized biosensors consisting of a polypeptide and a protein domain recognizing an active target molecule have also been successfully designed and applied for the detection of the small GTPases RhoA and Cdc42.25,56,64 However, for live cell imaging, genetically-encoded FRET biosensors are particularly appealing because these biosensors can be conveniently introduced into cells and targeted at subcellular compartments to continuously monitor the local molecular signals. EBFP and EGFP were originally chosen as the FRET pair for biosensor development.40,70 While EGFP has decent fluorescent properties, EBFP is in general dim and less photostable. At the current stage, ECFP and EYFP, including the variants of EYFP such as Venus and Citrine, are the most popular FRET pair.21,28,47,52,93 ECFP and YPet has also been shown to serve as a high-efficiency FRET pair for a variety of biosensors.61 However, the brightness of ECFP is still a limiting factor for cell imaging. Recently, new variants for EBFP and ECFP have been developed, including Azurite,42 EBFP2,63 Cerulean,69 and mTFP1.2 All these newly developed FP variants displayed significantly enhanced fluorescent properties, such as brightness and photostability. Future experiments are needed to provide more substantial evidence on how these new FPs can improve the FRET biosensors, particularly in terms of their dynamic ranges.
Typically, a pair of FRET FPs serving as a donor and an acceptor can be fused with two different target molecules to monitor the interaction of these target molecules. The interaction and separation of the target molecules can alter the distance between the donor and acceptor, thus affecting the FRET efficiency. As such, the FRET efficiency between the donor and acceptor can serve as an indicator of the interaction of the two target molecules. Indeed, this approach has been applied to monitor the activities of cAMP94,95 and G-proteins.29 However, there are two main limitations for this approach: (1) the endogenous counterparts of the target molecules may interfere with the interactions of FP-fused target molecules; (2) The acceptor/donor ratio at different subcellular locations may not be the same, which prevents a simple calculation of the acceptor/donor ratio to monitor the FRET efficiency. Usually more complicated analysis approaches are needed to assess the FRET signals.99
It becomes increasingly popular that single-molecule FRET biosensors are developed to monitor intracellular signals. A typical single-molecule FRET biosensor consists of a donor and an acceptor FP, concatenated in between or flanking two intramolecular domains capable of interacting with each other. For example, a FRET-based biosensor capable of detecting Src kinase activation has been developed consisting of an N-terminal ECFP, a SH2 domain derived from Src kinase, a flexible linker, a substrate peptide derived from p130cas and specifically sensitive to Src phosphorylation, and a C-terminal Citrine (EYFP).90 When the Src kinase is in its resting state, the ECFP and Citrine (EYFP) are positioned in proximity as a consequence of; (1) the tendency of the wild type ECFP and Citrine to form anti-parallel dimers; (2) the flexible linker; and (3) the juxtaposition of N- and C-terminals of the SH2 domain. Therefore, a strong FRET can occur and the excitation of ECYP at 433 nm leads to the emission of EYFP at 527 nm. When Src kinase is activated, it phosphorylates the designed substrate peptide, which displays a high affinity and binds to the bottom pocket of the SH2 domain. This action will lead to the separation of the Citrine from the ECFP and decrease the FRET efficiency between these two fluorescent proteins. The excitation of ECFP at 433 nm then results in the emission from ECFP at 476 nm as shown in Fig. 4A. Hence, the emission ratio of ECFP/EYFP with the excitation of ECFP should serve as a reliable indicator of the status of Src activation. However, ECFP and Citrine tend to form weak anti-parallel dimers,92 which may result in intermolecular FRET between different copies of biosensors. To eliminate this unintended FRET, A206K mutations have been introduced into ECFP and Citrine to generate monomeric mECFP and mCitrine.96 In HeLa cells, this monomeric version of the biosensor under-went a significant FRET change upon Src activation triggered by EGF, which was reversible following EGF washout (Fig. 4B).90 As such, Src activity can be continuously monitored in live cells. With the same strategy, a wide range of single-molecule biosensors based on FPs have been developed to successfully monitor different kinds of intracellular signals, e.g. Ca2+,46–48,53–55,70,80 proteases,23,38,39,45 cAMP,12 phosphor-lipids,4 membrane receptor integrins,15,16,33 Guanine exchange factor Wasp,37,72 small GTPases Ras and Rap1,49 RhoA,65,93 Rac1,28,35 Cdc42,28,56 and tyrosine/serine/threonine kinases.17,36,71,79,86,90,100,101
Fig. 4.

(A) A cartoon scheme depicting the activation mechanism of the Src biosensor. When Src kinase is inactive, ECFP and Citrine are positioned proximal to each other and have strong FRET. The excitation of the biosensor at 433 nm results in the emission from Citrine at 527 nm. When Src kinase is activated to phosphorylate the substrate peptide in the biosensor, the biosensor will undergo a conformational change and separate Citrine from ECFP, thus resulting in the decrease of FRET. The excitation of the biosensor at 433 nm then leads to the emission from ECFP at 476 nm. Hence, the FRET efficiency or ECFP/Citrine emission ratio of the biosensor represents the activation status of Src kinase. (B) HeLa cells transfected with the monomerized Src biosensor were stimulated with EGF (50 ng ml−1), washed with serum-free medium (washout). “−EGF” and “+EGF” represent the images before and after EGF stimulation. Color images represent the ECFP/Citrine emission ratio images of the monomeric Src biosensor in HeLa cells in response to EGF or washout. The color scale bar on the left corresponds to the level of ECFP/Citrine emission ratio, with cold colors indicating low Src activity and hot colors indicating high Src activity. These images are adapted from Wang et al., 2005.90
At the current stage, most of the FRET imaging only allows the visualization of one active molecular event in a single live cell. To visualize multiple molecular events in a simultaneous fashion, biosensors should be either physically separated or with distinct colors. Physically separated biosensors with similar colors have been successfully applied to detect a fast cAMP accumulation in the nucleus but a slow PKA activation upon the activation of β2-adrenergic receptors.12 A similar approach has also allowed the quantification of a delay between the PIP3 accumulation at the plasma membrane and the Akt activation in the nucleus.4 Recently, FPs with different excitation and emission wavelengths have been developed through directed evolutionary strategies.73,74 These new FPs can allow the simultaneous visualization of multiple signaling events in the same live cells without physical limitation. Indeed, integrating two pairs of FPs for FRET biosensors (pair 1: a GFP mutant mAmetrine as the donor and an orange FP tdTomato as the acceptor; pair 2: a cyan FP mTFP1 as the donor and a yellow FP mCitrine as the acceptor), the onset of apoptosis can be visualized by monitoring the caspase activities.1 In another case, by combining a new FRET pair, an orange mKO and a far red mCherry, with the standard cyan and yellow FPs, multiple signaling events, including the activities of protein kinase Iα and protein kinase C, and the calcium-dependent interaction between connexin and the plasma membrane can be simultaneously monitored in live cells.66 These new approaches of integrating multiple FRET biosensors with distinct colors allow the visualization of molecular hierarchy and networks in a single live cell.
III. The application of FRET biosensors in mechanobiology
In the past decade, a large number of studies have been published on the effects of mechanical forces on various cell functions. The key question that remains to be answered is how mechanical forces are transduced into biochemical signals in a living cell. FRET biosensors can provide powerful tools in this aspect by offering the spatiotemporal visualization of signaling maps in live cells. In this section, we will describe the application of FRET biosensors in monitoring the molecular signaling transduction in live cells.
III.1 The detection of mechanotransduction in response to global stimuli
It was conceived that membrane receptors may play crucial roles in sensing the mechanical cues. Therefore, a FRET sensor has been developed to monitor the activation of a G protein-coupled receptor (GPCR), the human B2 bradykinin receptor, by fusing it with ECFP and EYFP. The results revealed that shear stress resulted from a laminar flow can rapidly activate B2 bradykinin in bovine aortic endothelial cells (BAECs), which can be inhibited by B2-selective antagonist.7 It is therefore possible that this membrane B2 bradykinin GPCR may sense the shear stress and trigger downstream signaling pathways. A downstream signaling pathway in relation to NFκB has also been studied similarly utilizing FRET. NFκB is an important molecule involved in endothelial inflammatory responses. A separated pair of ECFP-fused relA and EYFP-fused IκBα has been developed to monitor the interaction of relA and IκBα, an upstream molecule binding to NFκB and controlling the activation of NFκB. The results indicate that shear stress can induce the dissociation of relA and IκBα, as evidenced by the decrease of FRET between ECFP-relA and EYFP-IκBα.18 Other signaling pathways in response to mechanical stimulation examined by FRET include the Rho small GTPase family members. Two independent approaches have been employed to visualize the activity of the Rac1 GTPase in response to shear stress. In one study, the biosensor consists of the EGFP-fused Rac and Alexa568-p21-binding domain of PAK1 (PBD) which is capable of binding to the activated Rac. As such, the FRET between EGFP and Alexa568 can be monitored to measure the Rac activation.35 With this biosensor, active Rac was observed to accumulate at the cell leading edge along the flow direction when BAECs were subjected to a laminar flow.84 In another study, the Rac activity was inhibited in the cellular regions facing the flow, as detected by a Rac FRET biosensor utilizing ECFP and Venus.97 As such, there is a polarity of Rac activity generated in cells subjected to the directional laminar flow. Therefore, both studies suggest a polarized distribution of active Rac under laminar flows. Similarly, a polarized distribution of active Cdc42, another small GTPase, was observed by the FRET signals between a EGFP-Cdc42 and an Alexa568-PBD, which binds to active Cdc42.85 Recently, a calcium FRET biosensor has been developed utilizing the ECFP and YPet pair.61 This biosensor has allowed the visualization of a calcium oscillation in human mesenchymal stem cells (HMSCs).34 Interestingly, this oscillation was significantly inhibited when HMSCs were cultured on soft substrates. Further experiments revealed that RhoA small GTPase is essential, but not sufficient in mediating this mechanical stiffness-regulated calcium oscillation.34 These results suggest that FRET biosensors can be very powerful in visualizing the spatiotemporal activation map of various signaling molecules and elucidating the related molecular hierarchy, particularly at subcellular levels.
III.2 The detection of mechanotransduction in response to local stimuli
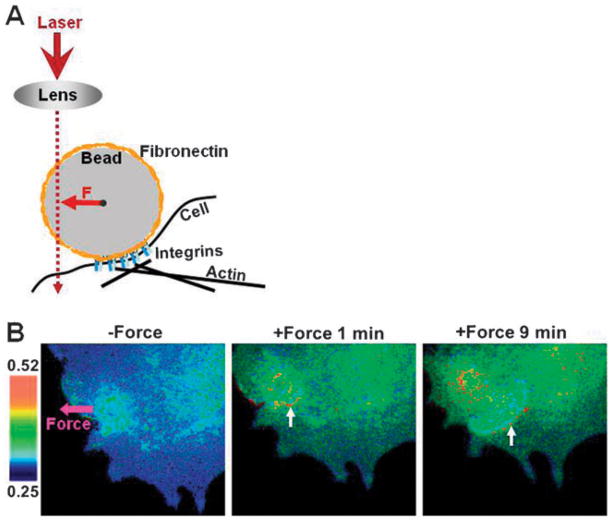
A membrane-tethered Src biosensor has further been developed to reside at the inner surface of the plasma membrane and monitor the membrane Src activity continuously.32 After a retrovirus version of this membrane-targeted Src biosensor was introduced into a human umbilical vein endothelial cell (HUVEC), a fibronectin-coated bead can be guided by a laser-tweezer to deposit on top of this cell surface and be coupled to cytoskeleton via the ligation between fibronectin and integrins32 (Fig. 5A). This laser-tweezer system can further apply traction force on the fibronectin-coated bead to mechanically perturb the cell at the local region. Upon the stimulation, a rapid distal Src activation and a slower activation wave of Src originated at the bead site and propagated in the opposite direction of the mechanical force can be observed90 (Fig. 5B). These results provided the evidence on how biochemical/molecular signals at the plasma membrane can be generated and transmitted in response to mechanical stimulation. However, it is still not clear whether mechanotransduction is unique or similar when compared with soluble factor induced signaling. This is not a trivial issue since at the present time it remains elusive how forces are different from chemical cues in controlling vital cellular functions. Over the last decade, several mechanotransduction models have been proposed.60,87 Of these models, one prevailing model states that mechanotransduction initiates at the cell surface, followed by cascades of signaling in the cytoplasm, similar to the signaling induced by soluble growth factors. Previously published reports are limited in timescale to test these models and to differentiate mechanotransduction from soluble factor-induced signal transduction. Combining the stress propagation mapping approach that we have developed with simultaneous measurements of biochemical activities, we have developed a useful method to map mechanochemical transduction in a living cell.51 We used a FRET-based cytosolic Src biosensor utilizing ECFP and YPet90 and quantified changes of Src activities in a living cell as a local surface stress was applied via integrin receptors using an Arg-Gly-Asp coated magnetic bead.26 The stress induced rapid (<0.3 s) activation of Src at remote cytoplasmic sites (>50 μm), which is at least 40 times faster than that induced by soluble epidermal growth factor (EGF)50 (Fig. 6A). This rapid, long-distant Src activation was dependent on stress magnitudes (Fig. 6B), integrin activation, substrate stiffness, and prestress.50 Nanometer scale cytoskeletal deformation analyses revealed that the strong activation sites of Src by stress co-localized with large deformation sites of microtubules (Fig. 7). Large microtubule deformation sites, in turn, co-localized with sites of large deformation of endosomal membranes50 onto which Src proteins anchor.19,30 Importantly, the size of the concentrated Src activation sites in Fig. 7 is ~0.5–0.8 μm in diameter, which is similar to the size of an endosome in a living cell. Disrupting microtubules with colchicine prevented stress-induced Src activation. Therefore, our results suggest that microtubules are essential structures for imparting deformation to Src-bound endosomes to activate cytoplasmic Src by directly changing their conformation. The Src activation at remote sites is too rapid to be explained by ion channel opening, diffusion, and/or translocation mechanisms. Therefore there appears to be major differences between force-induced Src activation and EGF-induced Src activation. A possible explanation of our data is the focal adhesion dependent, tensed actin bundle-mediated stress wave propagation along the cytoskeleton that can reach anywhere in the cytoplasm within 1 ms.89 More work is needed to examine if these findings can be extended to other cell types and to other molecules in the cytoplasm and in the nucleus.
Fig. 5.
(A) A cartoon scheme depicting the procedure of the laser-tweezer-traction experiment. A laser light is guided by an objective lens to focus o.-centered to exert force on a fibronectin-coated bead, which is adhered on a HUVEC and coupled to the cytoskeleton through integrin-ligation. (B) FRET response of a cell with clear directional wave propagation away from the site of mechanical stimulation introduced by laser-tweezers. The color bar on the left indicates ECFP/Citrine emission ratios, with cold color representing low ratios and hot color representing high ratios. The pink arrow represents the site of force application and the force direction. The white arrows point to the front edge of activated Src wave. This figure is adapted from Wang et al., 2005.90
Fig. 6.

(A) Stress-induced Src activation is rapid and EGF-induced Src activation is slow. Here is the time course of normalized CFP/YFP emission ratio, an index of Src activation in response to mechanical or soluble growth factor EGF stimulation. A local stress was applied to integrins using a 4-μm RGD-coated magnetic bead (17.5 Pa for 3 s step function). Src is activated at ~100 ms after stress but is not activated until ~12–20 s after EGF.50 n = 12 cells for +σ (stress), n = 8 cells for +EGF (40 ng ml−1). Error bars represent SEM. (B) Src activation is stress-magnitude dependent. Note that the stress threshold for Src activation appears to be ~1.8 Pa and the Src activation response is nonlinear. n = 12 cells for stress of 17.5 Pa; 4 for 8.8 Pa; 4 for 3.5 Pa; 4 for 1.8 Pa; 3 for 0.7 Pa. Error bars represent SEM. This figure is reproduced from Na et al.50 with permission.
Fig. 7.

Rapid (<300 ms) and strong Src activation (red/yellow spots) co-localizes with regions of large microtubule deformation (white arrows). A local stress was applied to integrins using a 4-μm magnetic bead (17.5 Pa for 3 s step) and Src activation was measured by CFP-YFP Src FRET. Then an oscillatory stress (17.5 Pa at 0.3 Hz) was applied for~10 s and microtubule deformation was quantified by tracking mCherry tubulin displacements induced by stress. For clarity, microtubule displacements <15 nm were omitted. Blue circle, bead–cell contact area (~6 μm2); blue arrow, bead movement direction; N = nucleus. The focal plane was ~1.5 μm above the cell base. Three other cells showed similar behaviors. 80% of strong Src activation co-localizes with regions of large microtubule deformation (>15 nm). In contrast, only ~10% of strong Src activation co-localizes with regions of large F-actin deformation.50 Reproduced from ref. 89 with permission.
IV. Conclusion and perspectives
It becomes increasingly clear that mechanical cues and environment play critical roles in regulating cellular physiology. For example, it has been shown that ECs subjected to disturbed flows, but not to laminar flows, tend to have a high and sustained permeability which facilitates the formation of atherosclerosis.44,58 Mesenchymal stem cells also tend to differentiate into osteoblasts or neuronal cells when cultured on stiff or soft substrates, respectively.14 While tremendous progress has been made during the past decades, it still remains unclear on how cells perceive the external mechanical cues and transmit them into biological/molecular signals to regulate the cellular functions. In many cases, technological development becomes a bottleneck in providing tools to address biological questions with the needed resolution in time and space. The development of FRET technology has provided the possibility of visualizing molecular activity in live cells with high spatiotemporal resolution. While the development of FRET biosensors has been exploding recently, the current scale of research activities in introducing and applying these biosensors for the study of mechanobiology is still limited. One limiting factor is that the dynamic range of most FRET biosensors available is still inferior, which have hindered their broad application for general biomedical studies. With the development of new technologies in high-throughput screening and computational modeling, more and more FRET biosensors with high dynamic ranges should become available in the near future. Consequently, it is expected that the integration of FRET and mechanobiology will become a main-stream practice in the research field of mechanotransduction and mechanobiology to advance our systematic and in-depth understanding on how cells perceive mechanical cues and coordinate signaling cascades accordingly to determine patho-physiological consequences.
Insight, innovation, integration
Mechanical forces play crucial roles in regulating cellular functions. However, it remains elusive on how cells perceive these mechanical cues and transduce them into biochemical signals. With the development of genetically-encoded biosensors based on fluorescence resonance energy transfer (FRET), it is possible to quantify the live-cell molecular activities in real time at subcellular levels when cells are subjected to mechanical stimulation. This article reviews the research on the integration of cutting-edge FRET biosensors and mechanobiology, with a special emphasis on the innovative imaging techniques in live cells. The integration of FRET and mechanobiology should advance our systematic and insightful understanding on how cells sense and adapt to different mechanical environments to yield concerted biological responses in a living organism.
Acknowledgments
We would like to thank people in our two labs for their contribution to the work mentioned in this review. P. W. appreciates the support of Drs Shu Chien and Roger Y. Tsien over the years. This work was supported by NIH grants NCI139272 and NS063405, NSF career award CBET0846429, NSF CMMI0800870, Walter H. Coulter Foundation and Beckman Laser Institute Inc. Foundation (to Y. W.), and NIH grant GM072744 (to N. W.).
Contributor Information
Yingxiao Wang, Email: yingxiao@uiuc.edu.
Ning Wang, Email: nwangrw@illinois.edu.
References
- 1.Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat Methods. 2008;5:401–403. doi: 10.1038/nmeth.1207. [DOI] [PubMed] [Google Scholar]
- 2.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ai HW, Shaner NC, Cheng Z, Tsien RY, Campbell RE. Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry. 2007;46:5904–5910. doi: 10.1021/bi700199g. [DOI] [PubMed] [Google Scholar]
- 4.Ananthanarayanan B, Ni Q, Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proc Natl Acad Sci U S A. 2005;102:15081–15086. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chachisvilis M, Zhang YL, Frangos JA. G protein-coupled receptors sense fluid shear stress in endothelial cells. Proc Natl Acad Sci U S A. 2006;103:15463–15468. doi: 10.1073/pnas.0607224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 9.Clegg RM. Fluorescence resonance energy transfer. In: Wang XF, Herman B, editors. Fluorescence Imaging Spectroscopy and Microscopy. John Wiley & Sons, Inc; New York: 1996. pp. 179–252. [Google Scholar]
- 10.Clegg RM. Nuts and bolts of excitation energy migration and energy transfer. In: Papageorgiou GC, Govindjee, editors. Chlorophyll a Fluorescence: A Signature of Photosynthesis. Springer; New York: 2005. pp. 83–105. [Google Scholar]
- 11.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 12.DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci U S A. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrig T, O’Kane DJ, Prendergast FG. Green-fluorescent protein mutants with altered fluorescence excitation spectra. FEBS Lett. 1995;367:163–166. doi: 10.1016/0014-5793(95)00557-p. [DOI] [PubMed] [Google Scholar]
- 14.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 15.Fu G, Wang C, Wang GY, Chen YZ, He C, Xu ZZ. Detection of constitutive homomeric associations of the integrins Mac-1 subunits by fluorescence resonance energy transfer in living cells. Biochem Biophys Res Commun. 2006;351:847–852. doi: 10.1016/j.bbrc.2006.10.127. [DOI] [PubMed] [Google Scholar]
- 16.Fu G, Yang HY, Wang C, Zhang F, You ZD, et al. Detection of constitutive heterodimerization of the integrin Mac-1 subunits by fluorescence resonance energy transfer in living cells. Biochem Biophys Res Commun. 2006;346:986–991. doi: 10.1016/j.bbrc.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Fujioka A, Terai K, Itoh RE, Aoki K, Nakamura T, et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281:8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 18.Ganguli A, Persson L, Palmer IR, Evans I, Yang L, et al. Distinct NF-kappaB regulation by shear stress through Ras-dependent IkappaBalpha oscillations: real-time analysis of flow-mediated activation in live cells. Circ Res. 2005;96:626–634. doi: 10.1161/01.RES.0000160435.83210.95. [DOI] [PubMed] [Google Scholar]
- 19.Gasman S, Kalaidzidis Y, Zerial M. RhoD regulates endosome dynamics through Diaphanous-related Formin and Src tyrosine kinase. Nat Cell Biol. 2003;5:195–204. doi: 10.1038/ncb935. [DOI] [PubMed] [Google Scholar]
- 20.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 21.Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 22.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 24.Hemsath L, Ahmadian MR. Fluorescence approaches for monitoring interactions of Rho GTPases with nucleotides, regulators, and effectors. Methods. 2005;37:173–182. doi: 10.1016/j.ymeth.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Hodgson L, Pertz O, Hahn KM. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 2008;85:63–81. doi: 10.1016/S0091-679X(08)85004-2. [DOI] [PubMed] [Google Scholar]
- 26.Hu S, Chen J, Fabry B, Numaguchi Y, Gouldstone A, et al. Intracellular stress tomography reveals stress focusing and structural anisotropy in cytoskeleton of living cells. Am J Physiol: Cell Physiol. 2003;285:C1082–C1090. doi: 10.1152/ajpcell.00159.2003. [DOI] [PubMed] [Google Scholar]
- 27.Inouye S, Tsuji FI. Aequorea green fluorescent protein. Expression of the gene and fluorescence characteristics of the recombinant protein. FEBS Lett. 1994;341:277–280. doi: 10.1016/0014-5793(94)80472-9. [DOI] [PubMed] [Google Scholar]
- 28.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 30.Kaplan KB, Swedlow JR, Varmus HE, Morgan DO. Association of p60c-src with endosomal membranes in mammalian fibroblasts. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 32.Katsumi A, Orr AW, Tzima E, Schwartz MA. Integrins in mechanotransduction. J Biol Chem. 2003;279:12001–12004. doi: 10.1074/jbc.R300038200. [DOI] [PubMed] [Google Scholar]
- 33.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 34.Kim TJ, Seong J, Ouyang M, Sun J, Lu S, et al. Substrate rigidity regulates Ca2+ oscillation via RhoA pathway in stem cells. J Cell Physiol. 2009;218:285–293. doi: 10.1002/jcp.21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 36.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J Biol Chem. 2004;280:5581–5587. doi: 10.1074/jbc.M411534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J. Imaging sites of N-wasp activity in lamellipodia and invadopodia of carcinoma cells. Curr Biol. 2004;14:697–703. doi: 10.1016/j.cub.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Luo KQ, Yu VC, Pu Y, Chang DC. Application of the fluorescence resonance energy transfer method for studying the dynamics of caspase-3 activation during UV-induced apoptosis in living HeLa cells. Biochem Biophys Res Commun. 2001;283:1054–1060. doi: 10.1006/bbrc.2001.4896. [DOI] [PubMed] [Google Scholar]
- 39.Mahajan NP, Harrison-Shostak DC, Michaux J, Herman B. Novel mutant green fluorescent protein protease substrates reveal the activation of specific caspases during apoptosis. Chem Biol. 1999;6:401–409. doi: 10.1016/s1074-5521(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan NP, Linder K, Berry G, Gordon GW, Heim R, Herman B. Bcl-2 and Bax interactions in mitochondria probed with green fluorescent protein and fluorescence resonance energy transfer. Nat Biotechnol. 1998;16:547–552. doi: 10.1038/nbt0698-547. [DOI] [PubMed] [Google Scholar]
- 41.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 42.Mena MA, Treynor TP, Mayo SL, Daugherty PS. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat Biotechnol. 2006;24:1569–1571. doi: 10.1038/nbt1264. [DOI] [PubMed] [Google Scholar]
- 43.Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 44.Miao H, Hu YL, Shiu YT, Yuan S, Zhao Y, et al. Effects of flow patterns on the localization and expression of VE-cadherin at vascular endothelial cell junctions: in vivo and in vitro investigations. J Vasc Res. 2005;42:77–89. doi: 10.1159/000083094. [DOI] [PubMed] [Google Scholar]
- 45.Mitra RD, Silva CM, Youvan DC. Fluorescence resonance energy transfer between blue-emitting and red-shifted excitation derivatives of the green fluorescent protein. Gene. 1996;173:13–17. doi: 10.1016/0378-1119(95)00768-7. [DOI] [PubMed] [Google Scholar]
- 46.Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 48.Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 49.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, et al. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 50.Na S, Collin O, Chowdhury F, Tay B, Ouyang M, et al. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci U S A. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Na S, Wang N. Application of fluorescence resonance energy transfer and magnetic twisting cytometry to quantify mechanochemical signaling activities in a living cell. Sci Signal. 2008;1:pl1. doi: 10.1126/scisignal.134pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 53.Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakai J, Ohkura M, Imoto K. A high signal-to-noise Ca(2+) probe composed of a single green fluorescent protein. Nat Biotechnol. 2001;19:137–141. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- 56.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 58.Noria S, Cowan DB, Gotlieb AI, Langille BL. Transient and steady-state effects of shear stress on endothelial cell adherens junctions. Circ Res. 1999;85:504–514. doi: 10.1161/01.res.85.6.504. [DOI] [PubMed] [Google Scholar]
- 59.Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 60.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev Cell. 2006;10:11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 61.Ouyang M, Sun J, Chien S, Wang Y. Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. Proc Natl Acad Sci U S A. 2008;105:14353–14358. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palm GJ, Zdanov A, Gaitanaris GA, Stauber R, Pavlakis GN, Wlodawer A. The structural basis for spectral variations in green fluorescent protein. Nat Struct Biol. 1997;4:361–365. doi: 10.1038/nsb0597-361. [DOI] [PubMed] [Google Scholar]
- 63.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 64.Pertz O, Hahn KM. Designing biosensors for Rho family proteins—deciphering the dynamics of Rho family GTPase activation in living cells. J Cell Sci. 2004;117:1313–1318. doi: 10.1242/jcs.01117. [DOI] [PubMed] [Google Scholar]
- 65.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 66.Piljic A, Schultz C. Simultaneous recording of multiple cellular events by FRET. ACS Chem Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 67.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 68.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 69.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 70.Romoser VA, Hinkle PM, Persechini A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J Biol Chem. 1997;272:13270–13274. doi: 10.1074/jbc.272.20.13270. [DOI] [PubMed] [Google Scholar]
- 71.Sato M, Ozawa T, Inukai K, Asano T, Umezawa Y. Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]
- 72.Seth A, Otomo T, Yin HL, Rosen MK. Rational design of genetically encoded fluorescence resonance energy transfer-based sensors of cellular Cdc42 signaling. Biochemistry. 2003;42:3997–4008. doi: 10.1021/bi026881z. [DOI] [PubMed] [Google Scholar]
- 73.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 74.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 76.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 77.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 78.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Truong K, Sawano A, Mizuno H, Hama H, Tong KI, et al. FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat Struct Biol. 2001;8:1069–1073. doi: 10.1038/nsb728. [DOI] [PubMed] [Google Scholar]
- 81.Tsien RY. Fluorescent probes of cell signaling. Annu Rev Neurosci. 1989;12:227–253. doi: 10.1146/annurev.ne.12.030189.001303. [DOI] [PubMed] [Google Scholar]
- 82.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 83.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 84.Tzima E, Del Pozo MA, Kiosses WB, Mohamed SA, Li S, et al. Activation of Rac1 by shear stress in endothelial cells mediates both cytoskeletal reorganization and effects on gene expression. EMBO J. 2002;21:6791–6800. doi: 10.1093/emboj/cdf688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tzima E, Kiosses WB, del Pozo MA, Schwartz MA. Localized cdc42 activation detected using a novel assay mediates microtubule organizing center positioning in endothelial cells in response to fluid shear stress. J Biol Chem. 2003;278:31020–31023. doi: 10.1074/jbc.M301179200. [DOI] [PubMed] [Google Scholar]
- 86.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- 88.Wang L, Jackson WC, Steinbach PA, Tsien RY. Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc Natl Acad Sci U S A. 2004;101:16745–16749. doi: 10.1073/pnas.0407752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 90.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 91.Webb MR. Development of fluorescent biosensors for probing the function of motor proteins. Mol BioSyst. 2007;3:249–256. doi: 10.1039/b614154d. [DOI] [PubMed] [Google Scholar]
- 92.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 93.Yoshizaki H, Ohba Y, Kurokawa K, Itoh RE, Nakamura T, et al. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol. 2003;162:223–232. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zaccolo M, De Giorgi F, Cho CY, Feng L, Knapp T, et al. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat Cell Biol. 2000;2:25–29. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- 95.Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 96.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 97.Zaidel-Bar R, Kam Z, Geiger B. Polarized downregulation of the paxillin-p130CAS-Rac1 pathway induced by shear flow. J Cell Sci. 2005;118:3997–4007. doi: 10.1242/jcs.02523. [DOI] [PubMed] [Google Scholar]
- 98.Zapata-Hommer O, Griesbeck O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 100.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 101.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]