Abstract
During the past ten years, remarkable progress has been made in understanding the transcriptional mechanisms that control the biology of stem cells. Given the importance of stem cells in development, regenerative medicine and cancer, it is no surprise that the pace of discovery continues to accelerate – paradigm-shifting models proposed only a few years ago are quickly giving way to even more sophisticated models of regulation. This review summarizes some of the major advances made in delineating the roles of two transcription factors, Sox2 and Oct-3/4, in stem cell biology. Additionally, unanswered questions related to their mechanisms of action are discussed. When viewed together, it is evident that Sox2 and Oct-3/4 exhibit the major properties expected of master regulators. They are each essential for mammalian development, they help regulate the transcription of other genes that are essential for development, and they influence their own transcription by both positive and negative feedback loops. Moreover, small changes in the levels of either Sox2 or Oct-3/4 trigger the differentiation of embryonic stem (ES) cells. Thus, each functions as a molecular rheostat to control the self-renewal and pluripotency of ES cells. Overall, understanding how Sox2 and Oct-3/4 function mechanistically will not only provide important insights into stem cells in general, but should also have a significant impact on our understanding of induced pluripotent stem cells, and, hence, the emerging field of regenerative medicine.
Keywords: Sox2, Oct-3/4, ES cells, stem cells, iPS cells, pluripotency, molecular rheostat, regenerative medicine
Introduction
Two transcription factor families, namely the Sox and the Octamer protein binding families, have taken center stage in our efforts to understand how the self-renewal and differentiation of stem cells are controlled. Members of the octamer protein binding family were first identified by virtue of their binding to an eight base pair DNA motif found within the regulatory regions of many genes. Each family member contains a bipartite POU domain (~150 to 160 amino acids), which is responsible for DNA binding. POU domains are highly conserved within the superfamily of POU transcription factors, which are represented by their prototypic members Pit-1, Oct-1 and Unc-86. Although Oct-1 is ubiquitously expressed, Oct-3/4 (also designated Oct-3, Oct4 and Pou5f1) has become the most widely studied octamer binding protein, because it is almost exclusively found in stem cells during development and in a growing list of tumors (1–3). Moreover, only two years ago Oct-3/4 was identified as one of four transcription factors required for the generation of induced pluripotent stem (iPS) cells (4). More recently, other combinations of transcription factors have been used to reprogram somatic cells to a pluripotent stem cell state (5–8). However, each combination included Oct-3/4 and one other transcription factor, Sox2; the only exception being cells that endogenously express one of these transcription factors, such as neural stem cells (9,10).
Like octamer binding proteins, the function of Sox proteins (SRY-related box proteins) during development has also been extensively studied over the past 10 years. In mammals, the Sox family of transcription factors comprises approximately 20 members (11). Sox proteins share highly conserved DNA binding domains of ~80 amino acids. These domains are referred to as HMG box domains, because of their sequence similarity to the DNA binding domains found in the superfamily of High Mobility Group DNA binding proteins. Since the cloning of the prototypic member of the Sox family, SRY (sex-determining region Y), the Sox family has been shown to play key roles during virtually all stages of mammalian development (11). During the past several years, Sox2 has been one of the most actively studied Sox proteins. As noted above, Sox2, like Oct-3/4, is required for the generation of iPS cells. Moreover, Sox2 and Oct-3/4 have each been shown by gene inactivation studies to be essential for mammalian development (12,13). Inactivation of the Sox2 gene or the Oct-3/4 gene results in embryonic lethality during the peri-implantation stage of development. In another parallel with Oct-3/4, knockdown of either Oct-3/4 or Sox2 in ES cells promotes their differentiation into trophectoderm-like cells (14,15). Thus, Oct-3/4 and Sox2 are both essential for the self-renewal and pluripotency of ES cells. In yet another parallel with Oct-3/4, expression of Sox2 is being reported in an expanding list of cancers (16,17).
In view of the importance of understanding how self-renewal and pluripotency of stem cells are controlled at the transcriptional level, this review focuses on Sox2 and Oct-3/4. Specifically, it discusses three main topics: 1) the evidence for a critical partnership between Sox2 and Oct-3/4, 2) the possible redundancy of Sox proteins in pluripotent stem cells, and 3) the need for both positive and negative feedback loops to control the expression levels of master regulators, such as Sox2 and Oct-3/4, in stem cells. In an effort to avoid an over lengthy discussion, this review does not address likely structural interactions between Sox2 and Oct-3/4, which were predicted by molecular modeling using X-ray crystallographic and nuclear magnetic resonance data generated from Sox2/Oct-1/DNA ternary complexes (18,19). In addition, this review does not address the interplay of Sox2 and Oct-3/4 with another master regulator, Nanog. Nor does this review address the rapidly expanding field of ES cell epigenetics. Readers interested in the latter two topics are referred to several excellent reviews (20–24).
Sox2 Partners with Oct-3/4
In the early 1990s, the FGF-4 gene became a model for deciphering how genes are regulated during embryogenesis and for determining how Sox2 and Oct-3/4 function in stem cells. This story began to unfold with the discovery that the FGF-4 gene contains a powerful enhancer located in the last exon of the gene (25). Soon thereafter, it was determined that Oct-3/4 could stimulate the promoter of the FGF-4 gene in F9 embryonal carcinoma (EC) cells by binding to a POU motif embedded within the FGF-4 enhancer (26). In 1995, this finding was complemented by the discovery that Oct-3/4 could act synergistically with a newly identified transcription factor, Sox2, and that these two factors, working in conjunction with one another, were likely to be essential for FGF-4 expression (27). Analysis of the FGF-4 enhancer identified closely spaced binding sites for Sox2 (an HMG motif) and Oct-3/4 (a POU motif), which together were eventually dubbed an HMG/POU cassette (28). The partnership between Sox2 and Oct-3/4 was initially shown in HeLa cells by determining that they could synergistically activate a heterologous promoter/reporter gene construct that contained multiple copies of the binding sites for Sox2 and Oct-3/4 found in the FGF-4 enhancer. Over the next several years, a wide range of experimental approaches generated increasing confidence in the conclusion that Sox2 and Oct-3/4 acted similarly on the FGF-4 gene in EC cells and ES cells.
The earliest evidence supporting the direct roles of Sox2 and Oct-3/4 in the regulation of the FGF-4 gene was derived from studies showing that several different heterologous promoter/reporter gene constructs containing HMG/POU cassettes were highly active in EC cells and ES cells, as well as in HeLa cells (27,29). Moreover, the activities of these promoter/reporter gene constructs were drastically reduced after EC and ES cells differentiated and the expression of Sox2 and Oct-3/4 was extinguished (25,26). Additionally, promoter/reporter gene constructs containing both the FGF-4 promoter and the FGF-4 enhancer were also found to be heavily dependent on the presence of the HMG/POU cassette. Disruption of the HMG/POU cassette reduced FGF-4 promoter activity to near background levels (27,29). Furthermore, the activity of an FGF-4 transgene during embryogenesis was found to be highly dependent on the presence of the FGF-4 enhancer, which contained the HMG/POU cassette (30). Other studies employed gel electromobility shift analysis to show that Sox2 and Oct-3/4 could bind cooperatively to a DNA probe containing the sequence of the HMG/POU cassette found in the FGF-4 enhancer (27,31). In addition, chromatin immunoprecipitation (ChIP) analyses demonstrated that Sox2 could bind to the FGF-4 enhancer in EC and ES cells (32,33). Together, these studies led to the generally accepted belief that expression of the FGF-4 gene in stem cells of the early embryo is heavily dependent on the direct action of Sox2 and Oct-3/4. However, as discussed below, recent studies suggest that a more complex regulatory model, involving Sox protein redundancy, must now be considered.
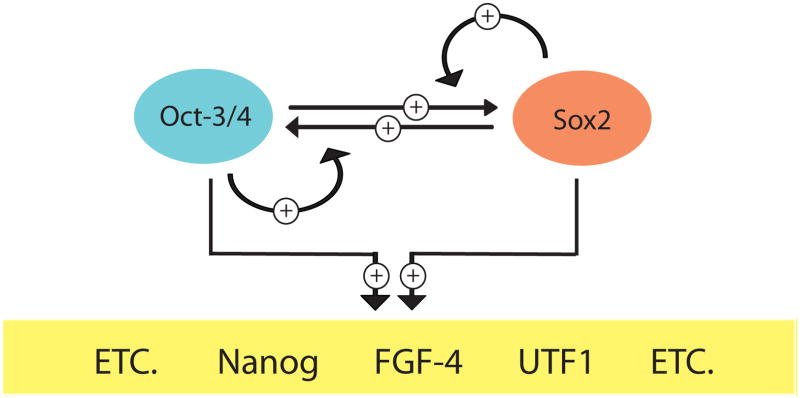
Work with the FGF-4 gene took on far greater interest with the discovery that other genes, including the regulatory regions of the Sox2 and the Oct-3/4 genes themselves, contain HMG/POU cassettes. As in the case of FGF-4 promoter/reporter gene constructs, the HMG/POU cassette present in Sox2 promoter/reporter gene constructs and in Oct-3/4 promoter/reporter gene constructs was found to be essential for promoter activity in each case. Disruption of their HMG/POU cassettes led to strong reduction in the activities of Sox2 and Oct-3/4 promoters in ES cells and also EC cells (15,34–36). The role of Sox2 and Oct-3/4 in the regulation of their own genes was bolstered strongly by ChIP studies showing that the regulatory regions, which contain their HMG/POU cassettes, are bound by both Sox2 and Oct-3/4 in ES cells (15). Again, the functions of the HMG/POU cassettes in the regulatory regions of these genes were found to be virtually eliminated when the expression of Sox2 and Oct-3/4 turns off as ES cells and EC cells differentiate. In combination, these findings laid the foundation for the hypothesis that Sox2 and Oct-3/4 control the expression of a network of genes required for embryogenesis and for the self-renewal and pluripotency of ES cells. More specifically, these findings supported a model in which Sox2 and Oct-3/4 work together cooperatively to regulate their own transcription by a positive feedback loop, while simultaneously working together to drive the transcription of essential downstream target genes, such as FGF-4, UTF1 and Nanog, which, like Sox2 and Oct-3/4, each contain an essential HMG/POU cassette (Figure 1).
Figure 1.
Sox2:Oct-3/4 partnership. Sox2 and Oct-3/4 work together cooperatively to regulate their own transcription and the transcription of a large set of downstream target genes in EC and ES cells.
Sox2 and Oct-3/4 cooperatively regulate large gene regulatory networks
Excitement surrounding the critical roles of Sox2 and Oct-3/4 in stem cells grew dramatically after the initial ChIP-chip and ChIP-PET analyses were performed on human and mouse ES cells (37,38). [These technologies employ chromatin immunoprecipitation followed either by partial sequencing of the immunopreciptiated DNA (ChIP-PET), or hybridization of the immunoprecipitated DNA to a microarray platform containing genomic DNA sequences (ChIP-chip).] The ChIP-chip study by Boyer et al. determined that Sox2 and Oct-3/4 co-occupy several hundred genes, a large percentage of which are expressed in ES cells (37). As in the case of earlier work, the importance of Sox2 and Oct-3/4 in the regulation of these genes was consistent with the finding that the vast majority of the genes co-occupied by Sox2 and Oct-3/4 and expressed in ES cells turn off at the RNA level when ES cells differentiate, presumably because Sox2 and Oct-3/4 expression is extinguished.
The ChIP-chip and ChIP-PET studies revealed several unexpected findings. The inital ChIP-PET study examined genome-wide binding of Oct-3/4 in mouse ES cells, and used bioinformatics to generate a consensus sequence for Oct-3/4 binding sites throughout the genome (38). Remarkably, the genomic regions bound by Oct-3/4 were found to contain DNA sequences that closely matched the consensus sequence for HMG/POU cassettes, which had been derived by comparing the HMG/POU cassettes found in a small set of genes reported to be regulated by Sox2 and Oct-3/4 (Figure 2). Subsequently, virtually the same consensus sequence was generated by bioinformatic analysis of ChIP-PET data from genome-wide binding of Sox2 in mouse ES cells (39). Thus, Sox2 and Oct-3/4 do not merely co-occupy a large number of genes, but in a high percentage of cases, they are bound to regions of DNA that contain adjacent binding sites for Sox2 and Oct-3/4. Additionally, a ChIP-reChIP study involving sequential Oct-3/4 and Sox2 ChIP determined that these two transcription factors are bound simultaneously to at least six different genes in mouse ES cells (38). In a separate study, in which in silico analysis was used to examine 50 of the genes identified in the Boyer et al. study, nearly three-fourths were found to possess sequences that match HMG/POU cassettes, and six of the seven cassettes examined in an in vivo based transcriptional assay were found to be functional (40).
Figure 2.
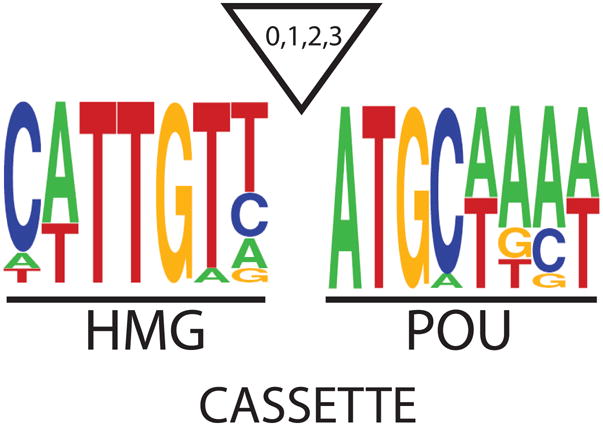
Consensus sequence for HMG/POU Cassettes. A consensus sequence for HMG/POU cassettes derived by comparing the HMG/POU sequences of six Oct-3/4:Sox2 target genes (FGF-4, Sox2, Oct-3/4, Nanog, Fbx15, and UTF1). The triangle represents possible inserts of up to 3 base pairs between the HMG and POU motifs. For example, the FGF-4 gene contains a 3 base pair insert between its HMG and POU motifs. For the remaining 5 genes, their HMG and POU motifs are directly adjacent to one another.
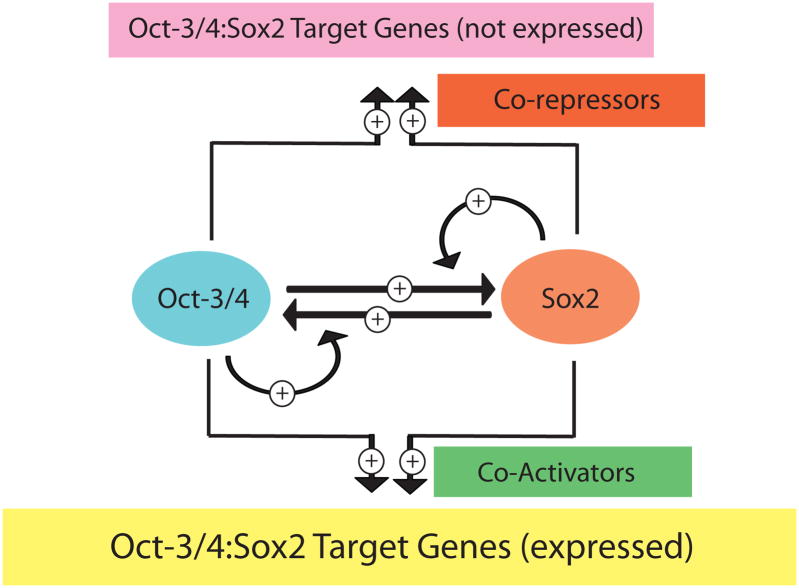
The ChIP-chip study by Boyer et al. led to another important insight (37). It established that Sox2 and Oct-3/4, in fact, co-occupy two classes of genes – those that are expressed in ES cells and a second class of genes that are only expressed after ES cells undergo differentiation. The co-occupancy of the second class of genes by Sox2 and Oct-3/4 does not appear to be a simple coincidence. Surprisingly, this class of genes is highly enriched in genes coding for transcription factors that play key developmental roles (37). This finding raised a new set of puzzling questions regarding the roles of Sox2 and Oct-3/4 in stem cells. In particular, do Sox2 and Oct-3/4 contribute in any way to the silencing of the second class of genes in ES cells, e.g. by helping to recruit repressive transcriptional machinery, such as polycomb repressor complex-2 (41,42), and, if so, how? Alternatively, do Sox2 and Oct-3/4 contribute to the expression of those genes that turn on rapidly when ES cells differentiate? Despite the new set of unanswered questions, these studies led to an expanded model to explain the coordinate regulation of a large set of genes that play prominent roles in self-renewal and pluripotency of ES cells (37). On the one hand, it was proposed that Sox2 and Oct-3/4 can activate the expression of a battery of genes, including their own genes, that are required for the self-renewal and pluripotency of ES cells. On the other hand, Sox2 and Oct-3/4 are also bound to genes that must remain silent in ES cells, yet are ready to be activated quickly in order to contribute to the rapid pace of cell differentiation and specification during mammalian development (Figure 3).
Figure 3.
An expanded model for the regulation of Oct-3/4:Sox2 target genes. Sox2 and Oct-3/4 work together cooperatively to regulate their own transcription and the transcription of a large set of downstream target genes. This includes genes expressed in ES cells and those not expressed in ES cells.
Sox protein redundancy in ES cells
A recent study by Masui et al. raised an additional set of questions and argued for a more complex regulatory model for Sox2 and Oct-3/4. Specifically, their work raised the possibility that Sox2 may not be the only Sox family member that partners with Oct-3/4 to regulate gene expression in ES cells. Using ChIP analysis, these workers demonstrated that several genes that possess HMG/POU cassettes, including the FGF-4 and Oct-3/4 genes, are bound by Sox4, Sox11, and Sox15, as well as by Sox2 (33). This suggested that some level of redundancy may exist between the four Sox proteins in ES cells. However, although all four Sox proteins are able to activate promoters driven by enhancers that contain HMG/POU cassettes when ectopically expressed in HeLa cells (33,43), only Sox2 has been found to be essential for the self-renewal of ES cells. Thus far, knockout and knockdown studies argue that Sox4, Sox11 and Sox15 are not essential for the self-renewal of ES cells or for the early stages of mammalian development (33,44–46). Thus, unlike Sox2 which is clearly essential for the self-renewal of ES cells (15), the roles played by Sox4, Sox11 and Sox15 in these cells, and their redundancy with Sox2, remain open questions. However, it is unlikely that each of these Sox proteins cooperates with Oct-3/4 to the same degree in ES cells (43,44).
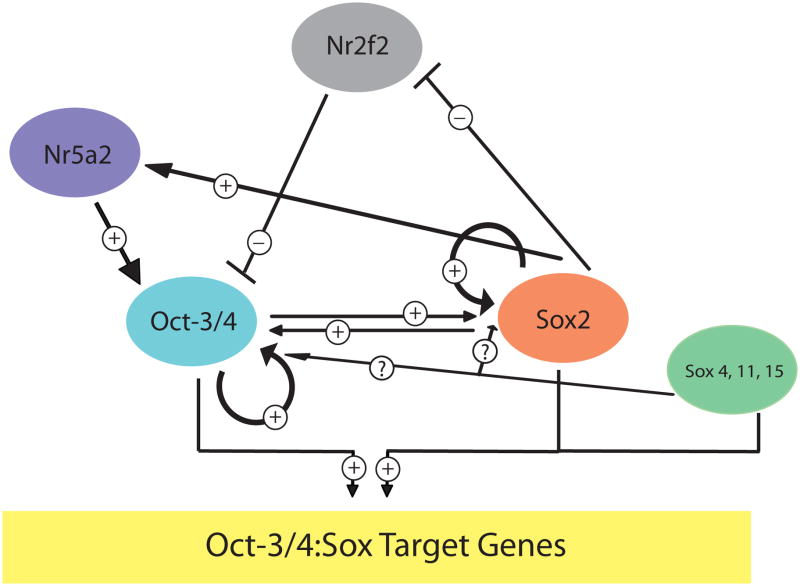
The study by Masui et al. also generated a more nuanced model for the critical roles of Sox2 in the self-renewal of ES cells. As noted earlier, knockdown of Sox2 in ES cells will normally trigger their differentiation. However, Masui et al. demonstrated that if the level of Oct-3/4 was maintained in these cells (with the aid of a transgene), one can rescue both the ability of these cells to self-renew as well as their ability to differentiate into cells derived from each of the three embryonic germ layers under appropriate conditions (33). Thus, these workers proposed that the primary role of Sox2 in ES cells is to maintain the proper levels of Oct-3/4, and further argued that Sox2 contributes to Oct-3/4 transcription both directly and indirectly. Their model argues that Sox2 regulates Oct-3/4 transcription indirectly by both up-regulating Nr5a2, which is able to activate Oct-3/4 expression (at least at the epiblast stage), and by down-regulating Nr2f2, which serves to repress Oct-3/4 transcription. On the other hand, Sox2 (as well as Sox4, Sox11 and/or Sox15) can directly activate Oct-3/4 expression and a large set of genes, by binding, along with Oct-3/4, to their HMG/POU cassettes (Figure 4). Again, many questions surround this model. The most obvious being, do Oct-3/4 and other members of the Sox protein family actually work cooperatively in ES cells? For example, would a ChIP-reChIP study demonstrate that Oct-3/4 and Sox proteins other than Sox2 bind to the same gene simultaneously in ES cells? Moreover, would the overexpression of Nr5a2 and the knockdown of Nr2f2 rescue the self-renewal of ES cells in which Sox2 had been knocked down?
Figure 4.
Sox protein redundancy and the regulation of Oct-3/4:Sox target genes. Oct-3/4 is proposed to work in conjunction with Sox2, as well as Sox4, Sox11 and Sox15, in ES cells to regulate the transcription of the Oct-3/4 and Sox2 genes as well as the transcription of a large set of downstream target genes. Sox2 is also proposed to indirectly regulate the transcription of the Oct-3/4 gene by upregulating Nr5a2 and downregulating Nr2f2, which are positive and negative regulators of the Oct-3/4 gene, respectively. As indicated by the dotted lines, it remains to be determined whether Sox4, Sox11 or Sox15 influence the expression of the Oct-3/4 and Sox2 genes.
Oct-3/4 and Sox2 function as molecular rheostats in ES cells
In 2000, Niwa et al. demonstrated that small changes in the levels of Oct-3/4 promote the differentiation of ES cells, and that the cell types formed were heavily influenced by the expression levels of Oct-3/4 (14). Specifically, reductions in the levels of Oct-3/4 promote the differentiation of ES cells into trophectoderm-like cells, whereas a small increase in Oct-3/4 levels (<2-fold) promotes their differentiation into cells that exhibit the markers for extraembryonic endoderm and mesoderm. This elegant study indicated that master regulators, such as Oct-3/4, must be regulated precisely in order to maintain the self-renewal of ES cells. Equally important, this work raised an intriguing set of questions when it was discovered in 2005 that the Oct-3/4 gene can be regulated by an enhancer that binds Sox2 and Oct-3/4 in ES cells (15). Specifically, if Sox2 and Oct-3/4 work together by a positive feedback loop to regulate Oct-3/4 expression, does a negative feedback loop exist to ensure the proper expression of Oct-3/4? In addition, does this negative feedback loop involve Sox2, and must the levels of Sox2 also be regulated precisely in ES cells? Both questions have been addressed over the past few years.
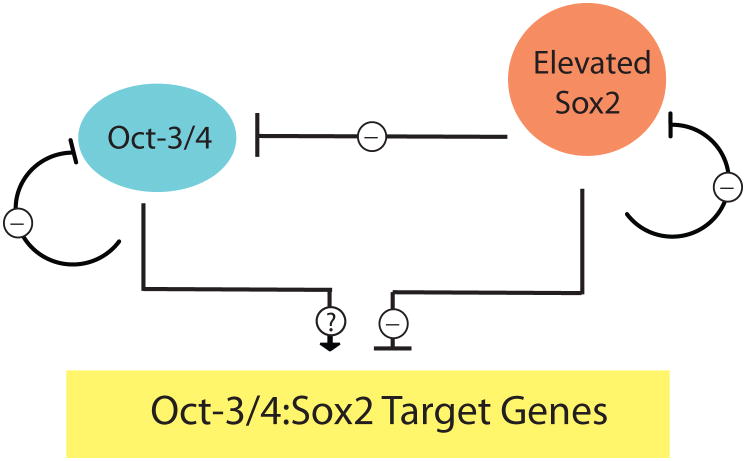
The first evidence for a negative regulatory loop surfaced with the finding that overexpression of Sox2, but not Oct-3/4, inhibited the activity of FGF-4 promoter/reporter gene constructs in EC cells (47). This finding was subsequently extended to the Sox2 promoter itself, as well as to the promoters for Oct-3/4, Nanog and UTF1 genes in EC cells and in ES cells (47,48). In both EC and ES cells, Sox2 overexpression strongly inhibited promoter/reporter gene constructs for all 5 genes (including FGF-4), and this inhibition was mediated by the HMG/POU cassette present in their promoter/reporter gene constructs. Importantly, in EC cells, Sox2 overexpression was also shown to inhibit the endogenous expression of the same five genes. Somewhat unexpectedly, the general inhibitory effect observed with Sox2 was not observed with Oct-3/4. Although Oct-3/4 overexpression appears to inhibit its own promoter and the Nanog promoter, it does not appear to inhibit the promoters of other genes inhibited by Sox2, in particular the Sox2 or FGF-4 promoters (48,49). Thus, Sox2 overexpression appears to exert an overarching inhibiting effect over Oct-3/4:Sox2 target genes (Figure 5). This finding led to the hypothesis that overexpression of Sox2 in ES cells would trigger their differentiation.
Figure 5.
Elevating the expression of Sox2 reduces the expression of genes required for the self-renewal of ES cells. Elevating the levels of Sox2 in EC and ES cells reduces the expression of the Oct-3/4 gene and the expression of downstream Sox:Oct target genes, such as FGF-4, Nanog and UTF1. As indicated by the dotted lines, it is unclear whether Oct-3/4 continues to positively influence the expression of Oct-3/4:Sox2 target genes when Sox2 is overexpressed.
To test the hypothesis that elevating Sox2 levels in ES cells would promote their differentiation, mouse ES cells were engineered for inducible overexpression of Flag-tagged Sox2. Using this model system, small increases in the levels of Sox2 (2-fold or less) were found to lead to rapid differentiation of ES cells into cells that express markers for neuroectoderm, mesoderm and trophectoderm (50). This contrasts with the formation primarily of trophectoderm when Sox2 is knocked down in ES cells (15) and differs from the cell types formed when Oct-3/4 is overexpressed in ES cells (14). As expected, overexpression of Sox2 in ES cells reduced the endogenous expression of several Sox:Oct target genes, including Nanog, FGF-4, UTF1 and the endogenous Sox2 gene itself. Remarkably, the expression of these genes at the RNA level was reduced within three hours after Flag-Sox2 protein was first detected (50). Hence, Sox2 overexpression acts rapidly to alter gene expression and bring about the differentiation of ES cells.
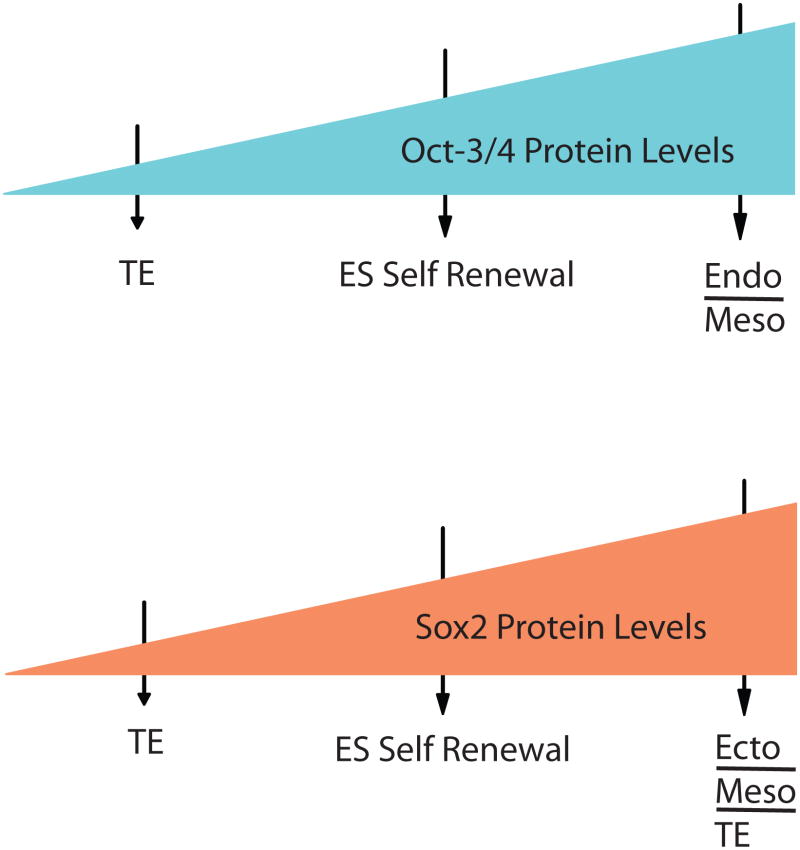
It is evident from these findings that Sox2 and Oct-3/4 both behave as molecular rheostats in the control of ES cell self-renewal and pluripotency. However, the mechanisms involved are far from clear. When these factors drop below a critical level, especially in the case of Oct-3/4, this may trigger the differentiation of ES cells into trophectoderm due to the action of the transcription factor Cdx2. Cdx2 can promote the differentiation of ES cells along the trophectoderm lineage, but its action in ES cells appears to be blocked by direct interaction with Oct-3/4 (51). [Recently, it has been reported that the knockdown of Oct-3/4 induces the formation of cells that express endoderm markers (e.g. GATA6), as well as cells that express trophectoderm markers (52). However, the predominant differentiated cell type formed during the knockdown of Oct-3/4 appears to be trophectoderm.] A different set of mechanisms, or at the very least a different set of genes, must exist for triggering differentiation when Sox2 and Oct-3/4 are knocked down, as opposed to when they overexpressed, because their over and under expression promote very different spectra of differentiated cells (Figure 6). Moreover, the mechanisms by which Oct-3/4 and Sox2 overexpression promote ES cell differentiation must also differ from one another, because their overexpression promotes the formation of different cell types. In the case of Oct-3/4 overexpression, cells expressing endodermal and mesodermal markers appear (14). However, in the case of Sox2 overexpression, cells expressing endodermal markers do not form (Figure 6) (50). Interestingly, one important clue exists regarding the action of Sox2 – the transactivation domain of Sox2 is required for its inhibitory activity (47,48). This has led to speculation that as the levels of Sox2 rise, Sox2 may begin to sequester one or more essential co-activators into incomplete complexes (“a squelching mechanism”) that are unable to transcribe genes required for the self-renewal of ES cells (48).
Figure 6.
Oct-3/4 and Sox2 function as molecular rheostats. Increasing or decreasing the levels of either Sox2 or Oct-3/4 in ES cells promotes their differentiation. However, the spectra of cell types that form vary. The cell types formed were determined by markers expressed in the differentiated cell populations. TE: trophectoderm, Endo: Endoderm, Meso: Mesoderm, Ecto: Ectoderm.
Conclusions and Future Perspectives
Research during the past 10 years has shown that Sox2 and Oct-3/4 exhibit at least four major properties expected of master regulators: 1) they are each essential for mammalian development, 2) they help regulate the transcription of other genes that are essential for development, 3) they influence their own transcription by both positive and negative feedback loops, and 4) they function as molecular rheostats in the control of the self-renewal and pluripotency of ES cells. This last property of master regulators often appears to be largely unappreciated. The fact that Sox2 and Oct-3/4 can function as molecular rheostats raises the intriguing possibility that increases, as well as decreases, in their expression directly influence stem cell differentiation during embryogenesis. Even if this proves not to be the case, it is reasonable to speculate that the levels of these two transcription factors are likely to heavily influence the efficiency of generating iPS cells. Not only should one expect that the correct levels of Sox2 and Oct-3/4 need to be achieved in iPS cells once generated, but that the levels of these two transcription factors during the reprogramming stage itself are likely to influence the overall efficiency of generating iPS cells. In this and other regards (e.g. Sox protein redundancy), issues discussed in this review pertain equally well to ES cells and iPS cells.
It is evident that significant progress has been made in understanding how Sox2 and Oct-3/4 influence stem cells. However, the mechanisms by which these two transcription factors act are poorly understood. In this connection, too little is known about the transcriptional machinery used by these two transcription factors to regulate gene expression. Aside from studies that have implicated the co-activator p300 in mediating the effects of Sox2 and Oct-3/4 (32) and recent studies examining chromatin modifying repressive complexes associated with Oct-3/4 and Nanog (53,54), the roles of specific co-activators and/or co-repressors in the action of these Sox2 and Oct-3/4 transcription factors remain largely unknown. Clearly, comprehensive proteomic analyses of the nuclear proteins with which Sox2 and Oct-3/4 interact would be an important step in understanding how they function mechanistically. In particular, a comprehensive proteomic study would help us understand how small changes in the levels of Sox2 and Oct-3/4 enable them to function as molecular rheostats. Overall, when viewed in the larger context of stem cell biology, understanding how Sox2 and Oct-3/4 function mechanistically will not only provide important insights into stem cells in general, but also could be very helpful in developing protocols that vastly improve the efficiency of generating iPS cells, including their development without the aid of viruses. This would have a very significant impact on the emerging field of regenerative medicine.
Acknowledgments
Members of the author’s laboratory are thanked for reading this review and for making helpful comments. Michelle Desler is thanked for assistance in the preparation of figures and Heather Rizzino is thanked for editorial assistance. Work in the author’s laboratory dealing with ES cells is supported by grants from the NIH (GM 080751) and the Nebraska Research Initiative.
References
- 1.Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990 Mar;344(6265):435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- 2.Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001 Jun;3(26):137–48. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- 3.Hu T, Liu S, Breiter DR, Wang F, Tang Y, Sun S. Octamer 4 small interfering RNA results in cancer stem cell-like cell apoptosis. Cancer Res. 2008 Aug;68(16):6533–40. doi: 10.1158/0008-5472.CAN-07-6642. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007 Dec;318(5858):1917–1920. doi: 10.1126/Science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnology. 2007 Nov;26(1):101–106. doi: 10.1038/nbt1374.. [DOI] [PubMed] [Google Scholar]
- 7.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnolol. 2008 Jul;26(7):795–7. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, Zhaohui Y, Hommond HH, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved Efficiency and Pace of Generating Induced Pluripotent Stem Cells from Human Adult and Fetal Fibroblasts. Stem Cells. 2008 Aug;26:1998–2005. doi: 10.1634/stemcells.2008-0346.. [DOI] [PubMed] [Google Scholar]
- 9.Eminli S, Utikal JS, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of Neural Progenitor Cells into iPS Cells in the Absence of Exogenous Sox2 Expression. Stem Cells. :16. doi: 10.1634/stemcells.2008-0317. Published online July 17, 2008. [DOI] [PubMed] [Google Scholar]
- 10.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M, Schöler HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008 Jul;454(7204):646–50. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 11.Kiefer JC. Back to Basics: Sox Genes. Developmental Dynamics. 2007 Aug;236(8):2356–2366. doi: 10.1002/dvdy.21218.. [DOI] [PubMed] [Google Scholar]
- 12.Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Schöler H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998 Oct;95(3):379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 13.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003 Jan;17(1):126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000 Apr;24(4):328–30. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 15.Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox 2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005 Jul;25(14):6031–46. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008 Jun;283(26):17969–78. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- 17.Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. Br J Cancer. 2008 Feb;98(4):824–31. doi: 10.1038/sj.bjc.6604193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reményi A, Lins K, Nissen LJ, Reinbold R, Schöler HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003 Aug;17(16):2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams DC, Jr, Cai M, Clore GM. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J Biol Chem. 2004 Jan;279(2):1449–57. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 20.Boiani M, Schöler HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005 Nov;6(11):872–84. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 21.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007 Jan;17(1):42–9. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 22.Sharov AA, Ko MSH Human ES. Cell Profiling Broadens the Reach of Bivalent Domains. Cell Stem Cell. 2007 Sep;1(3):237–8. doi: 10.1016/j.stem.2007.08.015.. [DOI] [PubMed] [Google Scholar]
- 23.Lorincz MC, Schübeler D. RNA Polymerase II: Just Stopping By. Cell. 2007 Jul;130(1):16–18. doi: 10.1016/j.cell.2007.06.040.. [DOI] [PubMed] [Google Scholar]
- 24.Jaenisch R, Young R. Stem Cells, the Molecular Circuitry of Pluripotency and Nuclear Reprogramming. Cell. 2008 Feb;132(4):567–582. doi: 10.1016/j.cell.2008.01.015. 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curatola AM, Basilico C. Expression of the K-fgf proto-oncogene is controlled by 3′ regulatory elements which are specific for embryonic carcinoma cells. Mol Cell Biol. 1990 Jun;10(6):2475–84. doi: 10.1128/mcb.10.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y-G, Rosfjord E, Huebert C, Wilder P, Tiesman J, Kelly D, Rizzino A. Transcriptional Regulation of the Murine k-FGF Gene in Embryonic Cell Lines. Developmental Biology. 1992 Nov;154(1):45–54. doi: 10.1016/0012-1606(92)90046-j. [DOI] [PubMed] [Google Scholar]
- 27.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995 Nov;9(21):2635–45. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 28.Boer B, Bernadt CT, Desler M, Wilder PJ, Kopp JL, Rizzino A. Differential Activity of the FGF-4 Enhancer in F9 and P19 Embryonal Carcinoma Cells. Journal of Cellular Physiology. 2006 Jul;208(1):97–108. doi: 10.1002/jcp.20635. 10.1002/jcp.20635. [DOI] [PubMed] [Google Scholar]
- 29.Nowling TK, Johnson LR, Wiebe MS, Rizzino A. Identification of the Transactivation Domain of the Transcription Factor Sox-2 and an Associated Co-activator. The Journal of Biological Chemistry. 2000 Feb;275(6):3810–3818. doi: 10.1074/jbc.275.6.3810. [DOI] [PubMed] [Google Scholar]
- 30.Fraidenraich D, Lang R, Basilico C. Distinct regulatory elements govern Fgf4 gene expression in the mouse blastocyst, myotomes, and developing limb. Dev Biol. 1998 Dec;204(1):197–209. doi: 10.1006/dbio.1998.9053. [DOI] [PubMed] [Google Scholar]
- 31.Ambrosetti D-C, Schöler HR, Dailey L, Basilico C. Modulation of the Activity of Multiple Transcriptional Activation Domains by the DNA Binding Domains Mediates the Synergistic Action of Sox2 and Oct-3 on the Fibroblast Growth Factor-4 Enhancer. The Journal of Biological Chemistry. 2000 July;275(30):23387–23397. doi: 10.1074/jbc.M000932200.. [DOI] [PubMed] [Google Scholar]
- 32.Nowling T, Bernadt C, Johnson L, Desler M, Rizzino A. The Co-activator p300 Associates Physically with and Can Mediate the Action of the Distal Enhancer of the FGF-4 Gene. The Journal of Biological Chemistry. 2003 Apr;278(16):13696–13705. doi: 10.1074/jbc.M207567200.. [DOI] [PubMed] [Google Scholar]
- 33.Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, Ko MSH, Niwa H. Nature Cell Biology. 2007 June;9(6):625–635. doi: 10.1038/ncb1589.. [DOI] [PubMed] [Google Scholar]
- 34.Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Research. 2002 Jul;30(14):3202–3213. doi: 10.1093/nar/gfk435.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 Regulate Oct-3/4 Gene in Embryonic Stem Cells. J Biol Chem. 2005 Feb;280(7):5307–17. doi: 10.1074/jbc.M410015200.. [DOI] [PubMed] [Google Scholar]
- 36.Mallanna SK, Boer B, Desler M, Rizzino A. Differential regulation of the Oct-3/4 gene in cell culture model systems that parallel different stages of mammalian development. Mol Reprod Dev. 2008 Aug;75(8):1247–57. doi: 10.1002/mrd.20871.. [DOI] [PubMed] [Google Scholar]
- 37.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells. Cell. 2005 Sept;122(6):947–56. doi: 10.1016/j.cell.2005.08.020.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong K-Y, Sung KW, Lee CWH, Zhao X-D, Chiu K-P, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei C-L, Ruan Y, Lim B, Ng H-H. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genetics. 2006 Apr;38:431–440. doi: 10.1038/ng1760.. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Log Y-H, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung W-K, Clarke ND, Wei C-L, Ng H-H. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008 Jun;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043.. [DOI] [PubMed] [Google Scholar]
- 40.Chakravarthy H, Boer B, Desler M, Mallanna SK, McKeithan TW, Rizzino A. Identification of DPPA4 and Other Genes as Putative Sox2:Oct-3/4 Target Genes Using a Combination of In Silico Analysis and Transcription-Based Assays. Journal of Cellular Physiology. 2008 Feb;216(3):651–662. doi: 10.1002/jcp.21440. [DOI] [PubMed] [Google Scholar]
- 41.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K-I, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of Developmental Regulators by Polycomb in Human Embryonic Stem Cells. Cell. 2006 Apr;125(2):301–313. doi: 10.1016/j.cell.2006.02.043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006 Apr;441:349–353. doi: 10.1038/nature04733.. [DOI] [PubMed] [Google Scholar]
- 43.Wiebe MS, Nowling TK, Rizzino A. Identification of Novel Domains within Sox-2 and Sox-11 Involved in Autoinhibition of DNA Binding and Partnership Specificity. The Journal of Biological Chemistry. 2003 May;278(20):17901–17911. doi: 10.1074/jbc.M212211200.. [DOI] [PubMed] [Google Scholar]
- 44.Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Differential Roles for Sox15 and Sox2 in Transcriptional Control in Mouse Embryonic Stem Cells. J Biol Chem. 2005 Jul;280(26):24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
- 45.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996 Apr;380(6576):711–4. doi: 10.1038/380711a0, Letter. [DOI] [PubMed] [Google Scholar]
- 46.Sock E, Rettig SD, Enderich J, Bösl MR, Tamm ER, Wegner M. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol Cell Biol. 2004 Aug;24(15):6635–44. doi: 10.1128/MCB.24.15.6635-6644.2004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bernadt CT, Nowling T, Rizzino A. Transcription Factor Sox-2 Inhibits Co-Activator Stimulated Transcription. Molecular Reproduction and Development. 2004 Nov;69(3):260–267. doi: 10.1002/mrd.20168.. [DOI] [PubMed] [Google Scholar]
- 48.Boer B, Kopp J, Mallanna S, Desler M, Chakravarthy H, Wilder PJ, Bernadt C, Rizzino A. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Research. 2007 Feb;35(6):1773–1786. doi: 10.1093/nar/gkm059.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan G, Li J, Zhou Y, Zheng H, Pei D. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. FASEB J. 2006 Aug;20(10):1730–32. doi: 10.1096/fj.05-5543fje. [DOI] [PubMed] [Google Scholar]
- 50.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small Increases in the Level of Sox2 Trigger the Differentiation of Mouse Embryonic Stem Cells. Stem Cells. 2008 Apr;26(4):903–911. doi: 10.1634/stemcells.2007–0951.. [DOI] [PubMed] [Google Scholar]
- 51.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 Determines Trophectoderm Differentiation. Cell. 2005 Dec;123(5):917–929. doi: 10.1016/j.cell.2005.08.040.. [DOI] [PubMed] [Google Scholar]
- 52.Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004 Mar;22(2):225–35. doi: 10.1634/stemcells.22–2-225.. [DOI] [PubMed] [Google Scholar]
- 53.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008 Jun;10(6):731–9. doi: 10.1038/ncb1736, Letters.. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006 Nov;444(7117):364–368. doi: 10.1038/nature05284.. [DOI] [PubMed] [Google Scholar]