Abstract
DNA mismatch repair corrects mispaired bases and small insertions/deletions in DNA. In eukaryotes, the mismatch repair complex MSH2–MSH6 binds to mispairs with only slightly higher affinity than to fully paired DNA in vitro. Recently, the high-mobility group box1 protein, (HMGB1), has been shown to stimulate the mismatch repair reaction in vitro. In yeast, the closest homologs of HMGB1 are NHP6A and NHP6B. These proteins have been shown to be required for genome stability maintenance and mutagenesis control. In this work, we show that MSH2–MSH6 and NHP6A modulate their binding to DNA in vitro. Binding of the yeast MSH2–MSH6 to homoduplex regions of DNA significantly stimulates the loading of NHP6A. Upon binding of NHP6A to DNA, MSH2–MSH6 is excluded from binding unless a mismatch is present. A DNA binding-impaired MSH2–MSH6F337A significantly reduced the loading of NHP6A to DNA, suggesting that MSH2–MSH6 binding is a requisite for NHP6A loading. MSH2–MSH6 and NHP6A form a stable complex, which is responsive to ATP on mismatched substrates. These results suggest that MSH2–MSH6 binding to homoduplex regions of DNA recruits NHP6A, which then prevents further binding of MSH2–MSH6 to these sites unless a mismatch is present.
INTRODUCTION
DNA mismatch repair (MMR) plays a key role in increasing replication fidelity and maintaining genome integrity by identifying and repairing DNA mismatches that have escaped the proofreading activity of replication polymerases. Defects on MMR lead to the accumulation of mutations and are the underlying cause of hereditary nonpolyposis colorectal cancer (HNPCC) (1) and various sporadic cancers.
Repair of mismatched DNA involves several proteins that carry out a multiple-step reaction. Eukaryotes possess two heterodimeric mismatch binding complexes: MSH2–MSH6, which preferentially binds base–base mismatches and 1-nt insertions/deletions loops (IDLs) (2), and MSH2–MSH3, which recognizes IDLs larger than 4 nt. The binding affinity of these complexes to mispaired DNA in vitro has been determined to be 10–30-fold higher than to fully paired DNA, a level that is inconsistent with the rate of misincorporation of replication polymerases that generate a mismatch every 105–106 nt. The mechanism involved in the discrimination of mismatches relative to fully paired DNA is not completely understood, but it is possible that additional factors are required to increase the binding affinity of MMR recognition complexes.
A previous study showed that purified human high-mobility group box1 protein (HMGB1) could complement a fractionated extract in an in vitro MMR reaction, and that HMGB1 could physically interact with MSH2–MSH6 (3). Furthermore, HMGB1 has been implicated to participate at the recognition and excision steps (4). HMGB1 belongs to the high mobility group (HMG) B family of abundant and ubiquitous nonhistone chromosomal DNA-binding proteins. At present four paralogs of HMGB1 exist (HMGB1-4) and the family comprises at least 39 members (www.uniprot.org). HMGB1 binds structurally modified DNA (5) with no sequence specificity and it has recently become the focus of many studies for its participation in proliferation, apoptosis, adhesiveness, migration and invasiveness (6,7). HMG proteins are highly conserved among eukaryotes. Structurally, they consist of one or two HMG-Box DNA-binding domains arranged into three α-helices, which fold into an L-shaped region, and an acidic C-terminal tail of variable length. Binding to distorted DNA is followed by the intercalation of one or two amino acids between DNA base pairs leading to further bending and unwinding of the DNA. HMGB1 has been shown to be involved in various DNA related processes such as transcription regulation, where it recruits the transcription machinery (8) and to enhance nonhomologous DNA repair through stimulation of the DNA–PK kinase activity by promoting its binding to DNA ends (9). HMGB1 has also been shown to be necessary for efficient and correct RSS cleavage hairpin processing in V(D)J recombination (10,11–13). In contrast, HMGB1 has been reported to negatively affect the repair of cisplatinated DNA by strongly binding to the adducts and protecting them from the NER machinery (14). Saccharomyces cerevisiae contains at least 8 HMGB1 homologs. The phylogenetically closest are NHP6A and NHP6B. NHP6A is 80% identical to NHP6B and 45% identical (62% homologous) to human HMGB1 (15–17). Knockouts of both genes are viable but have morphological defects and cannot grow at 37°C (18).
Like HMGB1, NHP6A and NHP6B bind DNA with no sequence specificity and are involved in chromatin remodeling and transcription. Recent work showed that NHP6A/B promote genome stability, as mutants of NHP6A/B display a higher rate of thymine dimers accumulation following UV irradiation, and higher gross chromosomal rearrangements than their isogenic counterparts (19).
In this work, we have cloned and purified yeast NHP6A to investigate the possible involvement of HMGB-like proteins in the yeast MMR pathway. NHP6A bound to DNA in an electrophoretic mobility shift assay. Addition of MSH2–MSH6 to the NHP6A DNA-binding assay showed that MSH2–MSH6 enhances recruitment of NHP6A onto the DNA, possibly through structural changes incurred by the DNA as a result of MSH2–MSH6 binding. Furthermore, we show that NHP6A binding to homoduplex DNA prevented MSH2–MSH6 binding. However, NHP6A did not affect MSH2–MSH6 binding to a heteroduplex, rather than the presence of NHP6A resulted in a reduction of MSH2–MSH6 nonspecific binding and the formation of a stable NHP6A-MSH2–MSH6-mismatched DNA complex. The MSH2–MSH6F337A mutant protein, which retains very low DNA-binding capability (20), was less effective at recruiting NHP6A onto DNA, suggesting that MSH2–MSH6 stimulatory effect on NHP6A DNA binding is mediated through its binding and bending of the DNA. Our data suggest that NHP6A may play a role in modulating the binding of MSH2–MSH6 to DNA affecting some of the DNA transactions for which these proteins are required.
MATERIALS AND METHODS
Strains and oligos
NHP6A was amplified using yeast chromosomal DNA from strain RKY3032. Escherichia coli BL21 (DE3) was used for transformation and expression of NHP6A harboring pET-28a. PCR amplification of NHP6A was carried out using oligos HFRO1263: 5′-TATATACCATGGTCACCCCAAGAGAACCTAAGAAGAGAACC-3′ and HFRO1264: 5′-TATATACTCGAGTTAGTGGTGGTGGTGGTGGTGAGCCAAAGTGGCGTTATATAAC-3′. Gel shift substrates: oligos were annealed to yield 37-mer and 50-mer substrates: HFRO1107: 5′-ATTTCCTTCAGCAGATAGGAACCATACTGATTCACAT-3′, HFRO1108: 5′-ATGTGAATCAGTATGGTTTCTATCTGCTGAAGGAAAT-3′, HFRO1109: 5′-ATGTGAATCAGTATGGTTCCTATCTGCTGAAGGAAAT-3′; HFRO1108 and HFRO1109 were annealed to HFRO1107, yielding a 37-mer G : T heteroduplex and a G : C homoduplex, respectively. HFRO1245: 5′-CTCATTCAGCATAACTTGATTTCTTTCAGCAGATAGAAACCATACTGATT-3′, HFRO1254 5′-AATCAGTATGGTTTCTATCTGCTGAAGGAAATCAAGTTATGCTGAATGAG-3′, HFRO1243 5′-AATCAGTATGGTTTCTATCTGCTGAAAGAAATCAAGTTATGCTGAATGAG-3′. HFRO1243 and HFRO1254 were annealed to HFRO1245 yielding 50-mer T : A heteroduplex and T : G homoduplex, respectively.
Cloning and protein purification
NHP6A was cloned into bacterial vector pET-28a (+) into NcoI and XhoI. The plasmid was then transformed into E. coli strain BL21 (DE3). Bacteria were grown in LB media with kanamycin and NHP6A expression was induced with 1 mM IPTG for 4 h. Lysis was performed in Buffer L (25 mM Tris–HCl pH 7.5, 100 mM NaCl, 0.01% IGEPAL and protease inhibitors). After centrifugation of the lysate, supernatant was passed through a nickel column previously equilibrated with wash buffer (100 mM Tris–HCl pH 7.5, 500 mM NaCl, 20 mM imidazole), and then eluted with elution buffer (100 mM Tris–HCl pH 7.5, 500 mM NaCl, 500 mM imidazole). Eluate was loaded on a sizing column (S-200) equilibrated in buffer A plus 100 mM NaCl (50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol and 0.01 IGEPAL). NHP6A was eluted using buffer A plus 500 mM NaCl.
MSH2–MSH6 was purified from S. cerevisiae by chromatography on PBE94, single-stranded DNA (ssDNA) cellulose and Q-Sepharose as described previously (2). MSH2–MSH6F337A purification was described earlier (20). Proteins purity was estimated to be over 90% by Coomassie-stained gels.
Substrate preparation
Annealing of oligos for substrate preparation was carried out by heating at 95°C for 5 min in 100-µl annealing buffer (0.5 M NaCl, 10 mM Tris–HCl pH 7.5, 1 mM EDTA) and slow cooling to 25°C. DNA was precipitated by adding 3 volumes of ethanol and centrifugation. Pellets were then resuspended in TE buffer containing 0.1 mM EDTA. Benzoylated naphthoylated DEAE cellulose (BND cellulose, Sigma) was used to remove single-stranded DNA (ssDNA). Labeling was performed at 37°C for 30 min in a reaction volume of 50 µl with [γ-32P]-ATP (Amersham) and T4 polynucleotide kinase (New England Biolabs). Reactions were stopped by addition of EDTA to 50 mM. Unincorporated [γ-32P]-ATP was removed by purification in a G-25-Sephadex column as described in manufacturer's protocol (Roche). Aliquots of the labeled duplex DNA were run alongside labeled single-stranded oligos to assess the removal of ssDNA.
Electrophoretic mobility shift assay
In this assay, 37-mer and 50-mer homoduplexes and heteroduplexes DNA substrates were used. Purified MSH2–MSH6, NHP6A or both were incubated on ice with 50 fmol of indicated radiolabeled substrate in the binding buffer (150 mM NaCl, 10 mM HEPES, 13 mM MgCl2, 0.05 mg/ml BSA) in a total volume of 20 µl for 15 min. Loading buffer (15% Ficoll type 4000, 0.25% bromophenol blue and 0.25% xylene cyanol) was added after incubation. No MgCl2 was present in the ATP-binding experiments to avoid ATP hydrolysis. For super shift assays, Tetra-His antibody (Qiagen) or rabbit serum raised against MSH6 were added, and incubation continued for another 30 min. Gel electrophoresis of the mixture was carried out under nondenaturing conditions in a 4.5% polyacrylamide gel containing 5% glycerol in TBE buffer (45 mM Tris–borate, 1 mM EDTA, pH 8.0) at 10 V/cm at 4°C. Gels were dried and exposed on Kodak BioMax film. For quantification, dried gels were exposed to PhosphorImager screen, detection was performed using PhosphorImager Storm Scanner and data was analyzed using ImageQuant software.
ATPase assay
Hydrolysis of [γ-32P]-ATP into ADP and Pi by the MSH2–MSH6 in presence of NHP6A was measured as previously described (21). Briefly, the reaction was carried out in a buffer containing 20 mM Tris–HCl pH 7.5, 2 mM MgCl2, 100 mM NaCl, 5% glycerol, 12 µg ml–1 100-mer homoduplex DNA and 2 mM [γ-32P]-ATP. NHP6A was incubated with DNA at 4°C for 20 min, MSH2–MSH6 was added and incubation continued at 30°C for 20 min. The reaction was stopped by addition of EDTA (20 mM final). Aliquots (1 µl) of each reaction were spotted onto a polyethyleneamide-TLC plate (Sigma). ATP and Pi were separated by chromatography in 1 M formic acid and 0.5 M LiCl. Products were analyzed in a PhosphorImager and quantified using the ImageQuant software. One unit of ATPase activity was defined as the amount of protein that hydrolyzed 1 pmol of ATP to ADP and Pi under the mentioned conditions above.
Statistical analysis
Data analysis and graphing were performed using the GraphPad Prism 4 software package. Specific analysis for each experiment is indicated in the figure legends. Linear regression was used for best curve fitting when necessary and is indicated.
RESULTS
NHP6A purification
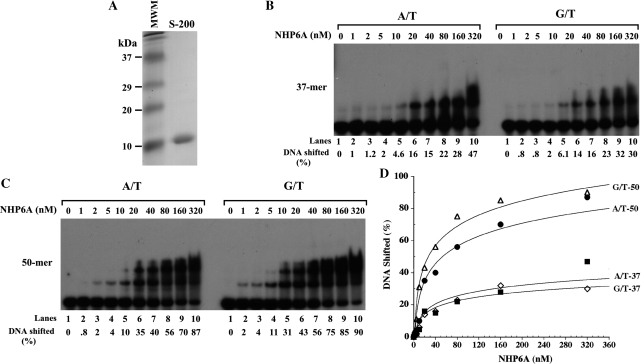
NHP6A was purified to near homogeneity by a two-step procedure carried out at high ionic strength (500 mM NaCl, see ‘Materials and Methods’ section) (Figure 1A). At this salt concentration NHP6A does not bind to DNA and the protein preparation is believed to be devoid of contaminating DNA as assessed by ethidium-bromide staining of a TCA-precipitated fraction (data not shown).
Figure 1.
Binding of NHP6A to homoduplexes and heteroduplexes substrates. Increasing amounts of NHP6A (nM) were incubated at 4°C for 15 min with homoduplex (A/T) and heteroduplex (G/T) substrates as described in ‘Materials and Methods’ section. (A) Coomassie-stained 14% SDS–PAGE gel of the purified NHP6A (S-200 pool). NHP6A runs as a 10-kDa protein. (B) The binding of NHP6A to substrates 37-bp long. (C) Binding of NHP6A to 50-bp long substrates. (D) Quantitation of the NHP6A DNA-binding data shown on (A) and (B).
NHP6A binds homoduplexes and heteroduplexes without preference
Using an electrophoretic mobility shift assay, we tested the ability of NHP6A to bind to homoduplex and heteroduplex substrates of different lengths (37 and 50 mers). As shown in Figure 1, NHP6A was competent at binding both 37-mer and 50-mer substrates irrespective of the presence or absence of a mismatch (Figure 1B–D). Increasing amounts of NHP6A resulted in the formation of higher-order complexes. Although the patterns of protein–DNA complexes formed on fully paired DNA were similar to those obtained on DNA harboring a G/T mismatch, NHP6A formed larger complexes with the 50-mer DNA compared to those assembled on the 37 mer. Discrete bands were observed, with three shifted DNA bands forming with the 50-mer compared to just two when a 37-mer substrate is used. This is due to the fact that the larger DNA can accommodate the binding of a higher number of molecules of NHP6A than the smaller DNA (Figure 1B and C).
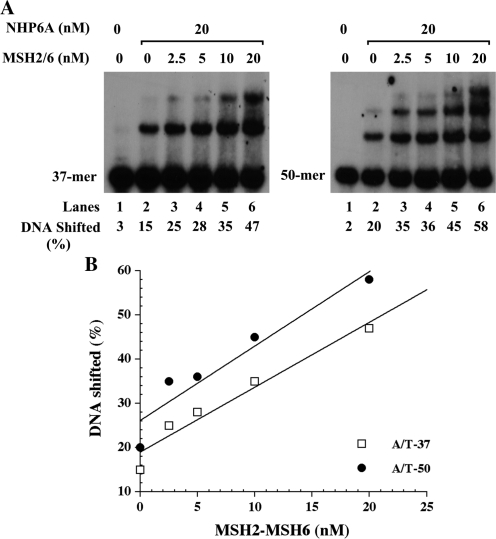
MSH2–MSH6 stimulates the binding of NHP6A to DNA
When homoduplex DNA is incubated with NHP6A at 20 nM concentration, a discrete band is observed in the 37-mer substrate as well as in the 50-mer substrate, which also presents low levels of a second slower migrating band. Interestingly, when NHP6A is incubated simultaneously with increasing amount of MSH2–MSH6 the formation of the NHP6A shifted band is significantly increased (Figure 2). At higher concentrations of MSH2–MSH6 (20 nM) a higher band is observed similar to that observed when NHP6A is incubated alone at higher concentrations with the homoduplex DNA substrate (Figure 2), for the 50-mer substrate, the slower migrating band intensifies in addition to the formation of a larger complex. The appearance of these additional bands is most likely the result of an enhancement in NHP6A capacity to bind to DNA since the total amount of bound DNA (percent of DNA bound) in the presence of 20 nM MSH2–MSH6 is twice that obtained with 20 nM NHP6A alone, and also because these bands migrate faster than a bona fide MSH2–MSH6–DNA complex (see Figure 3). Hence, we conclude that MSH2–MSH6 acts as an enhancer for NHP6A DNA-binding activity.
Figure 2.
Stimulation of the binding of NHP6A to DNA by MSH2–MSH6. (A) NHP6A (20 nM) was incubated at 4°C for 15 min with 37-mer and 50-mer homoduplex (A/T) substrates in the presence of increasing amounts of MSH2–MSH6 proteins as indicated at the top of the figure. (B) Quantitation of the data is shown in (A).
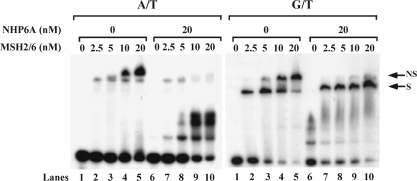
Figure 3.
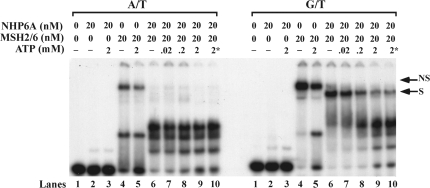
Effect of NHP6A on the binding of MSH2–MSH6 to homoduplex and heteroduplex DNA. Increasing amounts of MSH2–MSH6 were incubated at 4°C for 15 min with homoduplex (A/T) and heteroduplex (G/T) substrates, 50-bp long. Reaction products were resolved in a 4.5% PAA gel, dried and autoradiographed. The nonspecific band (NS) and specific band (S) are indicated in the figure. The specific band formed on G/T substrate in the presence of NHP6A migrates slightly slower than the corresponding band formed by MSH2–MSH6 alone.
Binding of NHP6A to DNA blocks MSH2–MSH6 binding to homoduplex regions but does not affect the binding to mismatches
When substrate DNA containing a mismatch is incubated with MSH2–MSH6, a complex is observed at low levels of the protein, which corresponds to the MSH2–MSH6 bound to the mismatch. As the concentration of MSH2–MSH6 is increased, a second slower migrating band is observed. This complex, which is also observed when MSH2–MSH6 is incubated with homoduplex DNA, corresponds to a nonspecific complex between MSH2–MSH6 and DNA. Interestingly, when MSH2–MSH6 is incubated with homoduplex DNA in the presence of NHP6A, the nonspecific band observed at high concentrations of MSH2–MSH6 (10 nM and 20 nM) is significantly reduced, with a concomitant increase in the NHP6A shifted band (Figure 3, A/T panel). This result suggests that as NHP6A binds to DNA, MSH2–MSH6 is prevented from binding to the homoduplex substrate. In contrast, when the heteroduplex DNA is used, at low concentrations of MSH2–MSH6 only the specific band is observed. As the concentration of MSH2–MSH6 increases so does the nonspecific complex, at the expense of the specific complex (Figure 3, G/T panel). However, in the presence of NHP6A, the nonspecific band is abrogated even at high concentrations of MSH2–MSH6 (20 nM). The specific band is not affected and intensifies as the concentration of MSH2–MSH6 is increased (Figure 3), and it appears to migrate slightly slower in the presence of NHP6A. These data suggest that NHP6A, despite preventing the binding of MSH2–MSH6 to homoduplex regions, does not block the recognition and binding of the mismatch by the repair complex. In addition, the NHP6A specific band observed with the homoduplex substrate does not form, suggesting that NHP6A is forming a complex with MSH2–MSH6 in the mismatch substrate.
MSH2–MSH6 and NHP6A coexist in complexes formed in mismatch containing substrates
To determine the composition of the complexes formed on homoduplex and heteroduplex DNA, we tested for the presence of NHP6A and MSH2–MSH6 by supershifting the bands with specific antibodies. Incubation of homoduplex with NHP6A results in small shifted species that can be supershifted by anti-his antibodies (against his-tagged NHP6A) but not by anti-MSH6 antibody (Figure 4, A/T panel lanes 2–4). When MSH2–MSH6 is incubated with the A/T substrate at high concentration (20 nM), the nonspecific band that is formed is supershifted by anti-MSH6 antibodies but not anti-his antibodies (Figure 4, A/T panel lanes 5–7). When both NHP6A and MSH2–MSH6 have been incubated with the homoduplex DNA, most of the shifted bands corresponds to the NHP6A complex, which can be completely supershifted with the anti-his antibody and present significant lower levels of MSH2–MSH6 (Figure 4, A/T panel lanes 8–10), indicating that MSH2–MSH6 has been excluded from the homoduplex substrate by NHP6A. When NHP6A is incubated with the heteroduplex DNA, the complex formed can be supershifted by anti-his antibodies and not anti-MSH6 antibodies (Figure 4, G/T panel lanes 2–4). When MSH2–MSH6 is incubated with the G/T substrate, the specific and nonspecific are formed which can be supershifted only by the anti-MSH6 antibodies (Figure 4, G/T panel lanes 5–7). Interestingly, when both NHP6A and MSH2–MSH6 are incubated with the heteroduplex DNA, the complex that is formed can be completely supershifted with both the anti-his and the anti-MSH6 antibodies (Figure 4, G/T panel lanes 8–10). Neither antibody bound DNA (data not shown). These data suggest that the composition of DNA–protein complexes formed depends on the presence or absence of mismatches in the DNA and that MSH2–MSH6 is excluded from homoduplex DNA by NHP6A but its binding to mismatches is not affected.
Figure 4.
Composition of complexes that form on homoduplex and heteroduplex substrates. Supershifting of the complexes was carried out using anti-His antibody against NHP6A and anti-MSH6 (rabbit polyclonal) specific antibodies. The proteins at the concentration indicated above, were incubated with either 50-mer A/T or 50-mer G/T substrates at 4°C for 15 min. Antibodies were added and incubation continued for another 30 min. Reaction products were resolved in a 4.5% PAA gel, dried and autoradiographed.
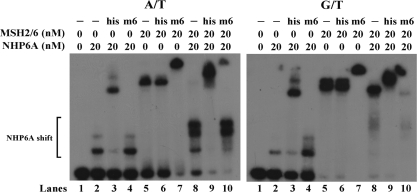
The DNA-binding activity of MSH2–MSH6 is required for the loading of NHP6A to DNA
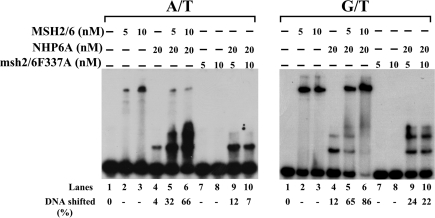
The observation that the MSH2–MSH6 complex stimulates the binding of NHP6A to DNA prompted us to determine if the DNA-binding activity of MSH2–MSH6 is required for this effect. For this purpose, we prepared a mutant of MSH2–MSH6, which has a F337A change on the MSH6 subunit. This mutation has been shown to have a reduced DNA-binding activity (22,20). When we tested this mutant mispair-binding complex in homoduplex DNA, we find that it has negligible amounts of DNA binding and that its ability to stimulate NHP6A binding to DNA is also considerably diminished (Figure 5, A/T panel lanes 7–10) when compared to wild-type MSH2–MSH6 (Figure 5, A/T panel lanes 2–6). Consistently, when substrates containing mismatches are tested the MSH2–MSH6F337A mutant protein does not bind efficiently to the substrates and does not form a tripartite complex with NHP6A and DNA (Figure 5, G/T panel lanes 7–10) when compared to the wild-type MSH2–MSH6 (Figure 5, G/T panel lanes 2–6). For both substrates, however, a slight stimulation of the NHP6A specific complex is still observed, suggesting that MSH2–MSH6F337A may interact with DNA, but not stably enough to remain bound to DNA.
Figure 5.
Effect of a DNA-binding mutation MSH6F337A on the stimulatory effect of MSH2–MSH6 on NHP6A binding. Increasing amounts of MSH2–MSH6 and the DNA-binding defective mutant, MSH2–MSH6F337A, were incubated with 50-mer homoduplex (A/T) or heteroduplex (G/T) substrates at 4°C for 15 min in the presence or absence of NHP6A (20 nM). Reaction products were resolved in a 4.5% PAA gel, dried and autoradiographed.
NHP6A does not affect the ability of MSH2–MSH6 to be released from DNA after ATP binding
A feature of MSH2–MSH6 binding to mismatches is its ability to be released from the DNA upon ATP binding and hydrolysis. To determine if the presence of NHP6A on the complex affects the ability of MSH2–MSH6 to slide off DNA, we tested if increasing amounts of ATP reduces the amount of preformed mismatch binding complex remaining bound to DNA. MSH2–MSH6 binding to homoduplex DNA results in a nonspecific complex, which has been shown to be partially refractive to ATP, as we also observed (Figure 6, A/T panel lanes 4 and 5). However, when NHP6 is present, this nonspecific complex is considerably reduced (Figure 6, A/T panel lane 6) and when increasing amounts of ATP are added to this preformed complex, the remaining nonspecific band is abolished (Figure 6, A/T panel lanes 7–9) to levels similar to those observed when ATP (2 mM) is present at the beginning of the reaction (Figure 6, A/T panel lane 10), leaving only NHP6A bound to the DNA. On substrates containing a mismatch, high concentrations of MSH2–MSH6 form specific and nonspecific complexes. Addition of NHP6A abolishes the nonspecific complex (Figures 3 and 6, G/T panel lane 6), leaving a band corresponding to the specific complex. Addition of increasing amounts ATP to the G/T–NHP6A–MSH2–MSH6 complex results in a reduction of the shifted band to levels similar to those observed when ATP is present at the beginning of the reaction, and as in the homoduplex substrate, only NHP6A remains bound to the DNA. When lower concentrations of MSH2–MSH6 are used that result only on a specific band, the complex formed in the presence of NHP6A is also responsive to ATP (data not shown). These results indicate that MSH2–MSH6 bound to a mismatch in the presence of NHP6A displays the same properties previously reported with respect to its sensitivity to ATP.
Figure 6.
Effect of ATP on complexes containing MSH2–MSH6 and NHP6A preassembled on homoduplex and heteroduplex substrates. Reactions containing MSH2–MSH6 (20 nM) and NHP6A (20 nM) were incubated with 50-mer homoduplex (A/T) or 50-mer heteroduplex (G/T) substrates at 4°C for 15 min. ATP was then added, where indicated, and incubation was continued for another 15 min at 37°C. Reactions products were then resolved in a 4.5% PAA gel, dried and autoradiographed. Asterisk indicates that ATP was present from the beginning of the incubation.
NHP6A reduces the homoduplex DNA-stimulated ATPase activity of MSH2–MSH6 but not its DNA-independent ATPase activity
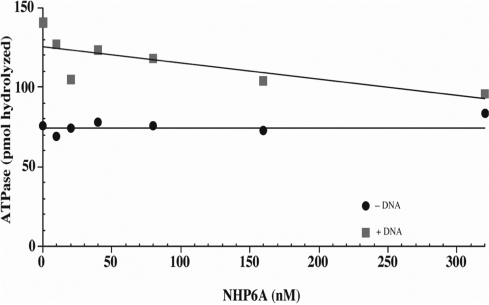
Since MSH2–MSH6 is an ATPase, we determined if NHP6A modifies its ATP hydrolytic activity. NHP6A was titrated in a reaction containing MSH2–MSH6 (25 nM) and ATP in the presence or absence of homoduplex DNA. ATP hydrolysis was determined as described in ‘Materials and Methods’ section. The DNA-independent ATPase activity was not affected by increasing amounts of NHP6A (Figure 7, black circles) even at the highest concentration of NHP6A (320 nM). When homoduplex substrate was present, the basal level of hydrolysis was twice as high as the DNA-independent ATPase activity. As NHP6A concentration was increased, the DNA-dependent ATPase activity was gradually reduced approaching that of the DNA-independent ATPase activity at high levels of NHP6A (Figure 7, gray squares). However, when a heteroduplex substrate was used, no significant reduction on the ATPase activity of MSH2–MSH6 was observed at the concentrations of NHP6A tested (data not shown). These data suggest that NHP6A coating of the homoduplex DNA prevents its access by MSH2–MSH6, which leads to a reduced ATP hydrolysis that corresponds mostly to the DNA-independent type.
Figure 7.
Inhibition of the homoduplex DNA-stimulated ATPase activity of MSH2–MSH6 by NHP6A. Increasing amounts of NHP6A were incubated with homoduplex DNA for 20 min at 4°C. MSH2–MSH6 was then added to the reaction and incubation continued at 30°C for 20 min. Aliquots (1 µl) of each reaction were spotted onto a polyethyleneamide-TLC plate (Sigma). ATP and Pi were separated by chromatography in 1 M formic acid and 0.5 M LiCl. Products were analyzed in a PhosphorImager and quantified using the ImageQuant software. Background has been subtracted.
DISCUSSION
HMGB1 has long been regarded as one of many chromatin remodeling and transcription proteins that bind DNA with little or no specificity. Recently, a growing evidence of its active involvement in the regulation of a number of DNA related and cell-fate processes has been reported. In addition to its involvement in nonhomologous DNA-end joining (NHEJ) and V(D)J recombination, another study showed that HMGB1 and HMGB2 were components of a larger sensing complex capable of recognizing DNA damage (23). Although HMGB1 binds DNA nonspecifically, it was shown to strongly bind DNA cisplatinated adducts. The role of HMG proteins in MMR has been recently reported (3,4). Although not an essential factor, HMGB1 stimulates the repair of mismatched substrate in an in vitro assay. The mechanism by which HMGB1 exerts its stimulation of MMR has not been yet established, although there is an evidence that it is required for the excision step (4). The yeast S. cerevisiae possesses several proteins that belong to the high mobility group family. The closest homologs to HMGB1 are NHP6A and NHP6B. Like HMGB1, NHP6A is involved in chromatin remodeling and transcription. NHP6A and NHP6B have been reported to facilitate genome stability as mutations in both genes, leads to accumulation of thymine dimers when exposed to UV light and to higher gross chromosomal rearrangements (GCRs) (19).
We have investigated the effect of NHP6A on the DNA-binding and ATPase activities of MSH2–MSH6. The main findings of this study are: (i) NHP6A can bind to DNA with no particular preference for the presence or absence of a mismatch; (ii) binding of MSH2–MSH6 to DNA stimulates the binding of NHP6A to DNA in a dose-dependent manner; (iii) NHP6A reduces the nonspecific binding of MSH2–MSH6 to DNA but does not affect its binding to mismatches; and (iv) NHP6A coexists with MSH2–MSH6 on complexes that form on mismatched DNA and these complexes are responsive to ATP.
NHP6A appears to have similar affinity for homoduplex and heteroduplex DNA, indicating that the presence of a mismatch does not result in a significantly distorted structure that is preferred substrate for HMGB types of proteins. However, addition of MSH2–MSH6 significantly stimulated the binding of NHP6A to DNA, both homoduplex and heteroduplex. A previous study of MutS binding to DNA using atomic force microscopy (24) found that MutS–DNA complexes form a single population of conformations in which the DNA is bent at homoduplex sites. Our data are in agreement with this report. We interpret the stimulation of NHP6A binding to DNA by MSH2–MSH6 as a consequence of DNA bending by MSH2–MSH6, which results in a substrate of higher affinity for NHP6A. Once the DNA is fully coated with NHP6A, these regions are no longer accessible to MSH2–MSH6, which results in exclusion of the repair complex from homoduplex DNA. This is noticeable in the disappearance of the non-specific band that forms when homoduplex DNA is used. However, a recent report (25) indicates that prokaryotic MutS has higher affinity to mismatches than previously reported, and that DNA end binding by MutS is a significant component of its nonspecific binding to DNA. It is possible that NHP6A prevents the nonspecific binding of MSH2–MSH6 to DNA ends as well. Current studies in our laboratory using circular substrates, however, indicate that MSH2–MSH6 does bind non-specifically to non-mismatched duplex DNA (Banerjee,S., Jaafar,L. and Flores-Rozas,H., unpublished data). The DNA-binding (and -bending) activity of MSH2–MSH6 appears to be critical for generating a substrate for NHP6A. When the mutant protein MSH2–MSH6F337A, which has defective DNA-binding activity, was tested, it showed a significant reduction in its ability to stimulate NHP6A binding to DNA as compared to the wild-type protein. Furthermore, interaction experiments revealed that MSH2–MSH6 and NHP6A do not associate in solution as determined by co-immunoprecipitation using anti-MSH2, anti-MSH6 and anti-His6 (NHP6A) antibodies (data not shown), suggesting that the loading of NHP6A on DNA upon MSH2–MSH6 binding is the result of the alteration of the DNA substrate (i.e. DNA bending). This also suggests that initial binding of MSH2–MSH6 to DNA is required for efficient binding of NHP6A to DNA. To our knowledge, this is the first report of a protein stimulating the DNA-binding activity of NHP6A. It is possible that other factors that bend DNA may have a similar effect and that this may constitute a conserved mechanism by which HMG proteins participate in DNA metabolism. Interestingly, if a mismatch is present, MSH2–MSH6 can efficiently recognize and bind to it, even if the homoduplex region of DNA is coated by NHP6A. In fact, we find that both MSH2–MSH6 can coexist on a DNA containing a mismatch and when ATP is added to the reaction, MSH2–MSH6 is released from the mismatched DNA, while NHP6A remains bound. The nonspecific band is also reduced by the presence of NHP6A confirming that this slower migrating complex corresponds to an interaction between MSH2–MSH6 and homoduplex regions (24).
NHP6A reduced the homoduplex–DNA-dependent ATPase activity of MSH2–MSH6. This effect, which is dose dependent, can be interpreted as a consequence of NHP6A coating the substrate DNA and making it unavailable to MSH2–MSH6. In fact, at high concentrations of NHP6A, ATP hydrolysis approaches that observed for the DNA-independent ATPase activity of MSH2–MSH6.
In addition to NHP6A, S. cerevisiae encodes NHP6B, which is highly homologous. Both NHP6A and NHP6B have regulated expression, and in the absence of one, the expression of the other protein is increased (26). We have tested NHP6B in combination with MSH2–MSH6 and found to have the same effect as with NHP6A (data not shown). Thus, it is possible that both proteins may play a role in regulating MSH2–MSH6 binding to DNA. It is also possible that other HMG-type proteins may play a role in MMR, since inactivation of NHP6A/B does not significantly increase the mutator phenotype, consistent with previous reports of modest increase in genomic instability (19) and suggesting the existence of redundant activities that can compensate for their absence as it also occurs with some MMR genes (27).
Taken together, our data show that HMGB-type protein NHP6A can modulate the binding of MMR complexes to DNA, affecting the activity of MSH2–MSH6 in a DNA transaction probably involved in recombination as previously suggested (25).
FUNDING
National Institutes of Health GM068536 (to H.F.-R.); and the Georgia Cancer Coalition (to H.F.-R.). Funding for open access charge: National Institutes of Health GM068536.
Conflict of interest statement. None declared.
REFERENCES
- 1.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J. Clin. Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 2.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 3.Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J. Biol. Chem. 2004;279:20935–20940. doi: 10.1074/jbc.M401931200. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 6.Riuzzi F, Sorci G, Donato R. The amphoterin (HMGB1)/receptor for advanced glycation end products (RAGE) pair modulates myoblast proliferation, apoptosis, adhesiveness, migration, and invasiveness. Functional inactivation of RAGE in L6 myoblasts results in tumor formation in vivo. J. Biol. Chem. 2006;281:8242–8253. doi: 10.1074/jbc.M509436200. [DOI] [PubMed] [Google Scholar]
- 7.Romine LE, Wood JR, Lamia LA, Prendergast P, Edwards DP, Nardulli AM. The high mobility group protein 1 enhances binding of the estrogen receptor DNA binding domain to the estrogen response element. Mol. Endocrinol. 1998;12:664–674. doi: 10.1210/mend.12.5.0111. [DOI] [PubMed] [Google Scholar]
- 8.Zappavigna V, Falciola L, Helmer-Citterich M, Mavilio F, Bianchi ME. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996;15:4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe F, Shirakawa H, Yoshida M, Tsukada K, Teraoka H. Stimulation of DNA-dependent protein kinase activity by high mobility group proteins 1 and 2. Biochem. Biophys. Res. Commun. 1994;202:736–742. doi: 10.1006/bbrc.1994.1992. [DOI] [PubMed] [Google Scholar]
- 10.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 12.Aidinis V, Bonaldi T, Beltrame M, Santagata S, Bianchi ME, Spanopoulou E. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mo X, Bailin T, Noggle S, Sadofsky MJ. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 2000;28:1228–1236. doi: 10.1093/nar/28.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arioka H, Nishio K, Ishida T, Fukumoto H, Fukuoka K, Nomoto T, Kurokawa H, Yokote H, Abe S, Saijo N. Enhancement of cisplatin sensitivity in high mobility group 2 cDNA-transfected human lung cancer cells. Jpn. J. Cancer Res. 1999;90:108–115. doi: 10.1111/j.1349-7006.1999.tb00673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolodrubetz D, Haggren W, Burgum A. Amino-terminal sequence of a Saccharomyces cerevisiae nuclear protein, NHP6, shows significant identity to bovine HMG1. FEBS Lett. 1988;238:175–179. doi: 10.1016/0014-5793(88)80251-5. [DOI] [PubMed] [Google Scholar]
- 16.Kolodrubetz D, Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem. 1990;265:3234–3239. [PubMed] [Google Scholar]
- 17.Yang H, Tracey KJ. High mobility group box 1 (HMGB1) Crit. Care Med. 2005;33:S472–S474. doi: 10.1097/01.ccm.0000187005.81616.a9. [DOI] [PubMed] [Google Scholar]
- 18.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giavara S, Kosmidou E, Hande MP, Bianchi ME, Morgan A, d'Adda di Fagagna F, Jackson SP. Yeast Nhp6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr. Biol. 2005;15:68–72. doi: 10.1016/j.cub.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 20.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 21.Flores-Rozas H, Hurwitz J. Characterization of a new RNA helicase from nuclear extracts of HeLa cells which translocates in the 5′ to 3′ direction. J. Biol. Chem. 1993;268:21372–21383. [PubMed] [Google Scholar]
- 22.Drotschmann K, Yang W, Brownewell FE, Kool ET, Kunkel TA. Asymmetric recognition of DNA local distortion. Structure-based functional studies of eukaryotic Msh2-Msh6. J. Biol. Chem. 2001;276:46225–46229. doi: 10.1074/jbc.C100450200. [DOI] [PubMed] [Google Scholar]
- 23.Krynetski EY, Krynetskaia NF, Bianchi ME, Evans WE. A nuclear protein complex containing high mobility group proteins B1 and B2, heat shock cognate protein 70, ERp60, and glyceraldehyde-3-phosphate dehydrogenase is involved in the cytotoxic response to DNA modified by incorporation of anticancer nucleoside analogues. Cancer Res. 2003;63:100–106. [PubMed] [Google Scholar]
- 24.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl Acad. Sci. USA. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Sass LE, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic Acids Res. 2005;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolodrubetz D, Kruppa M, Burgum A. Gene dosage affects the expression of the duplicated NHP6 genes of Saccharomyces cerevisiae. Gene. 2001;272:93–101. doi: 10.1016/s0378-1119(01)00568-6. [DOI] [PubMed] [Google Scholar]
- 27.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl Acad. Sci. USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]