Abstract
The X chromosome-linked inhibitor of apoptosis protein (XIAP) is the most potent intrinsic caspase inhibitor and plays an important role in the maintenance of intestinal epithelial integrity. The RNA binding protein, HuR, regulates the stability and translation of many target transcripts. Here, we report that HuR associated with both the 3′-untranslated region and coding sequence of the mRNA encoding XIAP, stabilized the XIAP transcript and elevated its expression in intestinal epithelial cells. Ectopic HuR overexpression or elevated cytoplasmic levels of endogenous HuR by decreasing cellular polyamines increased [HuR/XIAP mRNA] complexes, in turn promoting XIAP mRNA stability and increasing XIAP protein abundance. Conversely, HuR silencing in normal and polyamine-deficient cells rendered the XIAP mRNA unstable, thus reducing the steady state levels of XIAP. Inhibition of XIAP expression by XIAP silencing or by HuR silencing reversed the resistance of polyamine-deficient cells to apoptosis. Our findings demonstrate that HuR regulates XIAP expression by stabilizing its mRNA and implicates HuR-mediated XIAP in the control of intestinal epithelial apoptosis.
INTRODUCTION
In response to stressful environmental conditions, mammalian cells elicit rapid changes in gene expression patterns to regulate their survival, adapt to stress and maintain homeostasis (1). In addition to the stress-modulated gene transcription, changes in post-transcriptional regulation, particularly altered mRNA turnover and translation, also potently affect the steady state levels of many transcripts and the levels of the encoded proteins (2,3). An increasing body of evidence indicates that post-transcriptional fate of a given mRNA is primarily controlled by the interaction of specific mRNA sequences (cis elements) with specific trans-acting factors such as RNA binding proteins (RBPs) (3,4) and microRNAs (5). Ribonucleoprotein (RNP) associations regulate the intracellular transport of the mRNA and its association with the translation and decay machineries (6). Many labile mRNAs contain relatively long 3′-untranslated regions (UTRs) bearing U- and AU-rich elements (AREs) that function as determinants of mRNA stability and translation (7–10). Among the RBPs that regulate specific subsets of mRNAs, there are several RBPs that modulate mRNA turnover (HuR, NF90, AUF1, BRF1, TTP and KSRP) and RBPs that modulate translation (HuR, TIAR, NF90 and TIA-1), collectively known as translation and turnover regulatory (TTR)-RBPs (11–13). In cells responding to proliferative, immune and stress-causing stimuli, TTR-RBPs bind to the specific sequences in the 5′- and 3′-UTRs of collections of target mRNAs and govern their turnover and translation rates (1,3).
HuR is one of the best studied TTR-RBPs. It has two N-terminal RNA-recognition motifs (RRMs) with high affinity for AREs followed by a nucleocytoplasmic shuttling sequence and a C-terminal RRM that recognizes the poly(A) tail (14–16). Although the exact mechanism underlying target mRNA stabilization and translation by HuR is poorly understood, this process is intimately associated with the cytoplasmic presence of the HuR RNP. In unstimulated cells, HuR is predominantly located in the nucleus, but it rapidly translocates to the cytoplasm, where it directly interacts with and stabilizes specific mRNAs and/or variably affects their translation in response to various stimuli (17). Recent studies (18–21) have demonstrated that HuR plays a critical role in the regulation of normal intestinal mucosal tissue homeostasis by modulating intestinal epithelial cell (IEC) proliferation and apoptosis. These studies further revealed that the nucleocytoplasmic shuttling of HuR in IECs is tightly regulated by the levels of cellular polyamines (spermidine, spermine and their precursor putrescine). Decreasing cellular polyamines elevate cytoplasmic HuR levels, whereas elevating polyamines decrease cytoplasmic HuR; neither intervention alters whole cell HuR levels (21). HuR has been increasingly recognized as a key regulator of post-transcriptional gene regulation involved in maintaining the intestinal epithelial integrity, but its downstream target transcripts in IECs remain to be fully investigated.
The X chromosome-linked inhibitor of apoptosis protein (XIAP), a membrane of the IAP family, is associated with cell survival by protecting cells from caspase-mediated apoptosis (22–25). Eight human IAP isoforms have been identified to date, and the anti-apoptotic action of XIAP, cytosolic IAP (cIAP-1 and -2) and survivin have been characterized (22,26). XIAP is the most potent and best studied endogenous caspase inhibitor that blocks both the initiator and effector caspases. Besides regulating apoptosis, XIAP is also involved in other important cellular processes such as receptor-mediated signaling, cell cycle progression and protein ubiquitination (27,28). Given its distinct roles in various cellular functions, XIAP expression is tightly regulated at multiple levels, transcriptionally, post-transcriptionally and post-translationally. It has been shown that the stress-inducible transcriptional activator NF-κB enhances XIAP gene transcription in endothelial cells, and this mechanism also regulates expression of the other IAP genes (29,30). The levels of XIAP and other IAPs are further regulated by ubiquitin-mediated proteolysis (31,32). In addition, XIAP is also uniquely regulated at the level of protein synthesis (22,23). There is a strong internal ribosome entry site (IRES) within the XIAP 5′-UTR, and the IRES-dependent XIAP translation allows its protein to be continuously synthesized under conditions in which global translation is shut off (33). However, little is known about the regulation of XIAP mRNA stability.
An en masse search for HuR target mRNAs (34) identified the XIAP mRNA as a putative HuR target and computationally detected several hits of the HuR signature motif in the XIAP 3′-UTR and coding region (CR). Here, we set out to study if HuR binds the XIAP mRNA, and to elucidate the functional consequences of this interaction. Given our long-standing interest in understanding polyamine functions in gut mucosal homeostasis, we studied the influence of HuR on XIAP mRNA stability as a function of polyamine levels. HuR was found to associate with the XIAP mRNA and enhanced its stability. Moreover, polyamine depletion increased the cytoplasmic HuR levels, the abundance of [HuR/XIAP mRNA] complexes, the half life of XIAP mRNA and the steady state levels of XIAP. In turn, the HuR-elevated XIAP suppressed IEC apoptosis, as silencing XIAP or HuR sensitized IEC cells to apoptotic cell death.
MATERIALS AND METHODS
Chemicals and cell culture
Tissue culture medium and dialyzed fetal bovine serum were from Invitrogen (Carlsbad, CA, USA) and biochemicals were from Sigma (St Louis, MO, USA). The antibodies recognizing HuR and β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and the secondary antibody conjugated to horseradish peroxidase was purchased from Sigma. The antibody against XIAP was obtained from BD Bioscience, while anti-EGFP antibody was from Clontech (Mountain View, CA, USA). α-Difluoromethylornithine (DFMO) was from Genzyme (Cambridge, MA, USA).
The IEC-6 cell line, derived from normal rat intestinal crypt cells (35), was used at passages 15–20; and cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum. Stable Cdx2-transfected cells (IEC-Cdx2L1) were developed and characterized by Suh and Traber (36) and were a kind gift from Dr Peter G. Traber (Baylor College of Medicine, Houston, TX, USA). The expression vector, the LacSwitch System (Stratagene, La Jolla, CA, USA), was used for directing the conditional expression of the Cdx2 gene, and IPTG served as the inducer for the gene expression. Before experiments, IEC‐Cdx2L1 cells were grown in DMEM containing 4 mM IPTG for 16 days to induce cell differentiation as described earlier (37,38).
Recombinant virus construction and reporter plasmids
Recombinant adenoviral plasmids containing human HuR were constructed by using the Adeno-X Expression System according to the protocol provided by the manufacturer (Clontech). Briefly, the full-length cDNA of human wild-type HuR was cloned into the pShuttle by digesting the BamHI/HindIII and ligating the resultant fragments into the XbaI site of the pShuttle vector (19). pAdeno-HuR (AdHuR) was constructed by digesting the pShuttle construct with PI-SceI/I-CeuI and ligating the resultant fragment into the PI-SceI/I-CeuI sites of the pAdeno-X adenoviral vector. Recombinant adenoviral plasmids were packaged into infectious adenoviral particles by transfecting human embryonic kidney (HEK)-293 cells using LipofectAMINE Plus reagent (Gibco-Bethesda Res Lab, Gaithersburg, MD, USA). Titers of the adenoviral stock were determined by standard plaque-forming assay. Recombinant adenoviruses were screened for the expression of the introduced gene by western blot analysis using anti-HuR antibody. pAdeno-X, which was the recombinant replication-incompetent adenovirus carrying no HuR cDNA insert (Adnull), was grown and purified as described above and served as a control adenovirus. Cells were infected with AdHuR or Adnull, and expression of HuR was assayed at 24 or 48 h after the infection.
The EGFP reporter constructs, pEGFP-N1 and pEGFP-C1 (BD Bioscience), were used for measurement of XIAP CR and 3′-UTR activity. The complete XIAP CR without start codon was amplified by PCR using primers GCGAATTCACTTTTA-ACAGTTTTGAAGGATCT and CCGGTACCGCTTGAACGTAATGACTGTG, and the resulting PCR fragment was cloned upstream of the EGFP CR in plasmid pEGFP-N1. The XIAP 3′-UTR was amplified by PCR using primers GCGAATTCTTAATCCAG-CACCACAGTAGGCATGT and CCGGTACCTATACATAGTATATCTGTGAT, and the PCR fragment was cloned downstream of the EGFP CR in plasmid pEGFP-C1. Transient transfections were performed using the LipofectamineTM reagent following the manufacturer’s instructions (Invitrogen). The levels of EGFP expressed from these constructs were examined by western blot analysis.
RNA interference
The silencing RNA duplexes that were designed to specifically cleave HuR mRNA were synthesized and transfected into cells as described previously (21). The sequence of small interfering RNA (siRNA) that specifically targets HuR mRNA (siHuR) was AACACGCTGAACGGCTTGAGG; while the sequence of control siRNA (C-siRNA) was AAGTGTAGTAGATCACCAGGC. The siRNA that was designed to specifically cleave XIAP mRNA (siXIAP) was CAGCAGTCCTGTTTCAGCATCAACA, whereas the sequence of its C-siRNA was CAGCTGGTCTTTGACTACACCAACA. For each 60-mm cell culture dish, 15 µl of the 20 µM stock duplex siHuR, siXIAP or C-siRNA was mixed with 300 µl of Opti-MEM medium (Invitrogen). This mixture was gently added to a solution containing 15 µl of LipofectAMINE 2000 in 300 µl of Opti-MEM. The solution was incubated for 20 min at room temperature and gently overlaid onto monolayers of cells in 3 ml of medium, and cells were harvested for various assays after 48 h later.
Western blot analysis
Whole-cell lysates were prepared using 2% SDS, sonicated and centrifuged (12 000 r.p.m.) at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS‐PAGE (7.5% acrylamide). After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing XIAP, HuR or EGFP; following incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
Reverse transcription–PCR and real-time PCR analysis
Total RNA was isolated by using RNeasy mini kit (Qiagen, Valencia, CA, USA) and used in reverse transcription (RT) and PCR amplification reactions as described (19). PCR primers for detecting rat XIAP mRNA (CR) were 5′-CTTTCCCAACTGCTTCTTCG-3′ (sense) and 5′-CGCCTTAGCTGCTCTTCAGT-3′ (antisense). The levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PCR product were assessed to monitor the evenness in RNA input in RT‐PCR samples. Real-time quantitative PCR (qPCR) analysis was performed using 7500-Fast real-time PCR systems with specific primers, probes and software (Applied Biosystems, Foster City, CA, USA).
Biotin pull down assays
The synthesis of biotinylated transcripts and analysis of RBPs bound to biotinylated RNA were done as previously described (39). Complementary DNA from IEC-6 cells was used as a template for PCR amplification of the CR, 3′-UTR, 5′-UTR of XIAP. The 5′-primers contained the T7 RNA polymerase promoter sequence (T7): 5′-CCAAGCTTCTAATACGACT-CACTATAGGGAGA-3′. To prepare the XIAP CR template (spanning position 331–1822), oligonucleotides (T7): 5′-CAAGTTACATGTGAATTCTGCCAGGC-3′ and 5′-CGATCTGTGAAAGAGCAGGCTCTGTACTC-3′ were used. To prepare the XIAP 3′-UTR template (spanning position 1823–2469), oligonucleotides (T7): 5′-CAGCACCACAGTAGGCATGT-3′ and 5′-TCTGTGATGCTTTTCTATGTCAG-3′ were used. To prepare the XIAP 5′-UTR template (spanning position 1–330), oligonucleotides (T7): 5′-TCGGGCTGCATAATGAGGACTG-3′ and 5′-CTTCTCTTGAAAATACGACT-3′ were used. All sequences of oligonucleotides for preparation of various short RNA probes for mapping the XIAP CR and 3′-UTR were described in Supplementary Table S1. PCR-amplified products were used as templates to transcribe biotinylated RNAs by using T7 RNA polymerase in the presence of biotin-cytidine 5′-triphosphate as described in Xiao et al. (19). Biotinylated transcripts (6 µg) were incubated with 120 µg of cytoplasmic lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) and analyzed by western blot analysis.
RNP immunoprecipitation assays
To assess the association of endogenous HuR with endogenous XIAP mRNA, immunoprecipitation (IP) of RNP complexes were performed as described (34,21). Twenty million cells were collected per sample, and lysates were used for IP for 4 h at room temperature in the presence of excess (30 µg) IP antibody (IgG or anti-HuR). RNA in IP materials was used in RT followed by PCR and RT‐qPCR analysis to detect the presence of XIAP and GAPDH mRNAs.
Immunofluorescence staining
Immunofluorescence was performed as described in Vielkind and Swierenga (40) with minor changes (21). Cells were fixed using 3.7% formaldehyde, and the rehydrated samples were incubated overnight at 4°C with primary antibody against XIAP diluted 1:300 in blocking buffer and then incubated with secondary antibody conjugated with Alexa Fluor-594 (Molecular Probes, Eugene, OR, USA) for 2 h at room temperature. After rinsing, slides were incubated with 1 µM TO-PRO3 (Molecular Probes) for 10 min to stain nuclei, rinsed again, mounted and viewed through a Zeiss confocal microscope (model LSM410). Images were processed using PhotoShop software (Adobe, San Jose, CA, USA).
Assays for ODC enzyme activity and cellular polyamine content
ODC activity was determined by radiometric technique in which the amount of 14CO2 liberated from L-[1-14C]ornithine was estimated (41), and enzymatic activity was expressed as picomole of CO2 per milligram of protein per hour. The cellular polyamine content was analyzed by high-performance liquid chromatography as described previously (42). After 0.5 M perchloric acid was added, the cells were frozen at −80°C until ready for extraction, dansylation and HPLC analysis. The standard curve encompassed 0.31–10 µM. Values that fell >25% below the curve were considered as undetectable. The results are expressed as nanomoles of polyamines per milligram of protein.
Statistics
Values are mean ± SE from three to six samples. Immunoblotting and immunofluorescence staining were repeated three times. The significance of the difference between means was determined by analysis of variance. The level of significance was determined using Duncan’s multiple range test (43).
RESULTS
HuR directly interacts with the XIAP mRNA via AREs
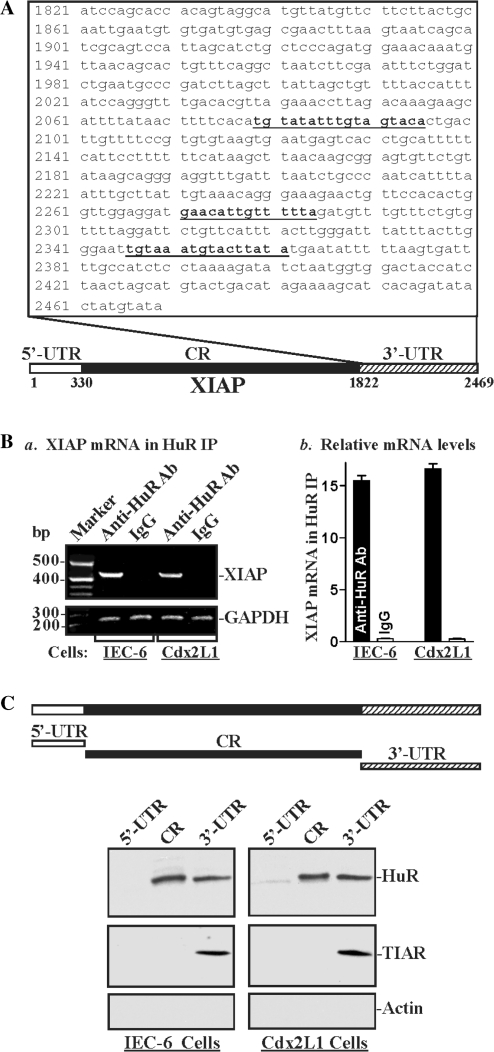
There are three computationally predicted hits of the HuR motif in the XIAP 3′-UTR (Figure 1A) and five potential HuR hits in the XIAP CR (Supplementary Table S2), suggesting that XIAP mRNA might be a direct target of HuR. We first examined if XIAP mRNA associated with HuR by performing RNP-IP assays using anti-HuR antibody under conditions that preserved RNP integrity (44). The interaction of XIAP mRNA with HuR was examined by isolating RNA from the IP material and subjecting it to RT, followed by either conventional PCR or real-time qPCR analyses. As shown in Figure 1B, the XIAP PCR products were highly enriched in HuR samples compared with control IgG samples in both parental IEC-6 and differentiated IEC-Cdx2L1 cells. The enrichment of c-Myc PCR product was also examined and served as a positive control (data not shown), since c-Myc mRNA is a target of HuR (18), while the amplification of GAPDH PCR products, found in all samples as low-level contaminating housekeeping transcripts (not HuR targets), served to monitor the evenness of sample input, as reported previously (39,45). HuR interacted specifically with XIAP mRNA, as mRNAs encoding other members of the IAP family (c-IAP1 and c-IAP2 mRNAs) were undetectable in HuR IP samples (data not shown). [HuR/XIAP mRNA] associations were further tested by using biotinylated transcripts that spanned the XIAP mRNA regions shown in Figure 1C (schematic). Following incubation with cytoplasmic lysates, the interaction between the biotinylated XIAP transcripts and HuR was examined by biotin pull down followed by western blot analysis (44,21). As shown, the XIAP 3′-UTR transcripts readily associated with HuR in parental IEC-6 cells and differentiated IEC-Cdx2L1 cells. HuR did not interact with a 330-bp region of the XIAP 5′-UTR immediately upstream of AUG in either cell line, but we could not rule out that HuR may interact further upstream, as the XIAP 5′-UTR is >5 kb in length. However, it did show prominent binding to the XIAP CR. The RBP TIAR also formed complexes with the XIAP 3′-UTR but not with its CR or 5′-UTR. Neither the XIAP 3′-UTR nor its CR interacted with β-actin, included here as a negative control.
Figure 1.
HuR binds the XIAP mRNA. (A) Schematic representation of the XIAP mRNA and the predicted hits of the HuR signature motif in its 3′-UTR. (B) Association of endogenous HuR with endogenous XIAP mRNA in IEC-6 cells and differentiated IECs (Cdx2L1 cells). After IP of RNA–protein complexes from cell lysates using either anti-HuR antibody (Ab) or control IgG1, RNA was isolated and used in RT reactions. (a) Representative RT–PCR products visualized in ethidium bromide-stained agarose gels; low-level amplification of GAPDH (housekeeping mRNA, which is not HuR targets) served as negative controls. (b) Fold differences in XIAP transcript abundance in HuR IP compared with IgG IP, as measured by RT–qPCR analysis. Values were mean ± SE from triplicate samples. (C) Representative HuR and TIAR immunoblots using the pull down materials by different fractions of XIAP mRNA. Top panel shows schematic representation of the XIAP biotinylated transcripts (5′-UTR, CR and 3′-UTR) used in this study. Cytoplasmic lysates prepared from IEC-6 and differentiated Cdx2L1 cells were incubated with 6 µg of biotinylated XIAP 5′-UTR, CR, or 3′-UTR for 30 min at 25°C, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. The presence of HuR or TIAR in the pull down material was assayed by western blotting. β-Actin in the pull down material was also examined and served as a negative control. Three experiments were performed that showed similar results.
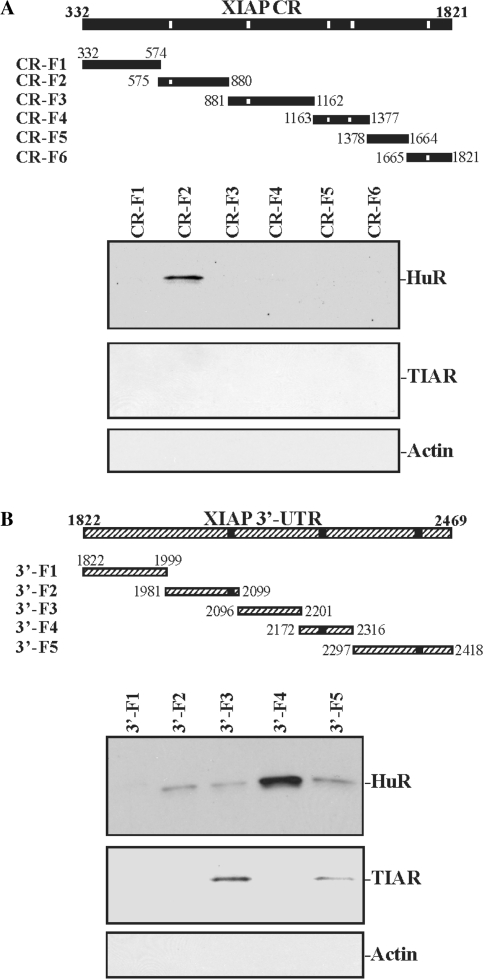
To determine if HuR binds to specific regions of the XIAP CR and 3′-UTR, we tested different segments of the XIAP CR and 3′-UTR. Partial biotinylated transcripts spanning the XIAP CR (spanning positions 332–1821) were prepared as shown in Figure 2A (schematic), and their associations with HuR were tested in pull down assays. HuR was found to specifically bind to the fragment CR-F2, the transcript which contained one hit of the HuR signature motif. In contrast, there was no detectable binding of HuR to fragments CR-F1 and CR-F5 (which lacked putative HuR hits) or fragments CR-F3, CR-F4 and CR-F6 (which contained several predicted HuR hits); in contrast, all CR fragments of XIAP mRNA failed to bind to TIAR. To identify the specific HuR binding sites in the XIAP 3′-UTR, different biotinylated transcripts spanning the XIAP 3′-UTR (spanning positions 1822–2469) were prepared (Figure 2B, schematic). HuR formed much more prominent complexes with the fragment 3′-F4 (containing one predicted HuR hit) than with other XIAP 3′-UTR fragment. HuR bound 3′-F2 and 3′-F5 fragments (containing predicted HuR hits) slightly, while HuR did not bind 3′-F1 or 3′-F3 fragments, which lacked potential HuR hits. This strong binding affinity of the fragment 3′-F4 for HuR was specific, because TIAPs were found to bind to 3′-F3 and 3′-F5 fragments but not to the 3′-F4. Although these assays do not demonstrate the interaction of HuR with XIAP in intact cells, they support the notion that HuR can specifically associate with the endogenous XIAP mRNA and with in vitro biotinylated XIAP RNA, and that these interactions occur at the XIAP 3′-UTR and CR segments containing predicted HuR interaction sites.
Figure 2.
HuR binding to different fractions of CR and 3′-UTR of the XIAP mRNA. (A) Representative HuR immunoblots in the materials pulled down by different biotinylated XIAP CR fractions (F); Top panel is a schematic representation of the XIAP CR biothinylated transcripts used in biotin pull down assays. After incubation of cytoplasmic lysates prepared from IEC-6 cells with various XIAP CR biotinylated fractions, the resulting RNP complexes were pulled down by using streptavidin-coated beads. The abundance of proteins HuR, TIAR and β-actin (which served as negative control) in the pull down material was examined by western blot analysis. (B) Representative HuR immunoblots in the material pulled down by different biotinylated fractions of the XIAP mRNA 3′-UTR. Top panel is a schematic representation of the XIAP 3′-UTR biotinylated transcripts used in this study. Three experiments were performed that showed similar results.
[HuR/XIAP mRNA] associations enhance the stability of XIAP mRNA
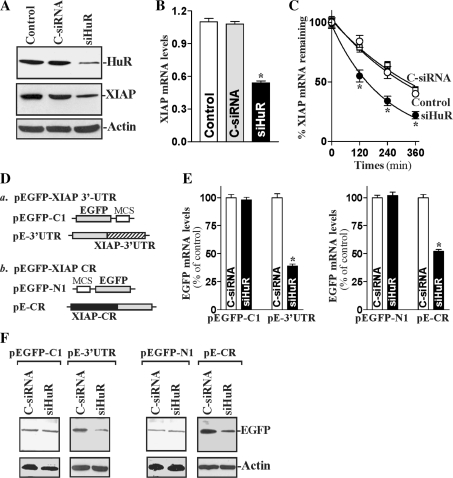
To determine the functional consequences of [HuR/XIAP mRNA] associations, HuR levels were reduced by siRNA targeting the HuR mRNA (siHuR). These specific siHuR nucleotides were designed to reduce rat HuR mRNA and to have a unique combination of specificity, efficacy and low toxicity (18,19). IEC-6 cells transfected with siHuR showed <20% of HuR levels as compared with the levels seen in the control group and cells transfected with C-siRNA (Figure 3A); this reduction was specific, as other RBPs such as TIAR were not affected in HuR-silenced populations (data not shown). Importantly, HuR silencing significantly reduced XIAP protein (Figure 3A, middle) and its mRNA levels (Figure 3B). To ascertain if the reduction in the XIAP expression in HuR-silenced populations was due to destabilization of XIAP mRNA, we first examined changes in the half life of XIAP mRNA after silencing HuR. As shown in Figure 3C, silencing HuR destabilized XIAP mRNA, because the XIAP mRNA half life was reduced in HuR-silenced populations (∼160 min) compared with those measured in controls (∼305 min) and in cells transfected with C-siRNA (∼318 min). Second, we determined if HuR regulates the stability of XIAP mRNA by interacting with its 3′-UTR, CR or both. We prepared reporter constructs that expressed chimeric RNA containing the EGFP and XIAP 3′-UTR or XIAP CR (Figure 3D) (32,33). HuR silencing reduced the XIAP mRNA levels by inhibiting the interaction of HuR with both the XIAP 3′-UTR and its CR, since the levels of EGFP mRNA (Figure 3E) and protein (Figure 3F) expressed from both pEGFP-XIAP 3′-UTR and pEGFP-XIAP CR were decreased in HuR-silenced populations of cells. We also examined changes in EGFP levels when cells were transfected with the control pEGFP vectors; as shown, HuR silencing failed to reduce levels of EGFP mRNA and protein expressed from the pEGFP-C1 (without XIAP 3′-UTR) and from pEGFP-N1 (without XIAP CR). Taken together, these results strongly support the notion that HuR regulates the stability of XIAP mRNA by interacting with both its 3′-UTR and CR.
Figure 3.
Changes in XIAP mRNA stability after HuR silencing. (A) Representative immunoblots of HuR and XIAP proteins in HuR-silenced cells. After IEC-6 cells were transfected with either siRNA targeting the HuR mRNA CR (siHuR) or C-siRNA for 48 h, whole-cell lysates were harvested for western blot analysis to monitor the expression of HuR and XIAP, and loading control β-actin. (B) Levels of XIAP mRNA in cells that were processed as described in (A). Total RNA from each group was harvested, and levels of XIAP mRNA were measured by RT–qPCR analysis. Data were normalized to amount of GAPDH, and values are mean ± SE of data from triplicate experiments. *P < 0.05 compared with controls and cells transfected with C-siRNA. (C) Half life of the XIAP mRNA in cells described in (A). Total cellular RNA was isolated at indicated times after administration of actinomycin D (5 µg/ml), and the remaining levels of XIAP and GAPDH mRNAs were measured by RT–qPCR analysis. Values were mean ± SE from triplicate experiments. *P < 0.05 compared with control cells and cells transfected with C-siRNA. (D) Schematic of plasmids: (a) chimeric EGFP-XIAP 3′-UTR; and (b) chimeric EGFP-XIAP CR. (E) Levels in reporter EGFP mRNA in cells that were processed as described in (A). Cells were transfected with the constructs described in (D), and 48 h later, the levels of EGFP mRNA expressed from control plasmids (pEGFP-C1; and pEGFP-N1) and pEGFP-XIAP 3′-UTR (pE-3′-UTR) or pEGFP-XIAP CR (pE-CR) constructs were tested by RT–qPCR analysis. Values are mean ± SE of data from triplicate experiments. *P < 0.05 compared with cells transfected with C-siRNA. (F) Levels in reporter EGFP protein as measured by western blot analysis in cells that were processed as described in (A). Three separate experiments were performed that showed similar results.
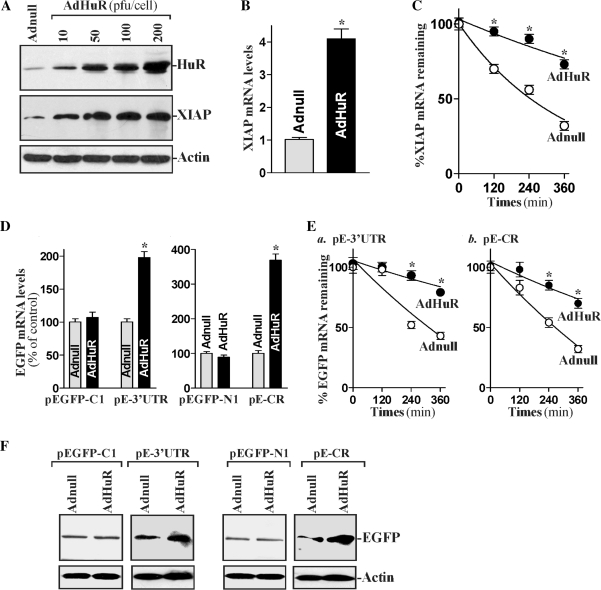
To further define the role of HuR in regulating XIAP mRNA stability, we examined the effect of overexpressing the wild-type HuR upon the half life of XIAP mRNA. The adenoviral vector containing the corresponding HuR cDNA under the control of the human cytomegalovirus immediate to early gene promoter (AdHuR) was generated as described in Abdelmohsen et al. (39). The adenoviral vectors used in this study infect IECs with nearly 100% efficiency (19,41 and data not shown). Infection with the AdHuR increased the levels of cytoplasmic and total HuR protein dramatically and also enhanced expression of XIAP (Figure 4A, middle, and B) significantly. Ectopic HuR expression increased the XIAP mRNA half life (Figure 4C), and this stimulatory effect is also mediated through HuR interaction with both the XIAP mRNA 3′-UTR and CR. HuR over expression increased reporter EGFP mRNA levels expressed from both the pEGFP-XIAP 3′-UTR and pEGFP-XIAP CR (Figure 4D), associated with enhanced EGFP mRNA stability (Figure 4E) and resulting in increased EGFP proteins (Figure 4F). In contrast, neither HuR protein levels nor XIAP expression were altered by infection with the control adenovirus (Adnull). These results indicate that HuR overexpression enhances XIAP expression by stabilizing the XIAP mRNA.
Figure 4.
Changes in XIAP mRNA stability after ectopic HuR overexpression. (A) Representative immunoblots of HuR and XIAP proteins after ectopic HuR expression. Cells were infected with the recombinant adenoviral vector encoding HuR cDNA (AdHuR, prepared as explained in ‘Materials and Methods’ section) or adenovial vector lacking HuR cDNA (Adnull) at a multiplicity of infection of 10–200 p.f.u. (plaque-forming units)/cell; the expression of HuR and XIAP proteins was analyzed 48 h after the infection. (B) Levels of XIAP mRNA as measured by RT–qPCR analysis in cells infected with the AdHuR or Adnull at the concentration of 100 p.f.u./cell for 48 h. Data were normalized to amount of GAPDH, and values are mean ± SE of data from triplicate experiments. *P < 0.05 compared with cells infected with Adnull. (C) Stability of the XIAP mRNA in cells that were processed as described in (B). The half life of the XIAP mRNA was measured as explained in the legend of Figure 3C. Values are the mean ± SE from triplicate samples. *P < 0.05 compared with cells infected with Adnull. (D) Levels of reporter EGFP mRNA in cells overexpressing HuR. After cells were transfected with one of the EGFP reporter constructs and infected with AdHuR or Adnull for 48 h, the levels of EGFP mRNA expressed from control plasmids (pEGFP-C1; and pEGFP-N1) and pEGFP-XIAP 3′-UTR (pE-3′-UTR) or pEGFP-XIAP CR (pE-CR) constructs were tested by RT–qPCR analysis. Values are the mean ± SE of data from triplicate experiments. *P < 0.05 compared with cells infected with Adnull. (E) The half life of the EGFP mRNA expressed from the pE-3′-UTR or pE-CR in cells described in (D). Data were normalized to GAPDH mRNA levels; *P < 0.05 compared with cells infected with Adnull. (F) Levels of reporter EGFP protein in cells described in (D). EGFP levels were examined by western blot analysis; equal loading was monitored by β-actin levels.
Polyamine depletion induces [HuR/XIAP mRNA] complexes enhancing XIAP mRNA stability
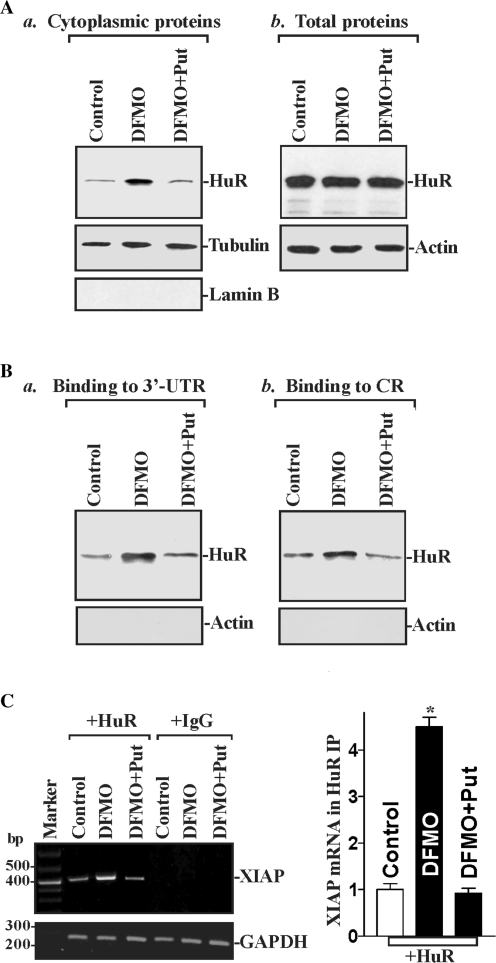
The natural polyamines (spermidine, spermine and their precursor putrescine) are ubiquitous, small basic molecules and are crucial for maintaining the intestinal epithelial integrity under biological conditions (47–49). Our previous studies showed that polyamines regulate XIAP expression in IECs (50), but the exact mechanism underlying this process remains unknown. Therefore, we examined if HuR plays a role in the regulation of XIAP expression by polyamines. Consistent with our previous studies (19,51), exposure of IEC-6 cells to 5 mM DFMO for 4 days completely inhibited ODC enzyme activity (the first rate-limiting step for polyamine biosynthesis) and almost totally depleted cellular polyamines. Putrescine and spermidine were undetectable by 4 days of continuous treatment with DFMO, and spermine had decreased by ∼60% (data not shown). As reported in our previous studies (20,21), polyamine depletion by DFMO also increased cytoplasmic HuR levels, although it did not change whole-cell HuR levels (Figure 5A). Supplementation with putrescine reversed the DFMO-triggered changes in HuR subcellular distribution, as spermidine supplementation did (data not shown). To monitor the quality of the cytoplasmic preparation, we examined the levels of β-tubulin (a cytoplasmic protein) and lamin B (a nuclear protein). Assessment of these markers revealed that there was no significant contamination of nuclear proteins in the cytoplasmic fractions. Based on these observations, we hypothesized that HuR association with the XIAP mRNA would increase in the cytoplasm following polyamine depletion. To test the possibility, two experiments were performed. First, we used biotinylated transcripts spanning the XIAP 3′-UTR or CR in RNA pull down assays using cell lysates prepared from either untreated or polyamine-deficient cells. Polyamine depletion increased HuR association with XIAP mRNA, and this induction was mediated through both XIAP 3′-UTR and CR (Figure 5B). The binding intensity increased significantly while using lysates prepared from cells that were treated with DFMO for 4 days, but was reduced when cells had been treated with DFMO plus putrescine. Second, we examined the in vivo association of endogenous XIAP mRNA with endogenous HuR following polyamine depletion as measured by RNP IP followed by RT‐qPCR analysis. As shown in Figure 5C, the association of endogenous XIAP mRNA with endogenous HuR was also increased in cells treated with DFMO, but decreased while testing lysates from cells treated with DFMO plus putrescine. The XIAP mRNA was undetectable in nonspecific IgG1 IPs in all three groups. These results indicate that decreasing cellular polyamines increase HuR association with the XIAP mRNA in IECs.
Figure 5.
Changes in HuR binding to the XIAP mRNA following polyamine depletion. (A) Representative immunoblots of cytoplasmic HuR (a) and total HuR (b) in control cells and cells treated with DFMO (5 mM) alone or DFMO plus putrescine (Put, 10 µM) for 4 days. Whole-cell and cytoplasmic lysates were prepared and subjected to SDS–PAGE. After detecting HuR, blots were reprobed to detect β-tubulin and lamin B in cytoplasmic and actin in whole-cell lysates to control for the quality of the fractionation procedure and the even loading of samples. (B) Representative HuR immunoblots using the biotin pull down materials from the XIAP 3′-UTR (a) or XIAP CR (b) in cells that were processed as described in (A). After cytoplasmic lysates were incubated with biotinylated XIAP 3′-UTR or CR, the resulting RNP complexes were pulled down; and levels of HuR and β-actin proteins in pull down materials were examined by western blotting. Three experiments were performed that showed similar results. (C) Association of endogenous HuR with endogenous XIAP mRNA in cells described in (A). Whole-cell lysates from all three groups were used for IP in the presence of anti-HuR antibody or nonspecific IgG1. Levels of XIAP and GAPDH mRNAs in the IP materials were examined by RT–PCR analysis (left) and RT–qPCR analysis (right). Values are the mean ± SE of data from three separate experiments. *P < 0.05 compared with control cells and cells exposed to DFMO plus Put.
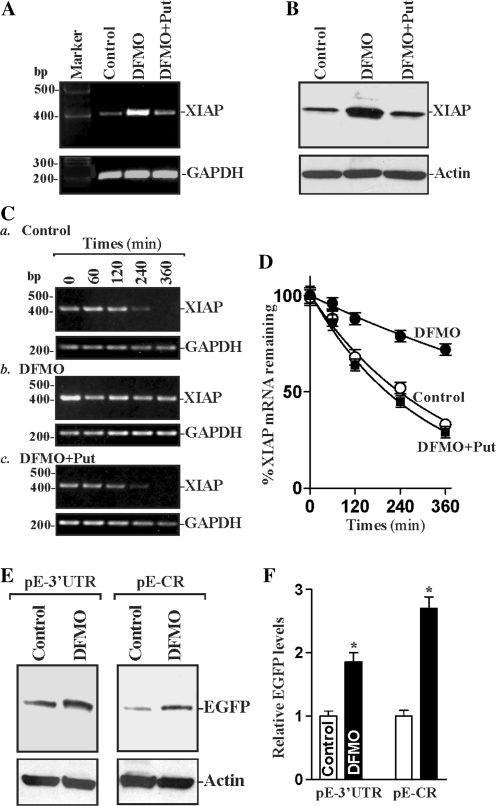
The induction in [HuR/XIAP mRNA] complexes in polyamine-deficient cells was associated with an increase in XIAP expression. The steady state levels of XIAP mRNA (Figure 6A) and protein (Figure 6B) increased significantly in cells treated with DFMO for 4 days, but this induction was completely prevented by addition of exogenous putrescine. To test if the increased XIAP mRNA levels by polyamine depletion were instead influenced by changes in mRNA turnover, we measured the half life of XIAP mRNA in different polyamine levels. As shown in Figure 6C, depletion of cellular polyamines by DFMO increased the stability of XIAP mRNA, which was prevented by exogenous putrescine. The half life of XIAP mRNA in cells exposed to DFMO plus putrescine was similar to that of control cells (Figure 6D). The results presented in Figure 6E and F further showed that polyamine depletion enhanced XIAP expression through a process involving both the XIAP 3′-UTR and CR. There were significant increases in the levels of reported EGFP expressed from the pEGFP-XIAP 3′-UTR and pEGFP-XIAP CR constructs in polyamine-deficient cells compared with those observed in control cells. On the other hand, there were no significant differences in the levels of EGFP expressed from the control vectors pEGFP-C1 and pEGFP-N1 between control cells and DFMO-treated cells (data not shown). These findings strongly suggest that polyamine depletion increases XIAP expression by inducing XIAP mRNA stability.
Figure 6.
Changes in XIAP expression and XIAP mRNA stability in polyamine-deficient cells. (A) PCR-amplified products displayed in agarose gels for mRNAs of XIAP and GAPDH. Total cellular RNA was harvested from control cells and cells treated with DFMO alone or DFMO plus Put for 4 days, and levels of XIAP and GAPDH mRNAs were measured by RT–PCR analysis. Three experiments were performed that showed similar results. (B) Representative immunoblots of XIAP protein as measured by western blotting in cells that were treated as described in (A). (C) Half life of XIAP mRNA in cells that were processed as described in (A). After cells were grown in control cultures and cultures containing DFMO alone or DFMO plus Put for 4 days, they were exposed to actinomycin D; and levels of XIAP and GAPDH mRNAs were measured at indicated times by RT–PCR analysis. (D) Percentage of XIAP mRNA remaining in the cells that were treated as described in (C). Values are mean ± SE of data from three separate experiments, and relative levels of XIAP mRNA were normalized to the amount of GAPDH (optical density of XIAP mRNA/optical density of GAPDH mRNA). (E) Levels in reporter gene EGFP protein in cell that were processed as described in (A). After cells were exposed to DFMO for 2 days, they were transfected with the pEGFP-XIAP 3′-UTR or pEGFP-XIAP CR constructs and then cultured for additional 48 h in the presence of DFMO. The levels of EGFP expressed from the pEGFP-XIAP 3′-UTR or pEGFP-XIAP CR were tested by western blotting. (F) Quantitative densitometric analysis of EGFP immunoblots described in (E). Values are mean ± SE of data from three separate experiments. *P < 0.05 compared with controls.
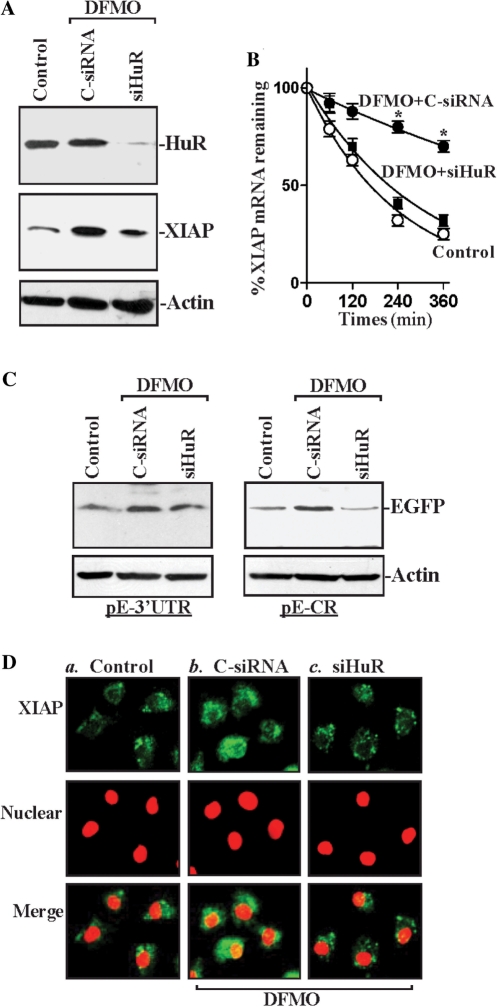
To further define the putative role of induced [HuR/XIAP mRNA] association in the induction of XIAP mRNA stability following polyamine depletion, siHuR was used to reduce HuR levels in DFMO-treated cells. Transfection with siHuR not only potently silenced HuR expression (Figure 7A) but also attenuated the increased stability of XIAP mRNA (Figure 7B) and its induced protein levels in polyamine-deficient cells. The half life of XIAP mRNA and XIAP protein content in DFMO-treated cells transfected with siHuR was similar to those observed in control cells (without DFMO). Results presented in Figure 7C also show that HuR silencing prevented the induced XIAP expression in polyamine-deficient cells by reducing the interaction of HuR with both the XIAP 3′-UTR and CR, as evidenced by a decrease in levels of reporter EGFP expressed from both the pEGFP-XIAP 3′-UTR and the pEGFP-XIAP CR. In addition, induced XIAP immunostaining following polyamine depletion was predominantly located in the cytoplasm (Figure 7D), whereas HuR silencing prevented the increase in XIAP signal, rendering the subcellular localization patterns identical to those seen in control cells. Together, these findings indicate that polyamine depletion enhances XIAP expression by inducing [HuR/XIAP mRNA] association.
Figure 7.
Effect of HuR silencing on XIAP expression and its mRNA stability in polyamine-deficient cells. (A) Representative HuR and XIAP immunoblots. After cells were grown in cultures containing DFMO for 2 days, they were transfected with either siHuR or C-siRNA, and whole-cell lysates were harvested 48 h thereafter. The levels of HuR and XIAP proteins were measured by western blot analysis, and equal loading was monitored by β-actin immunoblotting. (B) Half life of the XIAP mRNA in cells that were transfected and treated as described in (A). Total cellular RNA was isolated at the indicated times after administration of actinomycin D, and the remaining levels of XIAP and GAPDH mRNAs were measured by RT–qPCR analysis. Values are mean ± SE from triplicate samples. *P < 0.05 compared with control cells and DFMO-treated cells transfected with siHuR. (C) Levels of reporter EGFP protein in cells that were processed as described in (A). After cells were exposed to DFMO for 2 days, they were co-transfected with the siHuR or C-siRNA with the pEGFP-XIAP 3′-UTR or pEGFP-XIAP CR. Levels of EGFP were examined 48 h after co-transfection. Three separated experiments were performed that showed similar results. (D) Cellular distribution of XIAP in cells that were processed as described in (A): (a) control; (b) DFMO-treated cells trasfected with C-siRNA; and (c) DFMO-treated cells transfected with siHuR. Cells were permeabilized and incubated with the anti-XIAP antibody and then with anti-IgG conjugated with Alexa Fluor. Nuclei were stained with the TO-PRO3. XIAP was shown as green, while nuclei were shown as red color. Original magnification: ×1000.
Induced XIAP plays an important role in the induced resistance of polyamine-deficient cells to apoptosis
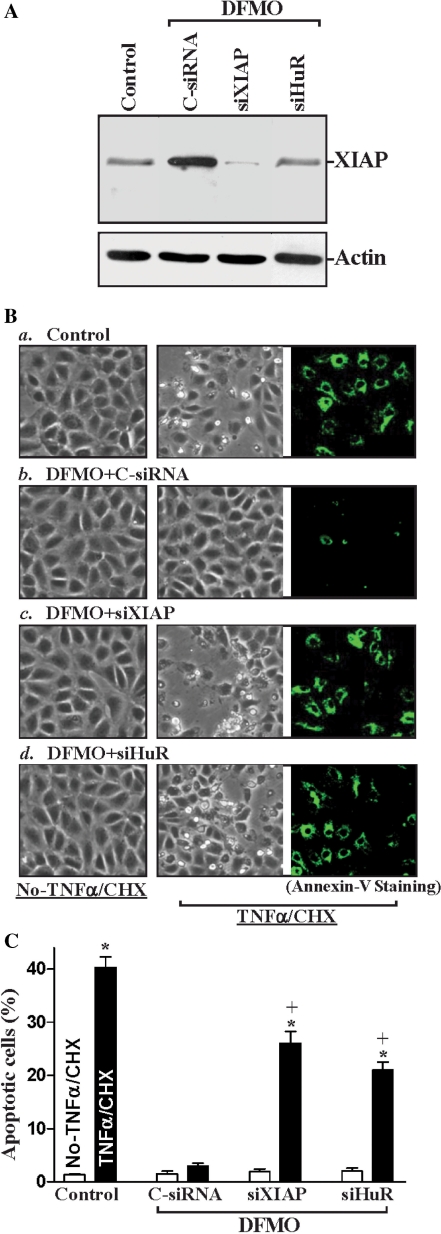
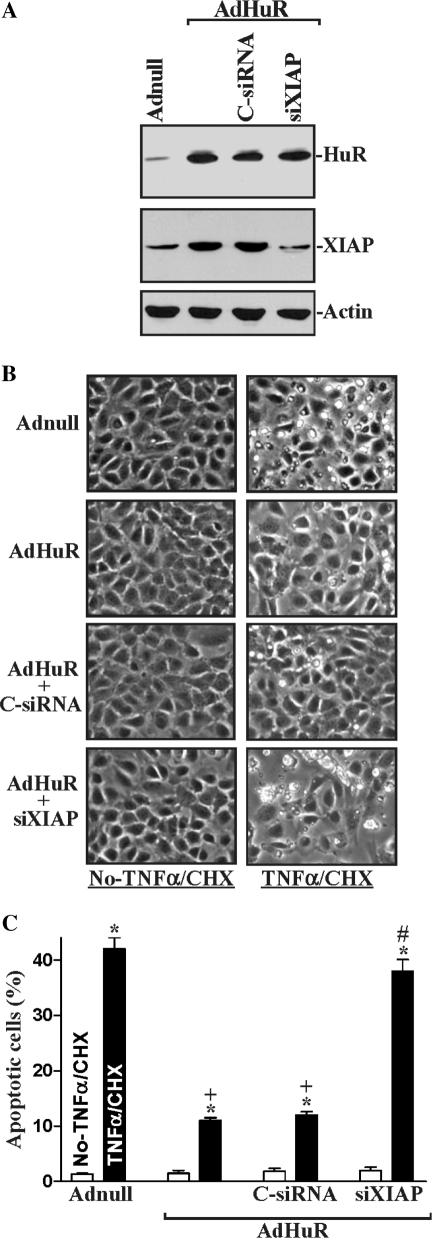
To investigate the physiological consequences of inducing endogenous XIAP by HuR following polyamine depletion, we examined the possible involvement of this process in regulating IEC apoptosis. Our previous studies (52,53) and the work of others (49,54,55) have shown that polyamines regulate apoptosis through multiple signaling pathways, and that polyamine depletion promotes resistance to apoptosis in normal IECs. Here, we first examined the spontaneous apoptotic cell death without any challenge of apoptotic stimulators after inhibition of XIAP expression by using siRNA targeting XIAP mRNA (siXIAP) or siHuR in the absence of cellular polyamines. Transient transfection with the siXIAP or siHuR prevented the increased expression of XIAP in polyamine-deficient cells (Figure 8A) but failed to directly induce apoptosis in polyamine-deficient cells (Figure 8B, left) or normal cells (data not shown). There were no apparent differences in cell viability in DFMO-treated cells transfected with C-siRNA compared with DFMO-treated cells transfected with siXIAP or siHuR. Second, we determined whether XIAP silencing altered the polyamine depletion-mediated resistance to apoptosis elicited by treatment with tumor necrosis factor-α (TNF-α) plus cycloheximide (CHX). This apoptotic model was chosen because TNF-α/CHX-induced apoptosis is widely accepted as a form of programmed cell death induced by a biological apoptotic inducer in IECs (49–54). TNF-α is a proinflammatory cytokine and it has a potent cytotoxic effect on IECs, especially when protein synthesis is inhibited (52,56). Because increased release of TNF-α in the gut mucosa occurs commonly together with reduced protein synthesis under various physiological and pathological conditions, this apoptosis model is especially applicable to the in vivo condition and was chosen for our study. As shown in Figure 8Ba, when control cells were exposed to TNF-α/CHX for 4 h, morphological features characteristic of programmed cell death were observed; and annexin V staining showed significant phosphatidylserine presence in the cell membrane, a classic indicator of apoptotic cells (Figure 8Ba, right). Consistent with our previous studies, exposure of polyamine-deficient cells to the same doses of TNF-α/CHX caused no apoptosis; likewise, there were no differences in the morphological features and percentages of apoptotic cells when comparing cells treated with DFMO alone and DFMO-treated cells exposed to TNF-α/CHX for 4 h (52,53 and data not shown). This increased resistance to TNF-α/CHX-induced apoptosis was not altered when polyamine-deficient cells were transfected with C-siRNA (Figure 8Bb), but it was lost when XIAP expression was silenced by siXIAP (Figure 8Bc) or siHuR (Figure 8Bd). The percentages of apoptotic cells (Figure 8C) in DFMO-treated cells transfected with siXIAP or siHuR were significantly increased compared with those observed in DFMO-treated cells transfected with C-siRNA after exposure to TNF-α/CHX. Third, we further examined changes in TNF-α/CHX-induced apoptosis after ectopic HuR overexpression in XIAP-silenced cells. As shown in Figure 9, HuR overexpression protected IEC-6 cells against TNF-α/CHX-induced apoptosis, but this protective effect was reduced by XIAP silencing. The numbers of apoptotic cells in HuR-expressing populations were lower than those observed in groups infected with Adnull after exposure to TNF-α/CHX. HuR-mediated protection was unaltered by transfection with C-siRNA (Figure 9B, middle), but it was reversed by XIAP silencing (Figure 9B, bottom). The percentage of apoptotic cells in XIAP-silenced cells infected with AdHuR increased remarkably after exposure to TNF-α/CHX (Figure 9C). In addition, inhibition of XIAP expression in normal cells (without DFMO) by siXIAP or siHuR also potentiated the TNF-α/CHX-induced apoptosis (Supplementary Figure S1). Taken together, these results indicate that the HuR-mediated increase in XIAP expression contributes to an increase in resistance to apoptosis following polyamine depletion.
Figure 8.
Effects of XIAP silencing or reduced XIAP by silencing HuR on apoptotic sensitivity in polyamine-deficient cells. (A) Representative immunoblots for XIAP protein. Cells were grown in the cultures containing DFMO for 2 days and then transfected with either siRNA specifically targeting XIAP mRNA CR (siXIAP), siHuR or C-siRNA. The levels of XIAP protein were measured by western blot analysis 48 h after transfection; equal loading was monitored by β-actin immunoblotting. Three separate experiments were performed that showed similar results. (B) TNF-α/CHX-induced apoptosis in cells described (A): (a) control cells; (b) DFMO-treated cells transfected with C-siRNA; (c) DFMO-treated cells transfected with siXIAP; and (d) DFMO-treated cells transfected with siHuR. Apoptosis was measured by morphological analysis (middle) and ApoAlert annexin V staining (right) 4 h after treatment with TNF-α/CHX. Original magnification: ×150. (C) Percentage of apoptotic cells as described in (B). Values are mean ± SE of data from six samples. *P < 0.05 compared with no-TNF-α/CHX. +P < 0.05 compared with DFMO-treated cells that were transfected with C-siRNA and then treated with TNF-α/CHX for 4 h.
Figure 9.
Effect of XIAP silencing on apoptotic sensitivity in cells overexpressing HuR. (A) Representative HuR and XIAP immunoblots. After cells were transfected with either C-siRNA or siXIAP for 24 h, they were infected with the AdHuR or Adnull (100 p.f.u./cell). HuR and XIAP proteins were examined by western blot analysis 24 h after the infection, and equal loading was monitored by β-actin immunoblotting. (B) TNF-α/CHX-induced apoptosis in cells described in (A). Apoptosis was measured by morphological analysis 4 h after exposure to TNF-α/CHX. Original magnification: ×150. (C) Percentage of apoptotic cells in cells described in (B). Values are the mean ± SE of data from six samples. *P < 0.05 compared with groups that were not treated with TNF-α/CHX. +P < 0.05 compared with cells infected with Adnull and then treated with TNF-α/CHX. #P < 0.05 compared with cells infected with AdHuR and then treated with TNF-α/CHX.
DISCUSSION
As XIAP is essential for cell survival by directly blocking caspase activity under conditions of cellular stress, its expression is tightly regulated by numerous factors at multiple levels (26,57,58). Our previous study (50) shows that XIAP is implicated in the regulation of apoptosis in normal IECs and plays a critical role in homeostasis in the intestinal epithelium, but the exact mechanisms that influence XIAP expression have not been investigated. This regulatory effect of XIAP on IEC apoptosis is particularly important because the intestinal epithelium has the most rapid turnover rate of any tissues in the body (59) and because maintenance of the intestinal epithelial integrity depends on a dynamic balance between cell proliferation and apoptosis (49,60,61). In the present study, we highlight a novel function of the RBP HuR in the regulation of XIAP expression and therefore in intestinal epithelial integrity. We found that HuR bound the XIAP mRNA by interacting with both its 3′-UTR and CR and affected the stability of the XIAP transcript. The results presented here also provide insight into the molecular regulation of XIAP expression by polyamines and show that decreasing cellular polyamines enhanced [HuR/XIAP mRNA] association and increased the steady state level of XIAP by stabilizing the XIAP mRNA.
An important conclusion from our analysis is that HuR can interact with the XIAP CR (Figure 2) at a segment spanning positions 575–880, which contains a predicted hit of the HuR signature motif. HuR also binds various segments of the XIAP 3′-UTR, which also bears several predicted hits of the HuR signature motif. It is important to note that we did not identify the specific XIAP CR or 3′-UTR nucleotides with which HuR interacts, which would require more specialized biochemical, crystallographic and molecular methods than those used here. Virtually in all HuR target mRNAs (8,9), binding has been localized to the 3′-UTR, with a few exceptions, such as the HIF-1α and p27 mRNAs, where binding was mapped to the 5′-UTR (62,63). In this regard, the binding of HuR to the XIAP CR and the consequences of this interaction on XIAP expression suggest that HuR could control the expression of a target mRNA through the CR. Prechtel and colleagues (64) reported that HuR specifically binds to a novel CR element in the CD83 mRNA and regulates its cytoplasmic accumulation, leading to its enhanced translation. Other RBPs have been shown to interact with CRs and 5′-UTRs present in other labile mRNAs, which also contribute to their post-transcriptional regulation. Plasminogen activator inhibitor type-2 (PAI-2) mRNA also has a CR ARE that interacts with the RBP tristetraprolin, resulting in PAI-2 mRNA degradation (65,66). Other functional mRNA stability determinants have also been reported in the CRs of many mRNAs, including c-Myc (67), vascular endothelial growth factor (VEGF) (68), u-PAR (69) and c-Fos (70).
Our results also indicate that HuR association with the XIAP mRNA stabilizes XIAP mRNA, because the half life of XIAP mRNA was decreased in HuR-silenced cells, but it was increased in cells overexpressing HuR. Our observations are consistent with studies demonstrating that HuR binds to the specific ARE motif in its target mRNAs and plays a general role as a mRNA stabilizer, as described for transcripts such as those encoding c-Fos, c-Myc, VEGF, cyclooxygenase-2, p21, p53, NPM, ATF2 and MKP-1 (2,19,21,45). The most significant finding reported here is that interaction of HuR with the XIAP CR also contributes to the regulation of XIAP expression. As shown in Figures 3 and 4, manipulating the levels of cellular HuR by its gene silencing or overexpression affected the activity of reporter gene EGFP expressed from not only the pEGFP-XIAP 3′-UTR but also the pEGFP-XIAP CR. In polyamine-deficient cells, HuR-induced XIAP expression was also mediated through HuR interaction with both XIAP 3′-UTR and CR (Figure 6). Although the precise mechanisms whereby HuR stabilizes its target mRNAs have not been fully elucidated, an increasing body of evidence support an emerging model whereby HuR binds to a given mRNA, likely assists in its nuclear export, and protects it from degradation in the cytoplasm (2,3). It is important to note that HuR and other Hu/ELAV members have also been shown to regulate the translation of numerous target transcripts (17,71–73). Thus, the possibility remains that HuR could also contribute to modulating XIAP translation. Studies are underway to test this hypothesis.
The data obtained in the present study further indicate that HuR association with the XIAP mRNA in IECs is highly regulated by cellular polyamines. Polyamines are organic cations found in all eukaryotic cells, and their intracellular levels are tightly regulated under physiological and pathological conditions (47). We recently demonstrated that decreasing cellular polyamines increased the cytoplasmic abundance of HuR by inhibiting the AMPK-regulated phosphorylation and acetylation of importin-α1, whereas increased levels of polyamines decreased cytoplasmic HuR concentration by activating the AMPK-driven importin-α1 pathway (20,21). Although the exact mechanism by which HuR regulates XIAP mRNA stability in polyamine-deficient cells is not fully understood, our current studies show that polyamine depletion by DFMO increased [HuR/XIAP mRNA] complexes, which was associated with an induction in XIAP levels. Polyamine depletion also increased the half life of XIAP mRNA, but this effect was completely abrogated in cells in which HuR expression was silenced by transfection with siRNA targeting HuR, which in turn resulted in a marked reduction of XIAP protein. Consistent with our current findings, increased cytoplasmic HuR also interacts with and stabilizes p53, NPM and ATF2 mRNAs in polyamine-deficient cells (19,21). Ongoing experiments are testing if the stabilizing effect of HuR on the XIAP mRNA depends on HuR phosphorylation, which was shown to regulate both its subcellular localization and its affinity for target mRNAs (74).
The HuR-mediated expression of XIAP is of physiological significance, because increased levels of endogenous XIAP by HuR could play an important role in the increased resistance to apoptosis observed after polyamine depletion. Previous studies from our laboratory and other groups (52–55) have demonstrated that polyamine depletion promotes the resistance of IECs to apoptosis, which is mediated through multiple signaling pathways. Polyamines downregulate NF-κB activity, and depletion of cellular polyamines increases NF-κB transcriptional activity, thus stimulating the expression of c-IAPs (50,52). Polyamines are also needed for the inhibition of focal adhesion kinase (FAK) (75) and Akt (53), as polyamine depletion induces the phosphorylation of FAK and Akt and increases their kinase activities. The data presented in Figure 8 further show that increased levels of endogenous XIAP by HuR also contribute to the increased resistance to apoptosis induced by treatment with TNF-α and CHX in polyamine-deficient cells, since this tolerance was significantly blocked by silencing XIAP or HuR. HuR is shown to have an anti-apoptotic influence (2,3), and this effect is implemented via the positive influence of HuR upon the expression of target mRNAs encoding anti-apoptotic factors such as prothymosin α, Bcl-2, Mcl-1, ATF2 and p21. In light of our results reported here, XIAP constitutes another critical downstream effector of the pro-survival program elicited by HuR.
In summary, HuR binds to the XIAP 3′-UTR and CR and regulates its mRNA stability in normal IECs. HuR silencing destabilizes XIAP mRNA, thus leading to an inhibition of XIAP expression, whereas HuR overexpression increases the stability of XIAP mRNA, leading to an induction in XIAP levels. The present study also shows that polyamines negatively regulate XIAP expression post-transcriptionally by altering the interaction of HuR with the XIAP mRNA. Polyamine depletion enhances [HuR/XIAP mRNA] association and increases the half life of the XIAP mRNA, thus inducing its expression. Increased levels of endogenous XIAP via HuR also promote the survival of IECs and elevate their resistance to TNF-α/CHX-induced apoptosis after polyamine depletion. Because polyamines are essential for maintaining the integrity of the intestinal epithelium and regulate HuR association with the XIAP mRNA, these findings suggest that polyamine-modulated XIAP expression through HuR is crucial for controlling intestinal mucosal homeostasis under physiologic conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Merit Review Grant from US Department of Veterans Affairs and National Institutes of Health (DK57819, DK61972, DK68491 to J.-Y.W.). Funding for open access charge: National Institutes of Health and Department of Veterans Affairs.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
J.-Y.W. is a Research Career Scientist, Medical Research Service, US Department of Veterans Affairs. M.G. is supported by the National Institute on Aging-Intramural Research Program, National Institutes of Health.
REFERENCES
- 1.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 2.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 6.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 7.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 9.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin. Cell Dev. Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 11.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 13.Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2005;2:11–13. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- 17.Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, Gorospe M, Wang JY. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Gastroenterology. 2008;134:A-237. doi: 10.1091/mbc.E09-07-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao L, Rao JN, Zou T, Liu L, Marasa BS, Chen J, Turner DJ, Zhou H, Gorospe M, Wang JY. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol. Biol. Cell. 2007;18:4579–4590. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1. Biochem. J. 2008;409:389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J. Biol. Chem. 2006;281:19387–193894. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 22.Holcik M. Translational upregulation of the X-linked inhibitor of apoptosis. Ann. NY Acad. Sci. 2003;1010:249–258. doi: 10.1196/annals.1299.043. [DOI] [PubMed] [Google Scholar]
- 23.Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5' UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;22:2605–2609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seligson DB, Hongo F, Huerta-Yepez S, Mizutani Y, Miki T, Yu H, Horvath S, Chia D, Goodglick L, Bonavida B. Expression of X-linked inhibitor of apoptosis protein is a strong predictor of human prostate cancer recurrence. Clin. Cancer Res. 2007;23:6056–6063. doi: 10.1158/1078-0432.CCR-07-0960. [DOI] [PubMed] [Google Scholar]
- 25.Ungureanu NH, Cloutier M, Lewis SM, de Silva N, Blais JD, Bell JC, Holcik M. Internal ribosome entry site-mediated translation of Apaf-1, but not XIAP, is regulated during UV-induced cell death. J. Biol. Chem. 2006;281:15155–15163. doi: 10.1074/jbc.M511319200. [DOI] [PubMed] [Google Scholar]
- 26.Dubrez-Daloz L, Dupoux A, Cartier J. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle. 2008;7:1036–1046. doi: 10.4161/cc.7.8.5783. [DOI] [PubMed] [Google Scholar]
- 27.Holcik M, Korneluk RG. XIAP, the guardian angel. Nat. Rev. Mol. Cell Biol. 2001;2:550–556. doi: 10.1038/35080103. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, Ueno N, Nishida E, Shibuya H, Matsumoto K. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashkar H, Deggerich A, Seeger JM, Yazdanpanah B, Wiegmann K, Haubert D, Pongratz C, Kronke M. NF-κB-independent down-regulation of XIAP by bortezomib sensitizes HL B cells against cytotoxic drugs. Blood. 2007;109:3982–3988. doi: 10.1182/blood-2006-10-053959. [DOI] [PubMed] [Google Scholar]
- 30.Stehlik C, de Martin R, Kumabashiri I, Schmid JA, Binder BR, Lipp J. Nuclear factor (NF)-κ-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor alpha-induced apoptosis. J. Exp. Med. 1998;188:211–216. doi: 10.1084/jem.188.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holley CL, Olson MR, Colón-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohn D, Totzke G, Essmann F, Schulze-Osthoff K, Levkau B, Jänicke RU. The proteasome is required for rapid initiation of death receptor-induced apoptosis. Mol. Cell Biol. 2006;26:1967–1978. doi: 10.1128/MCB.26.5.1967-1978.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis SM, Veyrier A, Hosszu Ungureanu N, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell. 2007;18:1302–1311. doi: 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell cultures from rat small intestine: characterization by morphologic and immunologic criteria. J. Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suh E, Traber PG. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell Biol. 1996;16:619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao JN, Liu L, Zou T, Marasa BS, Boneva D, Wang SR, Malone DL, Turner DJ, Wang JY. Polyamines are required for phospholipase C-γ1 expression promoting intestinal epithelial restitution after wounding. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G335–G343. doi: 10.1152/ajpgi.00282.2006. [DOI] [PubMed] [Google Scholar]
- 38.Rao JN, Liu SV, Zou T, Liu L, Xiao L, Zhang X, Bellavance E, Yuan JX, Wang JY. Rac1 promotes intestinal epithelial restitution by increasing Ca2+ influx through interaction with phospholipase C-γ1 after wounding. Am. J. Physiol. Cell Physiol. 2008;295:C1499–C1509. doi: 10.1152/ajpcell.00232.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vielkind U, Swierenga SH. A simple fixation procedure for immunofluorescent detection of different cytoskeletal components within the same cell. Histochemistry. 1989;91:81–88. doi: 10.1007/BF00501916. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, Guo X, Rao JN, Zou T, Marasa BS, Chen J, Greenspon J, Casero RA, Jr, Wang JY. Polyamine-modulated c-Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem. J. 2006;398:257–267. doi: 10.1042/BJ20060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Liu L, Rao JN, Esmaili A, Strauch ED, Bass BL, Wang JY. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through p21 after polyamine depletion. Gastroenterology. 2002;123:764–779. doi: 10.1053/gast.2002.35386. [DOI] [PubMed] [Google Scholar]
- 43.Harter JL. Critical values for Duncan’s new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]
- 44.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin α. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45α by genotoxic stress. Mol. Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Casero RA, Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 48.Gerner EW, Meyskens FL., Jr Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 49.Seiler N, Raul F. Polyamines and the intestinal tract. Crit. Rev. Clin. Lab. Sci. 2007;44:365–411. doi: 10.1080/10408360701250016. [DOI] [PubMed] [Google Scholar]
- 50.Zou T, Rao JN, Guo X, Liu L, Zhang HM, Strauch ED, Bass BL, Wang JY. NF-κB-mediated IAP expression induces resistance of intestinal epithelial cells to apoptosis after polyamine depletion. Am. J. Physiol. Cell Physiol. 2004;286:C1009–C1018. doi: 10.1152/ajpcell.00480.2003. [DOI] [PubMed] [Google Scholar]
- 51.Li L, Rao JN, Guo X, Liu L, Santora R, Bass BL, Wang JY. Polyamine depletion stabilizes p53 resulting in inhibition of normal intestinal epithelial cell proliferation. Am. J. Physiol. Cell Physiol. 2001;281:C941–C953. doi: 10.1152/ajpcell.2001.281.3.C941. [DOI] [PubMed] [Google Scholar]
- 52.Li L, Rao JN, Bass BL, Wang JY. NF-κB activation and susceptibility to apoptosis after polyamine depletion in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2001;280:G992–G1004. doi: 10.1152/ajpgi.2001.280.5.G992. [DOI] [PubMed] [Google Scholar]
- 53.Zhang HM, Rao JN, Guo X, Liu L, Zou T, Turner DJ, Wang JY. Akt kinase activation blocks apoptosis in intestinal epithelial cells by inhibiting caspase-3 after polyamine depletion. J. Biol. Chem. 2004;279:22539–22547. doi: 10.1074/jbc.M314337200. [DOI] [PubMed] [Google Scholar]
- 54.Bhattacharya S, Ray RM, Johnson LR. Decreased apoptosis in polyamine depleted IEC-6 cells depends on Akt-mediated NF-κB activation but not GSK3β activity. Apoptosis. 2005;10:759–776. doi: 10.1007/s10495-005-2943-3. [DOI] [PubMed] [Google Scholar]
- 55.Deng W, Viar MJ, Johnson LR. Polyamine depletion inhibits irradiation-induced apoptosis in intestinal epithelia. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289:G599–G606. doi: 10.1152/ajpgi.00564.2004. [DOI] [PubMed] [Google Scholar]
- 56.Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Li H, Derouet M, Filmus J, LaCasse EC, Korneluk RG, Kerbel RS, Rosen KV. ras Oncogene triggers up-regulation of cIAP2 and XIAP in intestinal epithelial cells: epidermal growth factor receptor-dependent and -independent mechanisms of ras-induced transformation. J. Biol. Chem. 2005;280:37383–37392. doi: 10.1074/jbc.M503724200. [DOI] [PubMed] [Google Scholar]
- 58.Liu Z, Li H, Wu X, Yoo BH, Yan SR, Stadnyk AW, Sasazuki T, Shirasawa S, LaCasse EC, Korneluk RG, et al. Detachment-induced upregulation of XIAP and cIAP2 delays anoikis of intestinal epithelial cells. Oncogene. 2006;25:7680–7690. doi: 10.1038/sj.onc.1209753. [DOI] [PubMed] [Google Scholar]
- 59.Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 60.Chen J, Xiao L, Rao JN, Zou T, Liu L, Bellavance E, Gorospe M, Wang JY. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol. Biol. Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCormack SA, Johnson LR. Role of polyamines in gastrointestinal mucosal growth. Am. J. Physiol. Gastrointest. Liver Physiol. 1991;260:G795–G806. doi: 10.1152/ajpgi.1991.260.6.G795. [DOI] [PubMed] [Google Scholar]
- 62.Galbán S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol. Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kullmann M, Göpfert U, Siewe S, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'U;TR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prechtel AT, Chemnitz J, Schirmer S, Ehlers C, Langbein-Detsch I, Stulke J, Dabauvalle MC, Kehlenbach RH, Hauber J. Expression of CD83 is regulated by HuR via a novel cis-active coding region RNA element. J. Biol. Chem. 2006;281:10912–10925. doi: 10.1074/jbc.M510306200. [DOI] [PubMed] [Google Scholar]
- 65.Tierney MJ, Medcalf RL. Plasminogen activator inhibitor type 2 contains mRNA instability elements within exon 4 of the coding region: sequence homology to coding region instability determinants in other mRNAs. J. Biol. Chem. 2001;276:13675–13684. doi: 10.1074/jbc.M010627200. [DOI] [PubMed] [Google Scholar]
- 66.Yu H, Stasinopoulos S, Leedman P, Medcalf RL. Inherent instability of plasminogen activator inhibitor type 2 mRNA is regulated by tristetraprolin. J. Biol. Chem. 2003;278:13912–13918. doi: 10.1074/jbc.M213027200. [DOI] [PubMed] [Google Scholar]
- 67.Yeilding NM, Lee WM. Coding elements in exons 2 and 3 target c-Myc mRNA downregulation during myogenic differentiation. Mol. Cell Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dibbens JA, Miller DL, Damert A, Risau W, Vadas MA, Goodall GJ. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the cooperation of multiple RNA elements. Mol. Biol. Cell. 1999;10:907–919. doi: 10.1091/mbc.10.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shetty S, Kumar A, Idell S. Posttranscriptional regulation of urokinase receptor mRNA: identification of a novel urokinase receptor mRNA binding protein in human mesothelioma cells. Mol. Cell Biol. 1997;17:1075–1083. doi: 10.1128/mcb.17.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kabnick KS, Housman DE. Determinants that contribute to cytoplasmic stability of human c-fos and β-globin mRNAs are located at several sites in each mRNA. Mol. Cell Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jain RG, Andrews LG, McGowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Millard SS, Vidal A, Markus M, Koff A. A U-rich element in the 5' untranslated region is necessary for the translation of p27 mRNA. Mol. Cell Biol. 2000;20:5947–5959. doi: 10.1128/mcb.20.16.5947-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang HM, Keledjian KM, Rao JN, Zou T, Liu L, Marasa BS, Wang SR, Ru L, Strauch ED, Wang JY. Induced focal adhesion kinase expression suppresses apoptosis by activating NF-κB signaling in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006;290:C1310–C1320. doi: 10.1152/ajpcell.00450.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.