Abstract
Gene editing mediated by oligonucleotides has been shown to induce stable single base alterations in genomic DNA in both prokaryotic and eukaryotic organisms. However, the low frequencies of gene repair have limited its applicability for both basic manipulation of genomic sequences and for the development of therapeutic approaches for genetic disorders. Here, we show that single-stranded oligodeoxynucleotides (ssODNs) containing a methyl-CpG modification and capable of binding to the methyl-CpG binding domain protein 4 (MBD4) are able to induce >10-fold higher levels of gene correction than ssODNs lacking the specific modification. Correction was stably inherited through cell division and was confirmed at the protein, transcript and genomic levels. Downregulation of MBD4 expression using RNAi prevented the enhancement of gene correction efficacy obtained using the methyl-CpG-modified ssODN, demonstrating the specificity of the repair mechanism being recruited. Our data demonstrate that efficient manipulation of genomic targets can be achieved and controlled by the type of ssODN used and by modulation of the repair mechanism involved in the correction process. This new generation of ssODNs represents an important technological advance that is likely to have an impact on multiple applications, especially for gene therapy where permanent correction of the genetic defect has clear advantages over viral and other nonviral approaches currently being tested.
INTRODUCTION
Manipulation of genomic sequences through gene trapping or gene targeting technologies has a wide spectrum of applications ranging from site-directed mutagenesis of bacterial vectors to development of animal models through DNA recombination technologies. Among those, gene editing can be used to target specific gene defects and restore protein expression for therapeutic applications (1–5). To date, however, the use of gene editing has been limited and is still at an early stage of development.
Single-stranded oligodeoxynucleotides (ssODNs) have been shown to be able to alter single nucleotides and induce stable alterations at the genomic level (3,6). Gene repair mediated by ssODNs takes advantages of specific repair mechanisms present in the cells that are able to recognize the presence of mismatches in genomic DNA. ssODNs complementary to the target sequence but containing a mismatch at the base targeted for modification are used as templates for the correction process. Once introduced into the cell, they have been shown to anneal to the genomic DNA sequence targeted for repair and initiate the repair process (7) leading to a single nucleotide exchange that is stably inherited throughout cell division. The technology has been successfully applied in different cell types including bacterial, yeast and mammalian cells (1,3,8–12).
Oligonucleotide-mediated gene correction has been investigated in several eukaryotic cell type as well as different models of genetic disorders. Correction has been successfully demonstrated in hepatocytes (13–16), retinal cells (17,18), bone marrow-derived cells (19) and muscle cells (5,9,10,20–22). The level of correction varies depending on the cell type being targeted suggesting that the repair process involved is differentially regulated in different cell types.
In skeletal muscle, the major focus of ssODN-mediated gene editing has been the treatment of genetic disease, in particular Duchenne muscular dystrophy (DMD). This disease is characterized by mutations in the dystrophin gene that lead to complete absence of dystrophin protein expression, progressive muscle degeneration and weakness, and in most of the cases death by the age of 30 years. Targeted single base alterations of the dystrophin gene has been successfully achieved both in vitro and in vivo, using mouse and dog models of DMD (5,9,10,20,21). Correction can be obtained in mature myofibers (5,10) as well as muscle progenitor cells (10), suggesting that oligonucleotides can target both postmitotic cells as well as replicating cells and can achieve frequencies of up to 10% depending on the strand targeted for repair and the type of mismatch created by the oligonucleotide (5,10,20,21).
The past few years have seen a considerable expansion of the application of gene editing to neuromuscular disorders. Oligonucleotides can be used not only to correct dystrophin gene defects due to single point mutations, but also to treat frameshift deletions by disrupting intron/exon consensus sequences to redirect mRNA assembly and splicing and induce the expression of novel in-frame transcripts (5,21). Along the same line, ssODNs have been shown to target and correct a splicing regulatory element in the SMN2 gene and to restore SMN protein expression in human fibroblasts (22), highlighting the potential of this technology for the treatment of spinal muscular atrophy, another debilitating neuromuscular disorder for which there is currently no effective treatments.
The mechanisms that lead to the repair process remain unclear, but appear to involve multiple steps. The first step requires the annealing of the ssODN to the targeted genomic sequence through the interaction of proteins such as Rad51, Rad54 and XRCC2 (23–25). The second step, the correction of the single base pair mismatch, is less well characterized. Initial data suggested that correction is mediated by mismatch repair (MMR) mechanisms that recognize the mismatch created by the annealing of the ssODN to the genomic DNA (26). Proteins involved in MMR appear to be crucial for the ssODN-mediated base exchange at least in yeast (7). In some mammalian cells, however, gene correction can occur even in the absence of MMR (27,28), suggesting that different mechanisms may operate to facilitate ssODN-mediated gene repair under different cellular contexts. Homologous recombination, nucleotide exchange repair and double-strand break repair have all been implicated as potential mechanistic pathways (29–31), and it is not clear if the different frequencies of gene correction detected in different cell types are in part due to different mechanisms being recruited. This hypothesis led us to investigate whether ssODN-mediated gene correction frequencies could be improved by designing ssODNs capable of activating specific repair mechanisms in the cells targeted for repair.
Deamination of 5-methylcytosine is among the most prominent genomic alterations in mammalian cells with rates up to 300 events per genome per day (32,33). Loss of the amino group leads the spontaneous conversion of 5-methylcytosine (m5C) into thymine which, if left unrepaired, would lead to the accumulations of C:G-to-T:A mutations (34,35). The presence of 5-methylcytosines and the frequency of mutation at such bases in the context of m5CpG sites contribute to genetic variation and are implicated in tumorigenesis (36,37). Repair of the mismatch created by the deamination of 5-methylcytosine requires the presence of specific repair mechanisms capable of recognizing the mismatch that arises as a result of the deamination. Recognition of deamination events in the context of m5CpG sites is mediated by the base excision repair (BER) mechanism through the specific activation of the methyl-CpG binding domain protein 4 (MBD4). MBD4 (also known as MED1) plays a crucial role in maintaining genome integrity by recognizing G:T or G:U mismatches at m5CpG sites on double-stranded DNA (38,39). MBD4 was originally identified in a yeast two-hybrid screen due to its interaction with MLH1, an important component of the MMR mechanism, which led to the hypothesis that the two systems could be linked (38,40). To date, however, the role of the interaction between MBD4 and MLH1 has yet to be clarified (41). In vitro studies have shown that MBD4 can efficiently recognize and hydrolyze G:T or G:U mismatches at hemi-methylated m5CpG sites. Furthermore, a m5CpG context, although preferred, is not absolutely necessary as G:T and G:U mismatches in nonmethylated CpG sequences can also be recognized and processed, although at a reduced rate (39). Binding of MBD4 to the mismatched T or U leads to glycosylation and removal of the base without altering the sugar phosphate backbone of the DNA. The apurinic site that is generated by MBD4 is then processed by specific endonucleases and ligases which are required to direct the addition of a new cytosine on this site and to complete the repair process (42,43).
We have designed a new generation of ssODNs containing a methyl-CpG modification and tested their ability to mimic, when annealed to the genomic sequence targeted for correction, the G:T mismatch that would occur upon deamination of 5′-methylcytosines (to become thymine) in the genomic DNA targeted for correction. Due to the high frequencies at which this process occurs in nature, we hypothesized that the use of these ssODNs could be much more efficient in directing stable single base alterations at the genomic level. Their ability to specifically activate the BER repair by recruiting MBD4 and to induce single base alterations was tested in muscle cells using a GFP reporter system. This system allowed us to determine the efficacies of gene correction of an m5CpG-containing ssODN and to compare its efficacy to targeting ssODNs either lacking any modified cytosines or containing a 5-methylcytosine but not in the context of a CpG dinucleotide, neither of which would be expected to recruit MBD4. We demonstrate that gene correction frequencies were consistently higher when an ssODN containing a methyl-CpG and capable of recruiting MBD4 was used. These studies expand the potential applications of ssODNs in inducing single base alterations at the genomic level and further advance this technology into therapeutic applications for the treatment of many disorders and in particular muscle diseases.
MATERIALS AND METHODS
Oligonucleotide synthesis
ssODNs were purchased from MWG Biotech Inc. (High Point, NC, USA). All ssODNs were labeled at the 5′-end with fluorescent CY3 and at the 3′-end with a tag (-cgcg) of phosphorothioate bases to increase stability of ssODNs from endonucleases. Oligonucleotides were HPLC purified and exhibited a single peak of the expected molecular weight as determined by MALDI-TOF mass spectroscopy analysis.
Plasmid constructs
The pGFPmut vector was generated using the QuickChange® II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA), following the manufacturer’s instructions. The pEGFP-N3 vector (BD Bioscience, Clontech, CA, USA) was annealed with a forward primer (GFPmut forward: 5′-GACCTACGGCGTGCAGTGATTCAGCCGCTACCCCGAC-3′) and a reverse primer (GFPmut rev: 5′-GTCGGGGTAGCGGCTGAATCACTGCACGCCGTAGGTC-3′) designed to induce the C-to-A transversion in position +213 of the GFP coding sequence. Initial denaturation was carried out at 95°C for 3 min, followed by a 20-cycle step of denaturation at 95°C for 3 s, annealing at 58°C for 1 min and extension at 68°C for 6 min. The PCR product (1 µl) was digested overnight at 37°C with DpnI and transformed in DH5α competent cells (Invitrogen, Carlsbad, CA, USA). Positive clones were selected by restriction endonuclease analysis and the C-to-A mutation was confirmed by direct sequencing using an Applied Biosystems ABI377 automated sequencer.
The pGEX3.MBD4 plasmid was generated by inserting the MBD4 coding sequence into the BamHI and XhoI sites of the pGEX3 vector (Amersham GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). The mouse cDNA encoding MBD4 was obtained by direct cloning of PCR products encoding the MBD4 cDNA (GenBank accession number NM_010774). The MBD4 gene encoding full-length cDNA was obtained by direct amplification of a cDNA library using a forward primer (MBD4-forw-BamHI: 5′-GCAGCAGGGATCCGAGAGCCC-3′) and a reverse primer (MBD4-rev-XhoI: 5′-CTCGAGATAGACTTAATTTTTCATG-3′). Amplification was carried out for 30 cycles in the presence of Pfu DNA polymerase (Stratagene) at an annealing temperature of 65°C for 1 min and an extension temperature of 68°C for 2 min. Amplification was terminated by an additional extension of 30 min at 72°C. Amplicons were digested with BamHI and XhoI. PCR products were run on a 1.2% agarose gel and purified using the gel extraction kit (Qiagen, Valencia, CA, USA) as previously described (5). Vector sequences were confirmed using an ABI377 automated sequencer.
Cell culture and transfection
Myoblasts were derived from limb muscle of neonatal C57 mice as previously described (44). For growth, cells were plated on dishes coated with 5 µg/ml laminin (Invitrogen Corp.) and maintained in growth medium (GM) consisting of Ham’s F10 nutrient mixture (Mediatech, Herndon, VA, USA) supplemented with 20% fetal bovine serum, penicillin and streptomycin.
Myoblasts were plated in wells of 6-well dishes (104 cells/well) 12 h prior to transfection. ssODNs (10 µg) were complexed with 1 µl of Lipofectamine 2000 (Invitrogen Corp.) for 30 min at room temperature in a total volume of 0.5 ml of Ham’s F10 nutrient mixture. The complex was then added to the wells containing 1.5 ml of GM. Transfections were stopped by replacing the transfection solution with fresh GM.
Expression of recombinant MBD4 protein
Expression constructs of wild-type MBD4 protein were propagated in Escherichia coli strain XL-1 Blue, as previously described (38,45). BL21(DE3)(pLysS) cells transformed with the expression vectors were grown to OD600 0.4 and induced with 1 mM IPTG at 37°C for 2 h. Cells were collected by centrifugation and lyzed in 10 mM Tris–HCl (pH 8.0), 500 mM NaCl, 0.1% Nonidet P-40 (NP-40), 10% glycerol and ‘Complete’ protease inhibitors (Boehringer-Mannheim Biochemicals, Indianapolis, IN, USA). After clarification by centrifugation at 12 000g, the soluble protein fraction was diluted in 150 mM NaCl and β-mercaptoethanol (7 mM) was added. Purification of recombinant proteins was conducted as previously earlier (46). Briefly, the sample was applied to a 5-ml Q Sepharose anion exchange column (Amersham) connected in series to a 5-ml SP Sepharose cation exchange column (Amersham). After disconnecting the Q Sepharose column, the SP Sepharose column was washed and elution was performed with buffer containing 1.5 M NaCl. The SP Sepharose eluate was applied to a 1.5-ml nickel-chelating Sepharose column (Amersham). Recombinant MBD4 protein was eluted with 5 ml of 10 mM Tris–HCl (pH 8.0), 300 mM NaCl, 10% glycerol, 2 mM β-mercaptoethanol and 150 mM imidazole. MBD4 protein was further purified by size-exclusion chromatography using a Superdex 200 PC 3.2/30 gel-filtration column with a SMART chromatography system (Pharmacia Laboratories, Piscataway, NJ, USA), as previously earlier (45,46). The final MBD4 preparation was estimated to be 95% pure by SDS–PAGE. Upon the addition of 10% glycerol, purified MBD4 fractions were frozen and stored in liquid nitrogen. Protein concentration was determined with the Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA), using bovine gamma globulin as a standard.
Flow cytometry
Fluorescence was measured using a Beckton Dickinson FACScalibur flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA). Cells were trypsinized and harvested at different time after transfection, resuspended in 0.5% BSA, 2 mM EDTA in PBS and processed immediately. For each analysis, a total of 2 × 104 cells were used. Experiments were repeated in triplicate for each time point analyzed and a total of three independent experiments were performed.
For DNA content analysis, cells were fixed in 70% ice-cold ethanol, washed in PBS and resuspended in 0.5 ml of PBS containing 50 µg/ml RNAse A, 1% FBS and 2.5 µg/ml propidium iodide. Analyses were performed using 2 × 104 cells.
Western blot analysis
Cells were lyzed in RIPA buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 0.5% deoxycholate and 1% Nonidet P-40] containing aprotinin (20 µg/ml), leupeptin (20 µg/ml), phenylmethylsulfonyl fluoride (10 µg/ml) and sodium orthovanadate (1 mM). Total protein in the extract was determined by the Bio-Rad protein assay. Immunoblot analysis was performed as previously described (10,47). Total protein from each sample (200 µg) was separated by electrophoresis (20 mA for 1.5 h) using 12% SDS-polyacrylamide gels, and then transferred (250 mA for 1 h) onto nitrocellulose membranes. Membranes were probed overnight at 4°C with an antibody directed toward the C-terminal region of the GFP protein (1GFP63; Santa Cruz Biotechnologies, Santa Cruz, CA, USA) or an antibody directed toward the N-terminal domain of the MBD4 protein (Abcam Inc., Cambridge, MA, USA) (21,47). The membranes were blocked with 5% milk in PBS for 1 h at room temperature. Blots were washed with 0.05% Tween-20 in PBS and then incubated with a horseradish peroxidase-coupled anti-mouse secondary antibody (Amersham). Specific antibody binding was detected using an enhanced chemiluminescent system (Amersham).
Genomic DNA and restriction endonuclease analysis
Cultured myoblasts were rinsed twice with PBS and genomic DNA was extracted using the Wizard Genomic DNA extraction kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Total genomic DNA (1 µg) was digested overnight at 37°C with 5 U of HinfI or BtsI, purified using Amicon Microcon®-PCR Centrifugal Filter Devices (Millipore Corporation, Bedford, MA, USA), and resuspended in 20 µl of H20. For each amplification reaction, 5 µl of digested genomic DNA was subjected to amplification using the forward primer (GFP-forw: 5′-GCTAGCGCTACCGGTCGCCAC-3′) specific to region −17 to −2 of the GFP open reading frame (ORF), and the reverse primer (GFP-rev: 5′-CCTTGAAGTCGATGCCCTTC-3′) complementary to the region +397 to +377 of the ORF.
Real-time PCR was performed using a My iQ single-color detection system (Bio-Rad) at the following thermal cycler conditions: 95°C for 10 min followed by 45 cycles of a 3-step reaction consisting of denaturation at 95°C for 30 s, annealing at 60°C for 1 min and extension and data collection at 72°C for 30 s. Amplicons were separated on 1.5% agarose gels and PCR products were purified using the Qiagen gel extraction kit (Qiagen). DNA sequencing was carried out using an Applied Biosystems ABI377 automated sequencer.
RT–PCR
Total RNA was extracted from cultured myotubes using TRI-REAGENT (Sigma, St. Louis, MO, USA). For each reaction, 2 µg of RNA was treated with 1 U of DNase I (GIBCO/BRL) at room temperature and reverse transcribed using the first strand cDNA synthesis kit (Invitrogen) in the presence of SuperscriptIII (Invitrogen) to a final volume of 20 µl. Of those, 10 µl were digested overnight with 5 U HinfI or BtsI in a final volume of 20 µl. The remaining 10 µl of undigested cDNA obtained after reverse transcription were brought to a final volume of 20 µl containing isomolar concentrations of salt and glycerol and were used to normalize transcript levels. PCR reactions were carried out using 4 µl of digested product in the presence of the GFP primers described earlier. Each amplification mixture contained 25 pmols of appropriate primer, 10% DMSO, 0.5 U Master Taq DNA polymerase (Takara, Panvera Corp., Madison, WI, USA), and 5 mM of each deoxyribonucleotide triphosphate. After an initial step of denaturation at 95°C for 5 min, amplification was performed for 35 cycles at 95°C for 1 min followed by annealing at 55°C for 3 min and extension at 72°C for 2 min. Amplification reactions were terminated by an additional extension step at 72°C for 10 min and products were fractionated on 1.5% agarose gels.
Electrophoretic mobility shift assay
An oligonucleotide homologous to the region of the GFP gene targeted for correction and ssODNs were synthesized by MWG and purified by HPLC. Targeting and control oligonucleotides were identical to the one previously described (Figure 1), but lacked the CY3 modification at their 5′-end. The probe was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Amersham).The ability of MBD4 to form a complex with the m5CpG ssODN alone was investigated using the m5CpGGFP labeled at its 5′-end with [γ-32P]ATP. Unincorporated label was removed with G25 spin columns (Amersham). Control and targeting ssODNs were annealed to equal amounts (∼100 000 c.p.m.) of radiolabeled probe using a thermal cycler at the following setting conditions: 95°C for 5 min for the initial denaturation step, followed by 80°C for 5 min, 75°C for 5 min, 70°C for 15 min, 55°C for 15 min, 37°C for 15 min and 20°C for 20 min. One microgram of recombinant MBD4 protein was incubated in binding buffer [50 mM Tris–HCl (pH 7.5), 2.5 mM dithiothreitol, 2.5 mM EDTA, 250 mM NaCl, 5 mM MgCl2 and 20% glycerol] in the presence of 1 µg of Poly(dI-dC):Poly(dI-dC) competitor (Amersham) for 15 min at room temperature. Double-stranded oligonucleotides were then added to the mixture and incubated for an additional 15 min. Samples were loaded on 5% polyacrylamide gels and resolved at 200 V for 2 h. Gels were dried for 1 h at 80°C prior autoradiographic exposure.
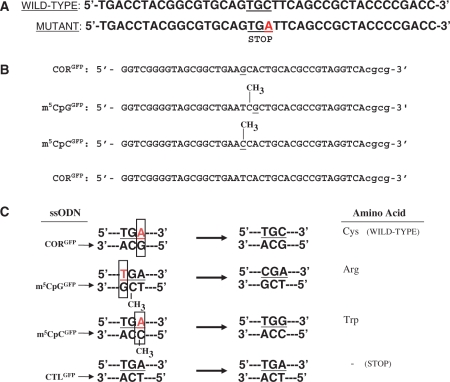
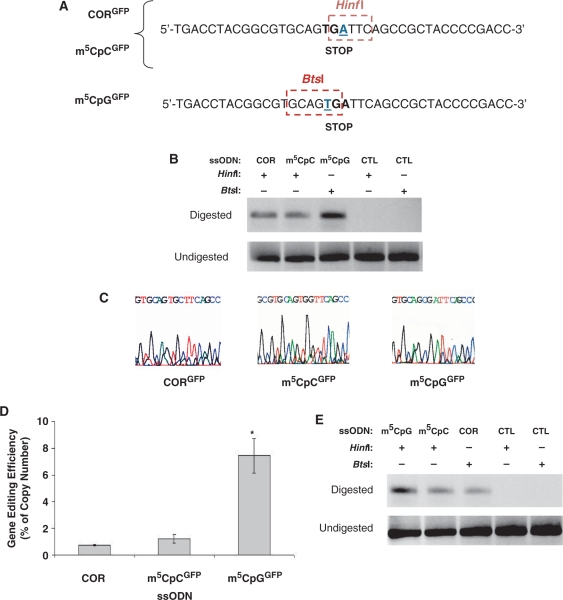
Figure 1.
Wild-type and mutant GFP sequences and ssODN design. (A) The pGFPmut vector contains a single point mutation at base pair +213 of the GFP coding sequence that creates a stop codon in this position. ssODNs were designed to target bases in the stop codon to restore GFP expression. (B) Sequences of targeting (CORGFP, m5CpGGFP, m5CpCGFP) and control (CTLGFP) ssODNs. (C) Annealing of the targeting ssODNs to the genomic sequence creates a mismatch (box), whereas the control ssODN pair perfectly. The CORGFP ssODN is designed to induce an A-to-C conversion that eliminates the stop codon and restores wild-type GFP expression. The m5CpGGFP ssODN creates a G:T mismatch in the context of a m5CpG, thus mimicking the delamination of m5C. This will induce a T-to-C substitution which will convert the stop codon into an arginine. The m5CpCGFP ssODN targets the same base as CORGFP but contains an m5C modification which will lead to an A-to-G conversion and result in a single amino acid substitution (a tryptophan in place of a cysteine). The CTLGFP ssODN perfectly complements the mutant sequence and therefore should not induce any genomic alterations.
siRNA treatment
The target sites were predicted in silico using on-line tools from Invitrogen Corp (http://bbiserv.techfak.uni-biecefeld.de/mfold/). In brief, the sequences targeted by the StealthTM siRNA were listed as residues 589–614 of the ORF with the following sequence: 5′-GCAGCCAAAUGAAACUGACGUUUCA-3′ and 3′-CGUCGGUUUACUUUGACUGCAAAGU-5′. The negative control StealthTM siRNA used were: 5′-GCAACAAGUAACAGUCAUGUCGUCA-3′ and 3′-CGUUGUUCAUUGUCAGUACAGCAGU-5′. No homologous sequences were found in mouse genome. The double-stranded siRNAs were synthesized chemically and modified into stealth siRNA (Invitrogen Corp) to enhance the stability in vitro. Transfection of the siRNAs was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Efficient silencing was demonstrated by real-time PCR in cells transfected with the siRNAs and was performed on RNA isolated at different time after siRNA transfection (Figure 9). First strand cDNA was prepared and reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time RT–PCR was performed using a My iQ single-color detection system (Bio-Rad) using the following thermal cycler conditions: 95°C for 10 min followed by 45 cycles of a 3-step reaction consisting of denaturation at 95°C for 30 s, annealing at 60°C for 1 min and extension and data collection at 72°C for 30 s. The primers RTMBD4-forw (5′-GATGTATACTTTATCAGCCCACAAG-3′) and RTMBD4-rev (5′-CTGGCGACTCTGAAGTACCAC-3′) were used to detect MBD4 mRNA. Transcripts levels were normalized to those of GAPDH using the following primers: GAPDH-forw: 5′-TGCGACTTCAACAGCAACTC-3′; GAPDH-rev: 5′-ATGTAGGCCATGAGGTCCAC-3′ as described earlier(48).
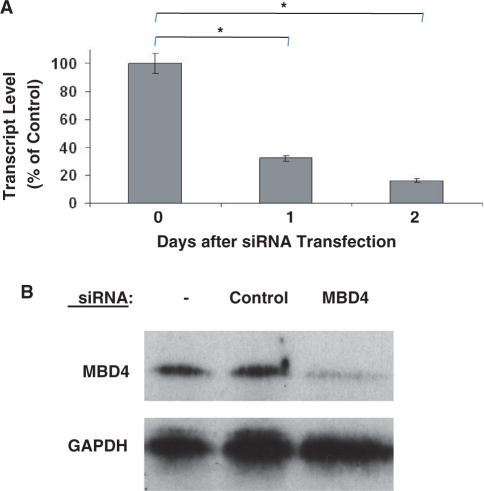
Figure 9.
Reduction of MBD4 expression by siRNA interference. (A) RNA interference was used to downregulate expression of MBD4 in myoblast cultures. Cells were transfected with a targeting or control siRNA and gene expression was followed over time by quantitative RT–PCR. Efficient downregulation of MBD4 expression was achieved by 24 h after siRNA transfection and the effect was maintained up to 48 h after transfection. *P ≤ 0.001. (B) Western blot analysis was carried out using total protein isolated 2 days after siRNA transfection. MBD4 protein was markedly reduced in myoblast cultures treated with MBD4 siRNA compared with untreated cells or cells treated with the control siRNA.
Statistical analysis
Data are presented as means and standard deviations. Comparisons between groups were done using Student’s t-test assuming two-tailed distribution and unequal variances.
RESULTS
The GFP reporter system and assay
We have established a reporter system and used it to follow gene repair over time in muscle cells. The pGFPmut vector contains a single point mutation (a C-to-A transversion at position +213 of the coding region), which creates a TGA stop codon and abolishes GFP protein expression in mammalian cells (Figure 2). The plasmid was transfected into murine myoblasts and a stable cell line was selected by G418 using the neomycin resistance gene encoded by the plasmid. Expression of GFP mRNA transcripts was confirmed using RT–PCR. Genomic DNA analysis performed using real-time PCR revealed the presence of 1.4 copies of integrated vector per cell. No GFP protein expression was detected either by western blot, flow cytometry or direct examination by fluorescence microscopy in those cultures.
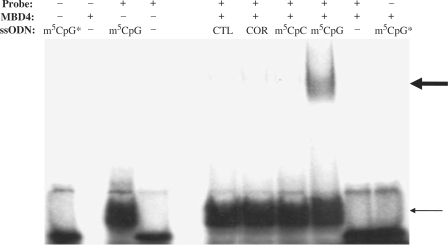
Figure 2.
Binding of GFP ssODNs to MBD4. A 45-nt oligonucleotide radiolabeled probe corresponding to the region of the pGFPmut vector targeted for repair was incubated with or without ssODNs and with and without recombinant MBD4 at 37°C for 30 min and analyzed by EMSA. The position of the ssODN/probe complexes is indicated by the thin arrow. The thick arrow above shows the position of the supershift achieved by the binding of MBD4 with the ssODN/probe complex. MBD4 was unable to form a complex with the target sequence in the absence of an ssODN bearing a mismatch. No binding of MBD4 to m5CpG [as assessed using a radioactively labeled m5CpG (m5CpG*)] was detected in the absence of the oligonucleotide sequence targeted for repair.
The ssODNs were designed to target and induce single base pair alterations within the early termination codon and to restore expression of GFP protein (Figure 1). The CORGFP ssODN was designed to correct the C-to-A transversion of the pGFPmut vector, thus restoring wild-type GFP protein expression. The m5CpGGFP ssODN contains a 5-methylcytosine adjacent to the base that creates the mismatch (the targeting base). Annealing of the m5CpGGFP ssODN to the mutant GFP sequence targets the T of the TGA stop codon, creating a G:T mismatch in the context of a m5CpG sequence, precisely mimicking the situation in which MBD4 recognizes the deamination of 5-methylcytosine. Correction of the mismatch will convert the TGA codon into a CGA codon, restoring the expression of a full-length GFP protein containing a single amino acid substitution (an arginine in place of a cysteine) compared with wild-type GFP protein. The effects of 5-methylcytosine modification of the targeting base itself, but not in the context of an m5CpG sequence, were analyzed using a targeting ssODN (m5CpCGFP) in which a 5-methylcytosine creates a mismatch with the mutant base in the GFP sequence. Correction of the mismatch induced by the m5CpCGFP should result in an A-to-G substitution, eliminating the stop codon and creating a missense mutation (a tryptophan in place of a cysteine) rather than a true restoration of the wild-type sequence (as with the CORGFP ssODN). A nontargeting (‘perfect match’) ssODN (CTLGFP), perfectly complementary to the mutant GFP sequence and therefore unable to activate any repair mechanism and induce any single base pair alteration, was used as a negative control (Figure 1).
Binding of MBD4 to ssODN/target sequence complex
We used electrophoretic mobility shift assays (EMSA) to test whether recombinant MBD4 would recognize specific mismatches and to determine if binding of ssODNs containing a methyl-CpG modification would efficiently recruit the MBD4 protein. ssODNs were annealed to an oligonucleotide 32P-labeled probe homologous to the region of the pGFPmut vector targeted for correction. Reactions were carried out in the presence or absence of recombinant MBD4 (Figure 2). Inclusion of ssODNs in the reaction mixture resulted in a complex with the labeled probe (Figure 2, lower arrow). A specific supershift, resulting from the binding of MBD4, was detected only when the probe was annealed to the m5CpGGFP ssODN and incubated with MBD4 but not when the recombinant protein was incubated with the probe annealed to any of the other ssODNs (Figure 2, upper arrow). No binding was detected when MBD4 was incubated with m5CpGGFP ssODN alone (Figure 2).
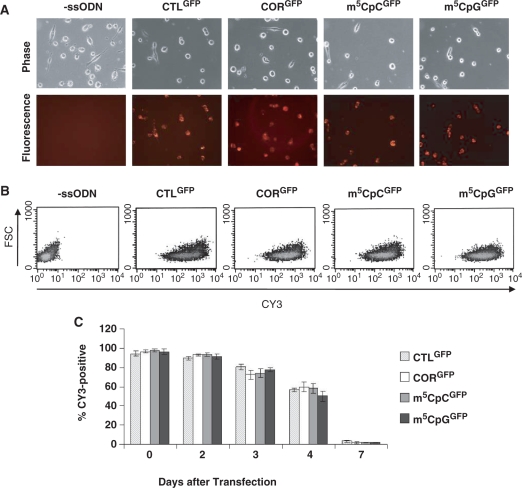
Transfection efficiencies of ssODNs
Myoblasts stably expressing pGFPmut were transfected with control or targeting oligonucleotides and ssODN uptake, assessed by immunofluorescence microscopy by the presence of the CY3 tag, was followed for up to 1 week after transfection (Figure 3). The fluorescent tag was detected in nearly 100% of cells in cultures treated with ssODNs 24 h after transfection (Figure 3A). FACS analysis demonstrated similar pattern of distribution of fluorescence in cells treated with control or targeting oligonucleotides suggesting equal transfection efficiencies among methylated and unmethylated ssODNs (Figure 3B). Fluorescence remained stable for up to 48 h after transfection and then steadily declined (Figure 3C). By 1 week, fluorescence had almost disappeared providing clues as to the stability of the oligonucleotides in the cells.
Figure 3.
Uptake of fluorescently labeled ssODNs. (A) Immunofluorescence analysis of myoblasts treated with control or targeting ssODNs 2 h after removal of the transfection reagent. Bright fluorescence could be detected virtually all cells transfected with ssODNs. (B) FACS dot-plots of CY3-positive cells 1 day after ssODN transfection. (C) No significant differences were detected in the number of CY3-positive cells transfected with different ssODNs and analyzed for up to 7 days after transfection.
Effects of ssODNs on cell viability
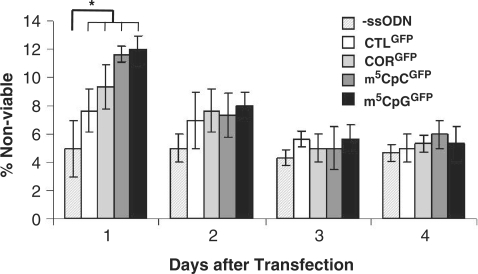
We used a cell viability assay to assess the toxicity of ssODNs on myoblast cultures for up to 4 days after transfection (Figure 4). A significant increase in non-viable cells was detected in all cultures treated with ssODNs 24 h after transfection. Oligonucleotides containing a 5-methyl cytosine appeared to be slightly more toxic than CORGFP, but those differences were not statistically significant. Those differences, however, were only transient and disappeared by 48 h after transfection. By 72 h, the number of apoptotic cells had returned to levels similar to those observed in sham transfected cells and remained stable through the duration of the experiment (Figure 4).
Figure 4.
Cell viability of myoblasts after ssODN transfection. Cell death was measured by propidium iodine staining for up to 4 days after transfection of ssODNs and compared with that measured in control transfected (‘-ssODN’) cells. Significant differences were observed in all cultures treated with ssODNs 1 day after transfection. *P ≤ 0.05. At this time, methylated ssODNs showed a higher percentage of propidium iodide-positive cells then cells transfected with CTLGFP and CORGFP. No significant differences were observed at later time points.
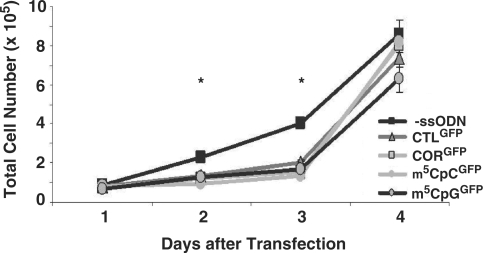
The rate of population growth was determined in cells treated with the control and targeting ssODNs and compared with those of cells transfected with vehicle only (Figure 5). Cell number doubled approximately every 18–20 h independently of the type of ssODN used. No significant differences in population growth were detected in cultures treated with targeting compared with control ssODNs.
Figure 5.
Proliferation rate of myoblasts after transfection. Myoblasts were seeded at equal density and cultured in growth media for 4 days after transfection with control and targeting ssODNs. Cells number was measured daily and growth rate of cells transfected with ssODNs was compared with that of control transfected (‘-ssODN’) cells at each time point analyzed. Cells transfected with ssODNs grew at slower rate compared with control transfected cells. No significant differences were detected in cultures transfected with nonmethylated and methylated ssODNs. *P ≤ 0.001.
We next analyzed the effects of ssODNs on cell cycle progression to determine whether ssODNs could affect the cell cycle. We did not observe any significant changes in the number of cells present at each phase of the cell cycle, even when the uptake of ssODNs was at maximal level within the first 48 h after transfection (Table 1). Furthermore, there were no changes related to the presence of methylated ssODNs compared with unmethylated ssODNs.
Table 1.
Cell cycle analysis of cells after transfection with ssODNs
| ‐ssODN |
CTLGFP |
CORGFP |
m5CpCGFP |
m5CpGGFP |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | S | G2 | G1 | S | G2 | G1 | S | G2 | G1 | S | G2 | G1 | S | G2 | |
| Day 1 | 44 ± 7 | 25 ± 1 | 30 ± 6 | 44 ± 7 | 25 ± 1 | 30 ± 7 | 41 ± 5 | 24 ± 1 | 34 ± 4 | 38 ± 4 | 24 ± 2 | 37 ± 5 | 44 ± 7 | 25 ± 1 | 31 ± 8 |
| Day 2 | 40 ± 3 | 26 ± 1 | 34 ± 3 | 41 ± 7 | 26 ± 1 | 33 ± 7 | 41 ± 5 | 30 ± 4 | 28 ± 7 | 40 ± 5 | 27 ± 3 | 33 ± 7 | 40 ± 6 | 27 ± 3 | 33 ± 7 |
| Day 3 | 41 ± 6 | 28 ± 3 | 31 ± 9 | 42 ± 4 | 27 ± 3 | 29 ± 5 | 45 ± 1 | 24 ± 3 | 31 ± 4 | 40 ± 6 | 29 ± 3 | 31 ± 4 | 45 ± 2 | 25 ± 4 | 30 ± 4 |
| Day 4 | 46 ± 5 | 26 ± 1 | 28 ± 5 | 43 ± 6 | 25 ± 3 | 31 ± 7 | 42 ± 1 | 27 ± 1 | 31 ± 1 | 36 ± 2 | 26 ± 2 | 38 ± 1 | 45 ± 8 | 28 ± 3 | 27 ± 8 |
Restoration of GFP expression after targeting ssODN treatment
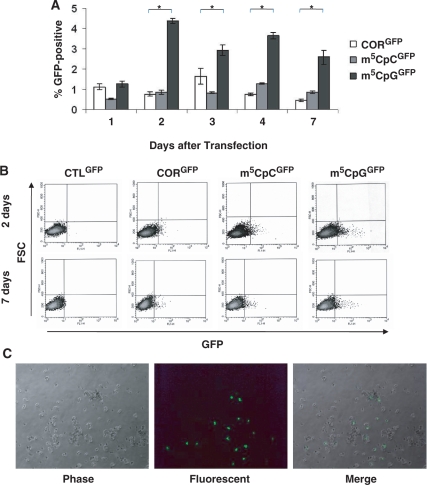
Flow cytometry was used to follow restoration of GFP expression over time in cultures transfected with ssODNs (Figure 6). No GFP-positive cells were detected at any time in untransfected cultures or in cultures transfected with the control ssODN. GFP-positive cells were detected as early as 1 day after transfection of targeting ssODNs. Two days after transfection, cultures transfected with m5CpGGFP showed up to 6-fold more GFP-positive cells than did cultures transfected with either m5CpCGFP or CORGFP (Figure 6A). GFP-positive cells were clearly detected in cultures treated with targeting oligonucleotides by both FACS and microscopic analyses (Figure 6B and C)
Figure 6.
Time course and stability of gene repair. (A) FACS analysis was used to follow GFP protein expression over time. GFP was detected as early as 1 day after ssODN transfection and did not increase significantly after 2 days. Cells transfected with m5CpGGFP showed a significantly greater number of GFP-positive cells as compared with cultures treated with targeting ssODNs, but lacking the methyl-CpG modification. *P ≤ 0.008. (B) FACS dot-plots of myoblasts treated with ssODNs 2 and 7 days after transfection of oligonucleotides. (C) Analysis of cultures by fluorescence microscopy after treatment with the m5CpGGFP ssODN and maintained in culture for 4 days. Bright fluorescence intensity was detected in a subset of replicating cells.
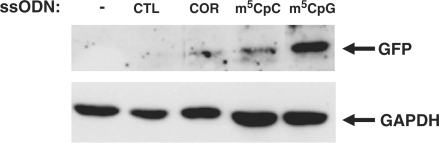
To confirm the stability of the repair process over time, myoblasts treated with ssODNs were analyzed by western blot 1 week after ssODN transfection. Expression of GFP protein was detected in cultures treated with targeting ssODNs but not with the control ssODNs (Figure 7). Myoblasts transfected with the m5CpGGFP ssODN showed 2.5-fold higher levels of GFP protein expression than cells treated with the other targeting ssODNs.
Figure 7.
GFP protein expression in muscle precursor cells in culture. Western blot analysis of C57 myoblasts stably expressing the GFP-mutant vector 1 week after ssODN transfection. Full-length GFP protein expression was higher in cells treated with m5CpGGFP compared with the other targeting ssODNs. No protein was detected in cells after transfection with the control ssODN.
Assessment of gene correction efficiency
To further confirm alteration of the GFP sequence targeted by the ssODNs and to have a precise estimate of frequencies of gene correction, we analyzed the genomic sequences by real-time PCR using DNA samples isolated 3 days after ssODN transfection. The GFP cDNA mutation in the pGFPmut vector creates a new restriction site (HinfI; Figure 8A). Using cells transfected with either CORGFP or m5CpCGFP, digestion of total genomic DNA with HinfI allowed us to cleave all copies of the mutant GFP sequence, leaving intact for amplification only that portion of DNA that had undergone single base pair alteration rendering it refractory to HinfI restriction digestion. Amplification of a specific DNA product was observed only in cells treated with the targeting ssODNs but not in untreated cells or cells transfected with the control ssODN (Figure 8B). For analysis of cells treated with m5CpGGFP, we used a similar strategy but a different restriction enzyme because of the different base targeted for correction (Figure 8A). In that case, digestion of total genomic DNA with BtsI should eliminate all copies not altered by ssODN-mediated gene modification. Again, amplification of a specific DNA product was observed only in cells treated with the targeting ssODN, but not in untreated cells or cells transfected with the control ssODN (Figure 8B). The amount of DNA amplified was higher in cells treated with m5CpGGFP, consistent with the data obtain at the protein level. Direct sequencing of the products obtained after restriction enzyme digestion and PCR amplification demonstrated the specificity of the PCR reaction and confirmed the presence of the predicted single base pair substitutions at the genomic level (Figure 8C).
Figure 8.
Evidence of ssODN-mediated single base modification at the molecular level. (A) The single base mutation on the GFP reporter vector (base underlined) generates a new restriction site for HinfI endonuclease. Promotion of an A-to-C transversion mediated by the CORGFP ssODN and an A-to-G transition mediated by the m5CpCGFP ssODN eliminate the restriction site and render the sequence refractory to HinfI digestion and cleavage. The T-to-C conversion mediated by m5CpGGFP eliminates the BtsI restriction site. (B) Genomic DNA was isolated from myoblasts stably expressing the mutant GFP 1 week after ssODN transfection and treated with either HinfI (for CORGFP, m5CpCGFP and CTLGFP) or BtsI (for m5CpGGFP and CTLGFP). Amplification was carried using a forward and a reverse primer encompassing the GFP mutation. No amplification was detected in samples treated with CTLGFP using either HinfI or BtsI. A specific PCR product of identical molecular weight to that obtained after amplification of a wild-type GFP plasmid was detected only in cells treated with targeting ssODNs. (C) Direct sequencing of PCR products obtained as in (B) by amplifying total genomic DNA having undergone HinfI or BtsI digestion demonstrated single base alterations at the genomic level and the specificity of the amplification products. (D) Real-time PCR was used to determine the relative amount of genomic DNA having undergone ssODN-mediated single base alteration. Cultures treated with m5CpGGFP showed 10-fold higher levels of gene editing compared with cultures treated with m5CpCGFP. No significant differences in efficiency were detected between cultures treated with the CORGFP and the m5CpCGFP ssODNs. The total amount of plasmid was determined by amplification of the undigested samples using the same primer combinations and was used as internal standard control to normalize DNA levels. *P = 0.001. (E) Total mRNA was isolated 1 week after ssODN transfection and reverse transcribed using an oligo dT primer. The cDNA was then digested overnight with HinfI or BtsI depending of the ssODN transfected (A) and amplified using the same primers used for the genomic DNA analysis. Modification of the mutant GFP sequence using the m5CpGGFP ssODN was more efficient than that observed in cultures treated with targeting ssODNs lacking the 5-methylcytosine (CORGFP) or the ssODN containing the m5C modification but not in a CpG context (m5CpCGFP).
The differences in fold changes of PCR products amplified after HinfI or BtsI digestion were determined using a standard delta-Ct method (48). The levels of gene correction in cells treated with m5CpGGFP were >10-fold higher than those detected using the CORGFP ssODN (Figure 8D) and corresponded to an average of 7% of the total number of copies of the pGFPmut vector present in culture. No significant changes in gene correction levels were detected when comparing the gene repair frequencies of m5CpCGFP with those achieved using the CORGFP ssODN (Figure 8D).
Endonuclease digestion analysis was also used to test for GFP mRNA expression as the result of the single base substitution at the genomic level. Total mRNA was isolated from cells 1 week after transfection and reverse transcribed prior to HinfI or BtsI digestion (see ‘Materials and Methods’ section). The control ssODN failed to produce any amplification of cDNA after restriction enzyme digestion (Figure 8E). The amount of GFP mRNA that was refractory to endonuclease digestion as a result of ssODN-mediated gene editing was consistently higher in cells treated with the MBD4-binding ssODN as compared with the other targeting ssODNs. Direct sequencing of the PCR products was used to confirm expression of GFP transcripts containing the desired single base alteration and to ensure specificity of the amplification product obtained after endonuclease digestion (data not shown).
Effects of knockdown of MBD4 expression on ssODN-mediated base alteration
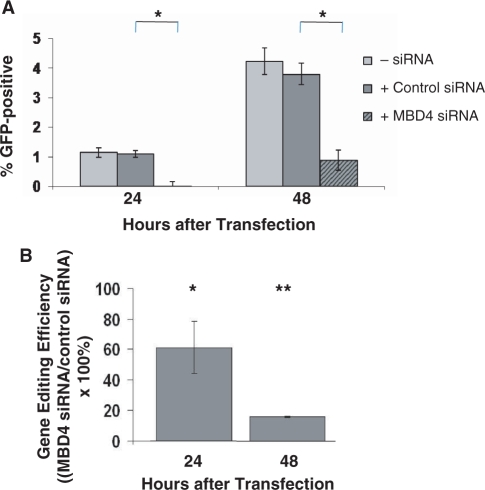
The involvement of MBD4 in the gene correction process mediated by methyl-CpG-modified ssODNs was investigated in cells by downregulating MBD4 expression. siRNA was used to knockdown expression of MBD4, leading to a significant reduction of MBD4 transcript levels as soon as 24 h after transfection and sustained for at least 48 h after transfection (Figure 9A). The level of MBD4 protein, as determined by western blot analysis, was reduced to <10% of control levels (i.e. levels observed in untransfected myoblasts or myoblasts transfected with a control siRNA) by 24 h after MBD4 siRNA transfection, and this knockdown at the protein level was maintained for at least another 24 h (Figure 9B).
We co-transfected the m5CpG ssODN with the control or MBD4 siRNA (or no siRNA) and followed restoration of GFP protein expression over time by flow cytometry. The number of GFP-positive cells detected was significantly reduced in cells treated with the MBD4 siRNA compared with those treated with control siRNA (Figure 10A). No changes in the percentage of GFP-positive cells were detected in cells co-transfected with CORGFP or m5CpCGFP and the MBD4 siRNA (data not shown). The efficiency of gene repair under each condition was confirmed at the DNA level using real-time PCR and after restriction enzyme digestion of total DNA using BtsI. The reduction in gene repair activity in cells in which MBD4 was knocked down was analyzed as a function of time after MBD4 siRNA transfection. A decrease in gene correction efficiency occurred as early as 24 h after co-transfection of m5CpGGFP and the MBD4 siRNA compared with that achieved after co-transfection of a control siRNA, and this effect was even greater at 48 h when the siRNA-mediated knockdown was most effective (Figure 10B).
Figure 10.
The influence of MBD4 knockdown on the efficiency of gene editing by a methyl-CpG-modified ssODN. (A) Expression of GFP protein was followed over time by FACS analysis. Myoblast cultures stably expressing the mutant GFP were co-transfected with m5CpGGFP ssODN and either a control siRNA or an siRNA capable of downregulating MBD4 expression (or with no siRNA). The percentage of GFP-positive cells was significantly decreased 24 and 48 h after transfection in cultures treated with the targeting siRNA compared with the control siRNA or no siRNA. *P ≤ 0.002. (B) Real-time PCR was used to determine gene editing efficiencies of cells transfected with the m5CpG ssODN and with either a control siRNA or the MBD4 siRNA. Genomic DNA was isolated 24 and 48 h after transfection and digested with BtsI. The levels of the amplification products in cultures treated with the control siRNA were set at 100%. Amplification products were significantly reduced in cells treated with the MBD4 siRNA at 24 h and further reduced at 48 h. *P ≤ 0.02; **P ≤ 0.004.
DISCUSSION
We have studied the ability of a modified-ssODN (m5CpGGFP), capable of binding MBD4 to direct a site-specific modification at the genomic level and to mediate enhanced gene repair compared with an older generation of correcting ssODN (CORGFP) unable to recruit the specific repair mechanism. Binding was demonstrated in vitro using recombinant MBD4 and was specific only to the ssODN containing the methyl-CpG modification (Figure 2). Gene correction was consistently more effective when the MBD4-binding ssODN was used as compared with an ssODN containing the 5-methylcytosine, but not in a CpG context or to an ssODN with no 5-methylcytosine modifications (Figure 8). These data were confirmed at the protein, transcript and genomic levels after treatment of cells with targeting ssODNs.
The differences in gene correction detected do not appear to be due to preferential uptake of ssODNs containing the m5CpG sequence nor to an increase in their stability after transfection, as suggested by FACS analysis of myoblasts transfected with fluorescently labeled control and targeting ssODNs and followed for up to 1 week after transfection (Figure 3). Although some toxicity was observed in ssODNs containing m5C, the increase in cell death was only transient and was not specific to MBD4-binding ssODNs as no significant differences were detected between cells transfected with m5CpGGFP and m5CpCGFP oligonucleotides. Finally, no differences in cell cycle regulation (Table 1) or number of cell divisions (Figure 5) were observed in cells treated with m5CpGGFP as compared with the other ssODNs or to sham transfected cells. Together, these data suggest that the differences in gene correction detected in myoblast cultures were not due to the differences in the ability of MBD4-binding ssODNs to influence cell replication, but were likely due to their ability to increase gene repair. The percentages of GFP-positive cells detected over time after ssODN transfection remained stable in all cultures treated with targeting oligonucleotides, but were significantly higher in cells treated with m5CpGGFP ssODN. These results clearly demonstrate that gene editing in myoblasts is stable over time, confirming the results previously obtained in our laboratory using gene editing strategies for the dystrophin gene (5,10,21). Furthermore, these data demonstrate that the use of methyl-CpG-modified ssODNs activating the BER through MBD4 binding can significantly enhance gene repair.
Restriction enzyme digestion analysis in combination with real-time PCR demonstrated that the use of ssODNs containing m5CpG and capable of binding MBD4 were able to induce up to 10-fold higher levels of gene correction than older generation oligonucleotides lacking any specific modification. In assessing the frequencies of gene correction, we have taken into consideration several parameters that are likely to yield different estimates of efficiency at the genomic, transcript and protein levels. Such variable include the number of copies of the pGFPmut plasmid integrated at the genomic level, level of expression of the plasmid after G418 selection, the possibility of silencing of plasmid gene expression in vitro and the sensitivities of the FACS and western blot analyses to detect low levels of protein expression. The use of quantitative PCR analysis allowed us to detect correction events occurring at the genomic level independently of possible silencing effects and to estimate frequencies of gene repair per total number of vectors integrated in the genome. The possibility that the results obtained were due to PCR artifacts can be excluded on several grounds. First, each real-time PCR performed had several internal controls, including DNA isolated from untransfected cells and cells transfected with the control ssODN and then subjected to PCR analysis after restriction enzyme digestion. The absence of amplification in those latter samples ensured us that the endonuclease digestion had reached completion. Second, PCR analyses following restriction enzyme digestion were repeated in triplicate experiments using different DNA samples isolated after transfection. Third, each run was loaded on agarose gels to confirm that no amplification product, other then the one expected in samples treated with the targeting ssODNs, would be present in the reaction. Finally, PCR products obtained after quantitative analysis were excised from the gel and sequenced to confirm the presence of the desired single base pair alteration.
MBD4 appears to be a major component of the mechanism of gene correction mediated by methyl-CpG-modified ssODNs. Downregulation of MBD4 alone was sufficient to prevent the restoration of GFP expression after modified ssODN treatment, as demonstrated by analysis at both the protein and the genomic DNA levels (Figure 10). These observations are supported by studies in MBD4 knockout models. Mice lacking MBD4 expression accumulate C-to-T mutations at CpG sites at a rate of 2- to 3-fold higher than wild-type mice (49,50). Other repair mechanisms, although important for maintaining genomic integrity in mammalian cells, do not appear to play an active role in recognizing deamination of 5-methylcytosine at m5CpG sites, nor can they efficiently compensate for the loss of MBD4 in mice. Therefore, the use of ssODNs containing methyl-CpG modifications and capable of annealing to the genomic sequence creating a mismatched T opposite the G in the methyl-CpG can be used to specifically recruit and activate the MBD4 pathway. The increase in the level of gene correction by ssODNs containing methyl-CpG modifications is likely the result of the efficacy of the repair mechanisms activated to precisely recognize and cleave the mismatched base on the genomic DNA targeted for repair and opposite to the ssODN.
These interpretations were also supported by EMSA assays which demonstrated that the binding of MBD4 occurred only when a G:T mismatch was created by the binding of an ssODN containing the m5CpG modification to the target sequence (Figure 2). The presence of a single methyl-CpG modification on the ssODN was sufficient to recruit MBD4 to the target sequence. Furthermore, the ability of MBD4 to recognize and bind the targeted sequence only in the presence of the methyl-CpG-modified ssODN annealed to the DNA and creating a G:T mismatch is a clear evidence that initiation of the process requires the ssODN to first anneal to the target sequence within the genomic DNA. No MBD4/DNA complexes were detected when full-length MBD4 protein was added to the reaction mixture containing the methyl-CpG-modified ssODN alone or when MBD4 was added to the reaction containing ssODNs without a methyl-CpG even in the presence of the target sequence (Figure 2).
For therapeutic applications, the levels of gene correction are likely to require higher efficiencies than those currently being achieved in different systems using ssODNs. The use of methyl-CpG-modified ssODNs is a technical advance that enhances gene repair efficiency as it directs the correction event specifically to the genomic sequence targeted for repair, diminishing the likelihood that the repair mechanism might instead alter the ssODN sequence. Understanding additional steps in the biochemical mechanisms that mediate the repair will also lead to further increases in gene repair efficiencies.
MBD4-binding ssODNs, although more effective than unmodified ssODNs, are sequence specific and can only direct conversion of a thymine into a cytosine. Furthermore, because MBD4 recognizes G:T mismatches only in the context of CpG sites, the sequence targeted by the ssODN requires the presence of a guanine immediately 3′ of the base targeted for repair. This further limits the number of mutations that can be targeted for repair in the context of human diseases. However, it should be noted that ssODN-mediated gene editing can be targeted to both the coding and noncoding strands (3,5,12,51,52), thus increasing the possible target sequences. In addition, methyl-CpG ssODNs may be useful in oligonucleotide-mediated exon skipping (21). This approach takes advantage of the ability of ssODNs to target and disrupt consensus sequences necessary for intron/exon splicing and assembly of mature mRNA. The alteration at the genomic level causes the skipping of one or more exons during the assembly of mRNA transcripts, and ssODNs designed to disrupt specific splicing regulatory elements can have therapeutic applications in cases where shorter, in-frame transcripts allow the production of partially functional proteins (21). This approach has already being employed in a mouse model of DMD using older generations of oligonucleotides with encouraging results (21). Although intron/exon are not specifically enriched in TG dinucleotides, that are the specific targets of methyl-CpG-modified ssODNs, such dinucleotides are frequently present in consensus sequences and other splicing control elements and are thus potential targets for altering splicing to restore functional protein expression from mutant genes. The ability of methyl-CpG-modified ssODNs to induce single base pair substitutions at the intron/exon boundaries of the dystrophin gene is currently being tested in our laboratory. The development of a successful approach using methyl-CpG-modified ssODNs in oligonucleotide-mediated exon skipping of the dystrophin gene would have a wide range of clinical applications. Its use could be applied to the majority of DMD patients (60–70%) in which large deletions or frameshift mutations could be corrected by restoring the dystrophin reading frame.
Random integration, mispairing and activation of homologous recombination at sites different from those targeted for repair might ultimately preclude this technology from entering a clinical scenario. For approaches aimed at repairing postmitotic muscles cells, those issues remain less of a concern since the effects are likely to be confined within a limited number of cells that have undergone repair. For those approaches aimed at targeting and correct muscle stem cells, a detailed analysis of cells undergone repair will be critical to demonstrate its safety and long-term effects. Those studies will have to focus on individual cells not just a pool of corrected cells so that the effects of the correction process can be determined in detail on a single cell base.
Future studies in our laboratory will be aimed at determining the safety of gene correction process in muscle cells and will be focused primarily on assessing the fate of cells that have undergone repair. The optimization of culturing conditions capable of maintaining in culture clones isolated after cell sorting will be critical to characterize those cells and study changes in gene expression profile in each individual cell that have undergone gene repair. To date, those studies have been problematic due to the inability to expand primary myoblasts isolated using sorting techniques. New technologies are now being developed that will allow us to study expression profiles from single cells and might enable us not only to study the effects of gene repair within each individual cell, but also to determine the effects of ssODNs immediately after transfection. Those technologies might ultimately hold the key in determining the applicability of ssODN-mediated gene correction for the treatment of many genetic defects.
Ultimately, safe and effective treatments of genetic diseases using ssODNs will necessitate synergistic efforts aimed not only at increasing the level of gene repair but also methods to insure efficient delivery of ssODNs to all the cells whose correction would be necessary for maximum therapeutic effect. For diseases in which the tissue being targeted undergoes continuous turnover either under normal physiological conditions or in the setting of the disease, gene correction of the stem or progenitor cells that are responsible for forming new tissue would be essential to insure sustained therapeutic effects. These delivery issues are faced by every form of gene therapy. However, especially for nonviral forms of gene therapy, the efficacy of the vector once delivered to the cell, both in terms of magnitude and duration, will be a critical determinant of the success of that therapeutic approach. The use of methyl-CpG-modified ssODNs is just such a method to enhance the efficacy of ssODN-mediated gene editing, and may have applications across a wide range of genetic disorders.
FUNDING
UCLA Human Gene Medicine Seed Grant (to C.B.); grants from the Muscular Dystrophy Association (USA) (to C.B. and T.A.R.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Cole-Strauss A, Yoon K, Xiang Y, Byrne BC, Rice MC, Gryn J, Holloman WK, Kmiec EB. Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science. 1996;273:1386–1389. doi: 10.1126/science.273.5280.1386. [DOI] [PubMed] [Google Scholar]
- 2.Chan PP, Lin M, Faruqi AF, Powell J, Seidman MM, Glazer PM. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J. Biol. Chem. 1999;274:11541–11548. doi: 10.1074/jbc.274.17.11541. [DOI] [PubMed] [Google Scholar]
- 3.Igoucheva O, Alexeev V, Yoon K. Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther. 2001;8:391–399. doi: 10.1038/sj.gt.3301414. [DOI] [PubMed] [Google Scholar]
- 4.Sangiuolo F, Bruscia E, Serafino A, Nardone AM, Bonifazi E, Lais M, Gruenert DC, Novelli G. In vitro correction of cystic fibrosis epithelial cell lines by small fragment homologous replacement (SFHR) technique. BMC Med. Genet. 2002;3:8. doi: 10.1186/1471-2350-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertoni C, Morris GE, Rando TA. Strand bias in oligonucleotide-mediated dystrophin gene editing. Hum. Mol. Genet. 2005;14:221–233. doi: 10.1093/hmg/ddi020. [DOI] [PubMed] [Google Scholar]
- 6.Agarwal S, Gamper HB, Kmiec EB. Nucleotide replacement at two sites can be directed by modified single-stranded oligonucleotides in vitro and in vivo. Biomol. Eng. 2003;20:7–20. doi: 10.1016/s1389-0344(02)00088-6. [DOI] [PubMed] [Google Scholar]
- 7.Gamper HB, Jr, Cole-Strauss A, Metz R, Parekh H, Kumar R, Kmiec EB. A plausible mechanism for gene correction by chimeric oligonucleotides. Biochemistry. 2000;39:5808–5816. doi: 10.1021/bi9921891. [DOI] [PubMed] [Google Scholar]
- 8.You HJ, Swanson RL, Harrington C, Corbett AH, Jinks-Robertson S, Senturker S, Wallace SS, Boiteux S, Dizdaroglu M, Doetsch PW. Saccharomyces cerevisiae Ntg1p and Ntg2p: broad specificity N-glycosylases for the repair of oxidative DNA damage in the nucleus and mitochondria. Biochemistry. 1999;38:11298–11306. doi: 10.1021/bi991121i. [DOI] [PubMed] [Google Scholar]
- 9.Bartlett RJ, Stockinger S, Denis MM, Bartlett WT, Inverardi L, Le TT, Man N, Morris GE, Bogan DJ, Metcalf-Bogan J, et al. In vivo targeted repair of a point mutation in the canine dystrophin gene by a chimeric RNA/DNA oligonucleotide. Nat. Biotechnol. 2000;18:615–622. doi: 10.1038/76448. [DOI] [PubMed] [Google Scholar]
- 10.Bertoni C, Rando TA. Dystrophin gene repair in mdx muscle precursor cells in vitro and in vivo mediated by RNA/DNA chimeric oligonucleotides. Hum. Gene Ther. 2002;13:707–718. doi: 10.1089/104303402317322276. [DOI] [PubMed] [Google Scholar]
- 11.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorensen CB, Krogsdam AM, Andersen MS, Kristiansen K, Bolund L, Jensen TG. Site-specific strand bias in gene correction using single-stranded oligonucleotides. J. Mol. Med. 2005;83:39–49. doi: 10.1007/s00109-004-0592-6. [DOI] [PubMed] [Google Scholar]
- 13.Kren BT, Bandyopadhyay P, Steer CJ. In vivo site-directed mutagenesis of the factor IX gene by chimeric RNA/DNA oligonucleotides. Nat. Med. 1998;4:285–290. doi: 10.1038/nm0398-285. [DOI] [PubMed] [Google Scholar]
- 14.Bandyopadhyay P, Ma X, Linehan-Stieers C, Kren BT, Steer CJ. Nucleotide exchange in genomic DNA of rat hepatocytes using RNA/DNA oligonucleotides. Targeted delivery of liposomes and polyethyleneimine to the asialoglycoprotein receptor. J. Biol. Chem. 1999;274:10163–10172. doi: 10.1074/jbc.274.15.10163. [DOI] [PubMed] [Google Scholar]
- 15.Kren BT, Chen Z, Felsheim R, Roy CN, Roy CJ, Steer CJ. Modification of hepatic genomic DNA using RNA/DNA oligonucleotides. Gene Ther. 2002;9:686–690. doi: 10.1038/sj.gt.3301762. [DOI] [PubMed] [Google Scholar]
- 16.Kren BT, Wong PY, Steer CJ. Short, single-stranded oligonucleotides mediate targeted nucleotide conversion using extracts from isolated liver mitochondria. DNA Repair (Amst) 2003;2:531–546. doi: 10.1016/s1568-7864(03)00022-3. [DOI] [PubMed] [Google Scholar]
- 17.Andrieu-Soler C, Casas M, Faussat AM, Gandolphe C, Doat M, Tempe D, Giovannangeli C, Behar-Cohen F, Concordet JP. Stable transmission of targeted gene modification using single-stranded oligonucleotides with flanking LNAs. Nucleic Acids Res. 2005;33:3733–3742. doi: 10.1093/nar/gki686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciavatta VT, Padove SA, Boatright JH, Nickerson JM. Mouse retina has oligonucleotide-induced gene repair activity. Invest. Ophthalmol. Vis. Sci. 2005;46:2291–2299. doi: 10.1167/iovs.04-1220. [DOI] [PubMed] [Google Scholar]
- 19.Flagler K, Alexeev V, Pierce EA, Igoucheva O. Site-specific gene modification by oligodeoxynucleotides in mouse bone marrow-derived mesenchymal stem cells. Gene Ther. 2008;15:1035–1048. doi: 10.1038/gt.2008.31. [DOI] [PubMed] [Google Scholar]
- 20.Rando TA, Disatnik MH, Zhou LZ. Rescue of dystrophin expression in mdx mouse muscle by RNA/DNA oligonucleotides. Proc. Natl Acad. Sci. USA. 2000;97:5363–5368. doi: 10.1073/pnas.97.10.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertoni C, Lau C, Rando TA. Restoration of dystrophin expression in mdx muscle cells by chimeraplast-mediated exon skipping. Hum. Mol. Genet. 2003;12:1087–1099. doi: 10.1093/hmg/ddg133. [DOI] [PubMed] [Google Scholar]
- 22.DiMatteo D, Callahan S, Kmiec EB. Genetic conversion of an SMN2 gene to SMN1: a novel approach to the treatment of spinal muscular atrophy. Exp. Cell Res. 2008;314:878–886. doi: 10.1016/j.yexcr.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Agarwal S, Kmiec E, Davis BR. Targeted beta-globin gene conversion in human hematopoietic CD34(+) and Lin(-)CD38(-)cells. Gene Ther. 2002;9:118–126. doi: 10.1038/sj.gt.3301610. [DOI] [PubMed] [Google Scholar]
- 24.Igoucheva O, Alexeev V, Yoon K. Nuclear extracts promote gene correction and strand pairing of oligonucleotides to the homologous plasmid. Antisense Nucleic Acid Drug Dev. 2002;12:235–246. doi: 10.1089/108729002320351557. [DOI] [PubMed] [Google Scholar]
- 25.Olsen PA, McKeen C, Krauss S. Branched oligonucleotides induce in vivo gene conversion of a mutated EGFP reporter. Gene Ther. 2003;10:1830–1840. doi: 10.1038/sj.gt.3302079. [DOI] [PubMed] [Google Scholar]
- 26.Parekh-Olmedo H, Kmiec EB. Progress and prospects: targeted gene alteration (TGA) Gene Ther. 2007;14:1675–1680. doi: 10.1038/sj.gt.3303053. [DOI] [PubMed] [Google Scholar]
- 27.Dekker M, Brouwers C, Te RH. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. 2003;31:e27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maguire KK, Kmiec EB. Multiple roles for MSH2 in the repair of a deletion mutation directed by modified single-stranded oligonucleotides. Gene. 2007;386:107–114. doi: 10.1016/j.gene.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferrara L, Kmiec EB. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004;32:5239–5248. doi: 10.1093/nar/gkh822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferrara L, Parekh-Olmedo H, Kmiec EB. Enhanced oligonucleotide-directed gene targeting in mammalian cells following treatment with DNA damaging agents. Exp. Cell Res. 2004;300:170–179. doi: 10.1016/j.yexcr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Radecke F, Peter I, Radecke S, Gellhaus K, Schwarz K, Cathomen T. Targeted chromosomal gene modification in human cells by single-stranded oligodeoxynucleotides in the presence of a DNA double-strand break. Mol. Ther. 2006;14:798–808. doi: 10.1016/j.ymthe.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 33.Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl Acad. Sci. USA. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barzilay G, Hickson ID. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays. 1995;17:713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 36.Laird PW, Jaenisch R. The role of DNA methylation in cancer genetic and epigenetics. Annu. Rev. Genet. 1996;30:441–464. doi: 10.1146/annurev.genet.30.1.441. [DOI] [PubMed] [Google Scholar]
- 37.Schmutte C, Jones PA. Involvement of DNA methylation in human carcinogenesis. Biol. Chem. 1998;379:377–388. doi: 10.1515/bchm.1998.379.4-5.377. [DOI] [PubMed] [Google Scholar]
- 38.Bellacosa A, Cicchillitti L, Schepis F, Riccio A, Yeung AT, Matsumoto Y, Golemis EA, Genuardi M, Neri G. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc. Natl Acad. Sci. USA. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 40.Bellacosa A. Role of MED1 (MBD4) Gene in DNA repair and human cancer. J. Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- 41.Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 42.Dianov G, Lindahl T. Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 1994;4:1069–1076. doi: 10.1016/s0960-9822(00)00245-1. [DOI] [PubMed] [Google Scholar]
- 43.Wilson SH. Mammalian base excision repair and DNA polymerase beta. Mutat. Res. 1998;407:203–215. doi: 10.1016/s0921-8777(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 44.Rando TA, Crowley RS, Carlson EJ, Epstein CJ, Mohapatra PK. Overexpression of copper/zinc superoxide dismutase: a novel cause of murine muscular dystrophy. Ann. Neurol. 1998;44:381–386. doi: 10.1002/ana.410440315. [DOI] [PubMed] [Google Scholar]
- 45.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Genuardi M, Karbowski M, Yeung AT, Matsumoto Y, Bellacosa A. Investigation of the substrate spectrum of the human mismatch-specific DNA N-glycosylase MED1 (MBD4): fundamental role of the catalytic domain. J. Cell Physiol. 2000;185:473–480. doi: 10.1002/1097-4652(200012)185:3<473::AID-JCP19>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 46.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J. Biol. Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 47.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 48.Bertoni C, Jarrahian S, Wheeler TM, Li Y, Olivares EC, Calos MP, Rando TA. Enhancement of plasmid-mediated gene therapy for muscular dystrophy by directed plasmid integration. Proc. Natl Acad. Sci. USA. 2006;103:419–424. doi: 10.1073/pnas.0504505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 50.Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, Fazzari M, Jin B, Brown AM, Lipkin M, et al. Mbd4 inactivation increases Cright-arrowT transition mutations and promotes gastrointestinal tumor formation. Proc. Natl Acad. Sci. USA. 2002;99:14937–14942. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kenner O, Kneisel A, Klingler J, Bartelt B, Speit G, Vogel W, Kaufmann D. Targeted gene correction of hprt mutations by 45 base single-stranded oligonucleotides. Biochem. Biophys. Res. Commun. 2002;299:787–792. doi: 10.1016/s0006-291x(02)02749-3. [DOI] [PubMed] [Google Scholar]
- 52.Brachman EE, Kmiec EB. Targeted nucleotide repair of cyc1 mutations in Saccharomyces cerevisiae directed by modified single-stranded DNA oligonucleotides. Genetics. 2003;163:527–538. doi: 10.1093/genetics/163.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]