Abstract
Archaeal family B polymerases bind tightly to the deaminated bases uracil and hypoxanthine in single-stranded DNA, stalling replication on encountering these pro-mutagenic deoxynucleosides four steps ahead of the primer–template junction. When uracil is specifically bound, the polymerase–DNA complex exists in the editing rather than the polymerization conformation, despite the duplex region of the primer-template being perfectly base-paired. In this article, the interplay between the 3′–5′ proofreading exonuclease activity and binding of uracil/hypoxanthine is addressed, using the family-B DNA polymerase from Pyrococcus furiosus. When uracil/hypoxanthine is bound four bases ahead of the primer–template junction (+4 position), both the polymerase and the exonuclease are inhibited, profoundly for the polymerase activity. However, if the polymerase approaches closer to the deaminated bases, locating it at +3, +2, +1 or even 0 (paired with the extreme 3′ base in the primer), the exonuclease activity is strongly stimulated. In these situations, the exonuclease activity is actually stronger than that seen with mismatched primer-templates, even though the deaminated base-containing primer-templates are correctly base-paired. The resulting exonucleolytic degradation of the primer serves to move the uracil/hypoxanthine away from the primer–template junction, restoring the stalling position to +4. Thus the 3′–5′ proofreading exonuclease contributes to the inability of the polymerase to replicate beyond deaminated bases.
INTRODUCTION
DNA polymerases from many species, including bacteria, viruses and eukaryotes, possess a 3′–5′ proofreading exonuclease activity which removes misincorporated bases from extending primers, thereby improving fidelity. The catalytic centres responsible for polymerase and exonuclease functions are well separated and co-crystal structures with DNA being extended (polymerase conformation) or subject to exonucleolytic proofreading (editing conformation) are distinct (1–6). The family-B DNA polymerases from the archaeal domain also demonstrate proofreading activity, and, consequently, are able to synthesize DNA with high accuracy. This feature, combined with extreme thermostability, makes these enzymes very useful in the PCR (7,8). Crystal structures of a number of archaeal polymerases, e.g. from Thermococcus gorgonarius (Tgo-Pol) have demonstrated, as expected, significant distance between the polymerase and exonuclease active sites (9).
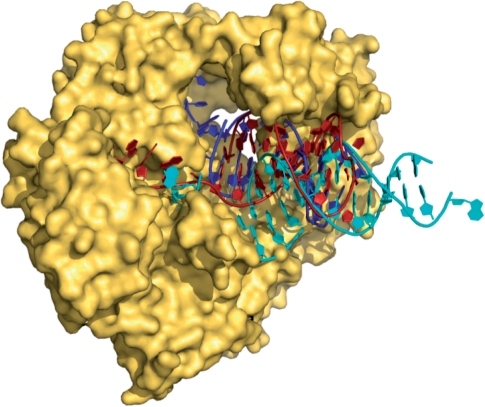
Archaeal DNA polymerases have an additional unusual property, binding tightly to the deaminated bases uracil and hypoxanthine and stalling DNA replication when these bases are encountered (10–16). During replication, these polymerases scan the template strand ahead of the replication fork and capture uracil/hypoxanthine in a specific pocket, when it is encountered four bases ahead of the primer–template junction (10,15). The subsequent cessation of replication stops the copying of uracil with adenine, and the conversion of a C:G to a T:A base-pair, in cases where the uracil resulted from cytosine deamination (10). Similarly, trapping of hypoxanthine (which can arise from adenine deamination) prevents A:T to G:C transitions. Recently, a crystal structure of Tgo-Pol, in complex with a primer-template containing uracil at the optimal +4 position in the template, has been solved (13). As anticipated (11), the uracil was flipped into a specific binding pocket located within the polymerase N-terminal domain. Comparison with a related family B DNA polymerase from the RB-69 bacteriophage, for which both editing and polymerization complexes have been determined (17,18), indicated that the archaeal polymerase bound the uracil-containing DNA in an editing mode (Figure 1). This observation was somewhat unexpected as the duplex region of the uracil-containing primer-template contained only bona fide Watson–Crick base-pairs.
Figure 1.
Structure of Tgo-Pol bound to a uracil-containing primer-template (red) (13). Superimposed are the expected positions of DNA bound in the polymerization mode (cyan) (17) and the editing mode (blue) (18) derived from structures of the family-B polymerase from bacteriophage RB69. The position of the uracil-containing primer-template clearly maps to the editing conformation more closely than to the polymerization.
Prior to the structure of Tgo-Pol with a uracil-containing primer-template, no link between deaminated base recognition, mediated by the N-terminal domain, and proofreading activity, catalysed by a distinct exonuclease domain, was suspected. Indeed, most mechanistic and structural investigations of deaminated base recognition have been conducted with polymerase mutants lacking exonuclease activity (exo−), to prevent unwanted degradation of primer-templates (10–15). With the observation that uracil capture places the DNA in an editing conformation, this study investigates any potential role of the 3′–5′ exonuclease activity in preventing replication beyond template strand deaminated bases. The family B polymerase from Pyrococcus furiosus (Pfu-Pol), which has ∼80% amino acid sequence identity to Tgo-Pol and has previously been employed extensively to characterize uracil recognition, has been used in all experiments.
MATERIALS AND METHODS
Exonuclease assays
The primer-templates (primers labelled at the 5′-end with Cy5) used for exonuclease assays are shown in Table 1. Reactions were carried out in 400 µl of 20 mM Tris–HCl (pH 8.5), 10 mM KCl, 20 mM MgSO4, 10 mM (NH4)2SO4, 20 nM primer-template, 100 nM Pfu-PCNA (19) and 100 nM Pfu-Pol (20), the last component being added for initiation. Exonuclease assays were carried out with both wild type Pfu-Pol (20) and V93Q, a deaminated base insensitive mutant (11,13). The assay temperature was 30°C and timed 40 µl aliquots were withdrawn and the reaction quenched by addition of 40 µl stop buffer (40% formamide, 0.1 M EDTA and orange G) and 1 µl of a 100 µM solution of ‘competitor DNA’ (an exact complement of the template strand under study but lacking Cy5). The samples were denatured by heating to 90°C for 10 min and then rapidly cooled on ice. The excess of the ‘competitor’ prevents any rehybridization of the Cy5 primer to the template and ensures all the Cy5 primer, and products derived from it, remain single stranded during analysis. Products were detected using denaturing polyacrylamide (15%) gel electrophoresis (even while using denaturing gels, we observed significant hybridization of the Cy5 primer to the template if the competitor was omitted, the resulting double-stranded structures interfering with the assay) followed by fluorescence detection using Typhoon scanner (GE Healthcare). For reactions with fast time courses, an RQF-63 rapid quench flow apparatus was used (Hi-Tech Scientific, Bradford on Avon, UK).
Table 1.
The primer-templates used in exonuclease and extension assays
| Designation | Sequence |
|---|---|
| Control | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTTCGTTCGAACAGAGG |
| Mismatch | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTCCGTTCGAACAGAGG |
| U-1 | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGUTCGTTCGAACAGAGG |
| U 0 | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTUCGTTCGAACAGAGG |
| U+1 | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTTUGTTCGAACAGAGG |
| U+2 | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTTCUTTCGAACAGAGG |
| U+3 | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTTCGUTCGAACAGAGG |
| U+4 | Cy5-GGGGATCCTCTAGAGTCGACCTGCAGGGCAA |
| (AA/TT) | CCCCTAGGAGATCTCAGCTGGACGTCCCGTTCGTUCGAACAGAGG |
| Control | Cy5-GGGGATCCTCTAGAGTCGACCTGC |
| (GC/CG) | CCCCTAGGAGATCTCAGCTGGACGACCGTTCGTTCGAACAGAGG |
| Mismatch | Cy5-GGGGATCCTCTAGAGTCGACCTGC |
| (GC/CG) | CCCCTAGGAGATCTCAGCTGGACAACCGTTCGTTCGAACAGAGG |
| U+2 | Cy5-GGGGATCCTCTAGAGTCGACCTGC |
| (GC/CG) | CCCCTAGGAGATCTCAGCTGGACGAUCGTTCGTTCGAACAGAGG |
| U+4 | Cy5-GGGGATCCTCTAGAGTCGACCTGC |
| (GC/CG) | CCCCTAGGAGATCTCAGCTGGACGACCUTTCGTTCGAACAGAGG |
| H+2 | Cy5-GGGGATCCTCTAGAGTCGACCTGC |
| (GC/CG) | CCCCTAGGAGATCTCAGCTGGACGAHCGTTCGTTCGAACAGAGG |
| H+4 | Cy5-GGGGATCCTCTAGAGTCGACCTGC |
| (GC/CG) | CCCCTAGGAGATCTCAGCTGGACGACCHTTCGTTCGAACAGAGG |
The top oligodeoxynucleotide serves as the primer strand (written in 5′–3′ direction) and the bottom as the template (3′–5′ direction). Emboldened and underlining is used to highlight bases that differ from the appropriate control template.
Primer–template extension assays
The conditions and analysis methods were identical to exonuclease assays except that 400 µM each of dATP, dCTP, dGTP and dTTP were added to the reaction mixture and the assay temperature was 50°C. Extension assays were carried out both with wild type Pfu-Pol and the D215A point mutant, which is disabled in 3′–5′ exonuclease activity (20). A second set of experiments used the deaminated base insensitive mutation V93Q (11,13) and the double mutant V93Q/D215A, disabled in both deaminated base recognition and 3′–5′ exonuclease activity.
Data analysis
For band-density analysis of gel images, ‘Image Quant’ software (GE Healthcare) was used to determine the percentages of substrate and products. Data was fitted to the equation for a first order reaction using ‘Grafit’ (Erithacus Software, London), allowing determination of the rate constant for selected reactions.
RESULTS
Primer–templates for Pfu-Pol exonuclease assays
The primer-templates shown in Table 1 have been exploited to investigate any coupling between deaminated base recognition and 3′–5′ exonuclease by Pfu-Pol. The series labelled AA/TT has, for the control, two A:T base pairs in the double-stranded region immediately adjacent to the primer–template junction. Within the AA/TT set, the first two entries are primer-templates, containing only the four standard bases, either completely Watson–Crick base-paired (control), or with a single mismatch at the primer–template junction (mismatch). The remaining nucleic acids all contain a single uracil in the template strand. In two cases, the uracil is located in the duplex region [at positions −1 and 0 (at the primer–template junction)], in both instances being correctly base-paired with adenine. For the remainder, the uracil is located in the single-stranded region of the template spanning positions +1 to +4. A second series, documented GC/CG, has a slightly shorter primer strand which, with the control, places two G:C base pairs at the primer–template junction. This set consists of a fully base-paired control, a single mismatch at the primer–template junction and uracil or hypoxanthine at locations +2 and +4.
Exonuclease activity of Pfu-Pol in the presence of template strand deaminated bases
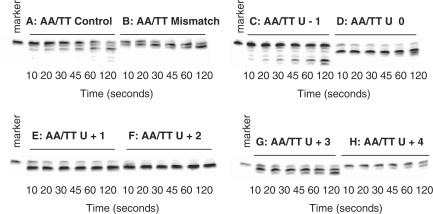
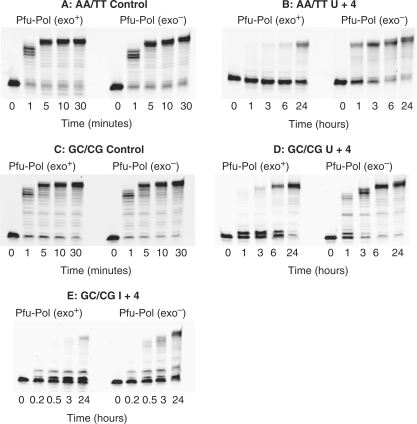
Any influence of the polymerase 3′–5′ exonuclease activity on the recognition of deaminated base was initially investigated using the AA/TT set under single turnover kinetic conditions. The polymerase binds tightly to uracil/hypoxanthine-containing primer-templates, resulting in complete binding of the DNA with a slight excess of protein at achievable concentrations of both components (15,19). Unfortunately, the poor interaction of the polymerase with standard primer-templates (e.g. DNA labelled control and mismatch in Table 1) makes full binding of the DNA difficult to achieve at reasonable concentrations of the two macromolecules. Fortunately, the presence of PCNA, the processivity clamp for Pfu-Pol, significantly improves the interaction of primer-templates with the polymerase. Therefore, all reactions contained Pfu-Pol (100 nM), PCNA (100 nM) and primer-template (20 nM), conditions previously shown to result in full binding of primer-templates, including those lacking uracil/hypoxanthine, to the polymerase and, hence, single turnover conditions (19). Figure 2 shows the exonucleolysis of the AA/TT primer-templates listed in Table 1, under the single turnover conditions detailed above, and over a time course of two minutes. The fully base-paired control is degraded with a half life of about 1 min and, as expected, the mismatched substrate is hydrolysed more rapidly, with most of the starting material removed at about 45 s. Remarkably, the primer-templates containing uracil at positions 0, +1, +2 and +3 are broken down very rapidly with the majority of the initial primer removed within 10 s. In contrast, when uracil is at +4, exonucleolysis is slow and most of the primer persists for the entire 2 min reaction time. The primer-template with uracil at −1 behaves in a similar manner to the control and is broken down neither especially rapidly nor slowly, as seen with the other uracil-containing substrates.
Figure 2.
Exonuclease assay gel images for the AA/TT primer-templates listed in Table 1 observed with Pfu-Pol (exo+). The primer-templates are identified above each of the panels. The marker is the primer itself, showing the position of the starting material.
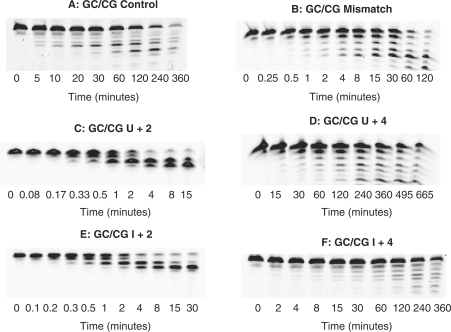
To confirm the generality of the above results, a second set of experiments has been undertaken with the GC/CG series of primer-templates. The essential difference is that the first set contains two A:T base-pairs at the primer–template junction, the second two G:Cs. The A:T series is expected to be more susceptible to ‘fraying’ than the G:C, giving rise, more easily, to single-stranded regions at the immediate 3′-terminus of the template. The proofreading 3′–5′ exonuclease activity of DNA polymerases requires unwinding of the template, with the generation of single strands, for activity (17,18). Therefore, the nature of the bases at the junction may influence the role the exonuclease plays on encountering deaminated bases. However, as shown in Figure 3, the exonuclease acts similarly on uracil, regardless of the base composition at the primer–template junction. Thus, for the GC/CG set the presence of uracil at +2 results in rapid exonucleolyis, faster than that observed for the mismatch. With uracil at +4, hydrolysis is slightly slowed, relative to the fully base-paired control. Uracil at other positions (U+1, U+3) causes the polymerase to behave in an analogous manner to that seen with the AA/TT primer-templates, with rapid exonuclease activity (data not shown). Figure 3 also shows the behaviour seen with hypoxanthine, the deamination product of adenine, which is also recognized by the polymerase (15,19). As with uracil, the presence of hypoxanthine at the +2 and +4 locations results in very rapid and marginally reduced exonucleolytic degradation, respectively.
Figure 3.
Exonuclease assay gel images for the GC/CG primer-templates listed in Table 1 observed with Pfu-Pol (exo+). The primer-templates are identified above each panel. Time 0 is the observation prior to adding enzyme and gives the position of the starting materials.
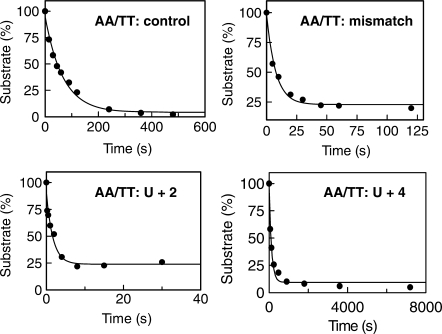
To put the data observed in Figures 2 and 3 on a more quantitative basis, the first order rate constants for the exonucleolysis of selected primer-templates have been determined. For the AA/TT set, experiments were carried out with the control, mismatch, U+2 (as representative of U0, +1, +2 and +3) and U+4 primer-templates. The same representatives (control, mismatch U+2 and U+4) were used with the GC/CG primer-templates, and in this case, H+2 and H+4 were also investigated. Rate constants were determined by carrying out the hydrolysis reactions over appropriate time spans, and the gels obtained (not shown) were of similar quality to those in Figures 2 and 3. Fits to single exponential decay plots, obtained for the AA/TT series, are shown in Figure 4 and the first order rate constants obtained are summarized in Table 2. All members of the AA/TT series are degraded more rapidly than their corresponding GC/CG partners, presumably because the ‘weaker’ primer–template junction is more prone to ‘fraying’ and the formation of single strands needed for exonucleolysis. Thus, for example, the control AA/TT has a rate constant for degradation some 75-fold higher than the corresponding GC/CG control. Similar ratios are obtained from other comparable pairs and, therefore, the influence of both mismatches and deaminated bases are similar in the two sets. The mismatched primer-template is hydrolysed about 5 (GC/CG)–10 (AA/TT) fold faster than the control. Most striking is the very rapid degradation of primer-templates containing a deaminated base at +2, maintained for both GC/CG and AA/TT and occurring with both uracil and hypoxanthine. Depending on the precise primer-template, exonucleolysis takes place about two orders of magnitude faster than controls, at even more rapid rates than seen for mismatches. Sequences containing a deaminated base at +4 are hydrolysed more slowly than the appropriate control, albeit by rather small (<2) factors.
Figure 4.
Determination of the rate constants for the exonucleolysis of primer-templates. Data, obtained from gels similar to those shown in Figures 2 and 3, were fitted to a first order decay to show the disappearance of substrate. Only the data obtained with the AA/TT series is shown (similar quality fits were obtained with the GC/CG set). For U+2, a rapid quench apparatus was used; for the other three primer-templates, manual stopping of the reactions was sufficient. The rate constants found are given in Table 2.
Table 2.
The rate constants, determined under single turnover conditions, for the 3′–5′ proofreading exonuclease of Pfu-Pol with different primer-templates
| Primer-templatea | Rate constantb (min−1) | Rate (relative to appropriate control) |
|---|---|---|
| Control (AA/TT) | 0.84 ± 0.06 | 1 |
| Mismatch (AA/TT) | 9 ± 2 | 10.7 |
| U+2 (AA/TT) | 48 ± 12 | 57 |
| U+4 (AA/TT | 0.42 ± 0.06 | 0.5 |
| Control (GC/CG) | 0.011 ± 0.001 | 1 |
| Mismatch (GC/CG) | 0.061 ± 0.007 | 5.5 |
| U+2 (GC/CG) | 1.8 ± 0.3 | 163 |
| U+4 (GC/CG) | 0.008 ± 0.0007 | 0.7 |
| H+2 (GC/CG) | 1.5 ± 0.3 | 136 |
| H+4 (GC/CG) | 0.009 ± 0.004 | 0.8 |
aThe full primer–template sequences are given in Table 1.
bEach rate constant is the average of three values ± SD.
Primer–template extension by Pfu-Pol exo+ and exo− with template strand deaminated bases
To determine the influence of the 3′–5′ exonuclease activity on the ability of Pfu-Pol to replicate beyond template strand deaminated bases, extension assays have been carried out with the control and U+4 primer-templates, in both the AA/TT and GC/CG series, listed in Table 1. Experiments were also recorded with I+4 in the GC/CG context. In contrast to the exonuclease experiments (performed at 30°C), extensions were carried out at 50°C in order to produce measurable results with deaminated base-containing templates. The results are given in Figure 5, which shows that replication using control templates (i.e. lacking deaminated bases) is the same for the exo+ and exo− variants of Pfu-Pol (Figure 5, panels A and C). The extension ladders are identical with full length product appearing in 5 min. In agreement with earlier results (10,15,19), copying beyond uracil is extremely slow and requires extended times of up to 24 h. Nevertheless, it is apparent that the exo− variant is more proficient at extension, with product clearly visible at 1 and 3 h and most of the starting material consumed after 6 h. With the wild type exo+ Pfu-Pol barely any product can be seen after 1 and 3 h and the majority of the substrate is still present after 6 h (Figure 5, panels B and D). Similar results were seen with I+4 (Figure 5, panel E), where the exo− variant produced more full length product then exo+.
Figure 5.
Gel images for the extension of primer-templates using wild type Pfu-Pol (exo+) and Pfu-Pol D215A, a 3′–5′ proofreading exonuclease deficient mutant (exo−). Unlike exonuclease assays (performed at 30°C), extensions were carried out at 50°C in order to produce observable incorporation with deaminated base-containing templates. The primer-templates are identified above each image. Note the much longer time courses needed to observe extension with deaminated bases present as compared to controls.
Stimulation of exonuclease activity is dependent on deaminated base binding
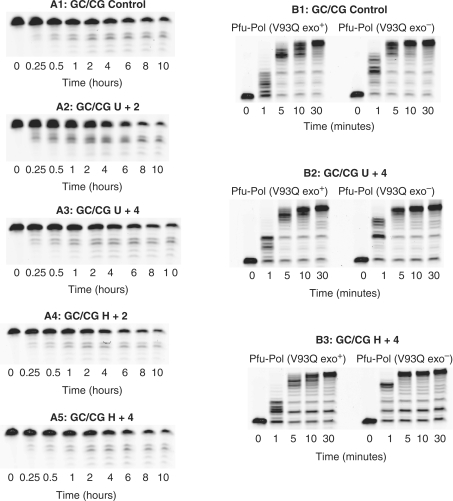
To verify that the marked increase in exonuclease rates seen on encountering uracil/hypoxanthine is a consequence of their specific binding, a deaminated base insensitive mutant, V93Q, has been used. Valine 93 makes a hydrophobic stacking interaction with uracil and its replacement with glutamine sterically occludes binding (13). As a result, V93Q does not bind strongly to deaminated bases and the stalling response is largely abolished. As shown in Figure 6 (panels labelled with an A), the gel patterns seen for exonucleolysis of the GC/CG set are largely independent of the presence of a deaminated bases. Confirmation comes from Table 3 , which illustrates that there is little difference in the rate constants observed for the exonuclease activity of V93Q with the control, U+2, U+4, H+2 and H+4. In particular, there is no marked acceleration with a deaminated base at +2, as seen with the wild type polymerase. A slight (2.6-fold) increase is seen with V93Q and U+2, but this is very much less than the factors measured with the wild type and may be accounted for by residual binding ability of the mutant. It is also noted that V93Q has a less powerful endonuclease activity than the wild type (compare the GC/CG controls in Tables 2 and 3); at present, the origin of this difference is unclear. Figure 6 (panels marked B) also shows that the exo− variant of V93Q is no more proficient at reading beyond deaminated bases than exo+, again in contrast to the wild type. V93Q exo− is slightly better at extension than exo+ (again why this arises is unclear), but this is a general change seen with the controls as well as the uracil/hypoxanthine containing primer-templates.
Figure 6.
Gel images for the exonucleolyis and polymerization by a deaminated base insensitive mutation, Pfu-Pol V93Q. The figures on the left (designated with an A) are 3′–5′ exonuclease assays seen with V93Q for the primer-templates identified on the top of each of the panels. The rate constants obtained from these gels are given in Table 3. The figures on the right (designated with a B) are polymerization assays for both V93Q exo+ and V93Q exo− using the primer-templates identified above each of the panels. All assays were carried out at 30°C.
Table 3.
The rate constants, determined under single turnover conditions, for the 3′–5′ proofreading exonuclease of Pfu-Pol V93Q with different primer-templates
| Primer-templatea | Rate constantb (min−1) | Rate (relative to control) |
|---|---|---|
| Control (GC/CG) | 0.0026 ± 0.0006 | 1 |
| U+2 (GC/CG) | 0.0067 ± 0.002 | 2.6 |
| U+4 (GC/CG) | 0.0034 ± 0.001 | 1.3 |
| H+2 (GC/CG) | 0.003 ± 0.0003 | 1.1 |
| H+4 (GC/CG) | 0.0027 ± 0.0001 | 1 |
aThe full primer–template sequences are given in Table 1.
bEach rate constant is the average of three values ± SD.
CONCLUSIONS
This article elucidates the role that the 3′–5′ proofreading exonuclease activity of archaeal family-B DNA polymerases plays in uracil recognition. Control experiments established the background rates for fully base-paired primer-templates and mismatch substrates. The increase in rate constant by about an order of magnitude when a mismatch is present at the primer–template junction is typical for DNA polymerases (3,21,22). However, we note that primer-templates terminated in two A:T base pairs are acted on much more rapidly than those ending G:C, assuredly due to the greater ease of unwinding the former. Using a fully base-paired primer-template with uracil or hypoxanthine at the +4 position results in slight inhibition of the exonuclease activity and such nucleic acids are also extended extremely slowly (10–12,14,15,19). A crystal structure of Tgo-Pol, complexed with a U+4 primer-template, shows that the DNA sequence adopts an editing conformation but the 3′ base of the primer remains double-stranded and does not protrude into the exonuclease catalytic centre (Figure 1) (13). A primer-template bound in such a mode would be acted upon poorly by both the exonuclease and polymerase activities, explaining the observed results. Perhaps the most remarkable finding is the strong stimulation of exonuclease activity, by about two orders of magnitude, with primer-templates containing uracil at +3, +2, +1 and 0 and also with hypoxanthine at +2. No structural data are available for an archaeal polymerase in complex with DNA containing a deaminated base at any of these locations. We would suggest that the DNA binds in an editing conformation but, additionally, the 3′-terminus of the primer strand unwinds to give a single-stranded region that enters the exonuclease active site (Figure 7). Although archaeal polymerases bind most tightly to primer-templates containing uracil at +4, significant affinity is still seen when uracil is positioned at +3, +2 and +1 (12). Previously it was unclear how the enzyme could accommodate shifts in uracil location without significant relative re-positioning of the uracil-binding pocket and the active site amino acids responsible for interaction with the primer–template junction. Linking the degree of melting of the primer strand with the position of uracil would maintain the effective separation between uracil and the primer–template junction at around 4 bases. It seems clear that the binding energy available from the polymerase–uracil/hypoxanthine interaction can, in certain circumstances, result in the unwinding of fully base-paired primer-templates, leading to rapid exonuclease activity. We note that the stimulation of the exonuclease ceases with uracil at −1 and the polymerase is no longer able to recognize the base once this position is reached. Further, the effect is observed for both ‘weak’ and ‘strong’ primer-templates, terminated in A:T and G:C base-pairs, respectively. Thus interaction of the polymerase with deaminated bases appears to be powerful enough to unwind even robustly base-paired junction.
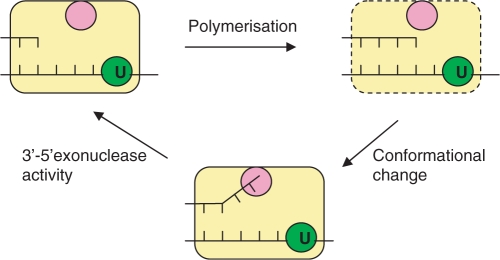
Figure 7.
Idling by Pfu-Pol on encountering a template strand deaminated base. The polymerase (yellow) captures uracil four bases ahead of the primer–template junction using a specific binding pocket (green). Further, extremely slow, polymerization can add two bases to give a strained conformation (indicated by hatched borders) in which the primer–template junction and the uracil binding site are too close, with a separation of two bases. A conformational change restores the spacing to four bases by unwinding the terminal two bases in the primer and, hence, relieves the strain. This conformational change places the single-stranded bases in the 3′–5′ exonuclease site (rose) enabling their removal.
It is well established that the main role of the 3′–5′ proofreading exonuclease activity is to excise bases aberrantly incorporated by the polymerase, thereby correcting replication errors and increasing fidelity (1–6). Additionally, the exonuclease appears to play a role in handling any non-canonical bases that a polymerase may encounter on template strands during replication. Many polymerases have been observed to ‘idle’ on running into template strand damage, making sequential use of their polymerase and exonuclease activities to repeatedly add and remove a standard base opposite the lesion (3,22–25). During ‘idling’, repeated dNTP to dNMP turnover is not accompanied by net DNA synthesis or degradation. Critically, the polymerase is prevented from progressing beyond the damage and the potential for permanently fixing a mutation is, thereby, avoided. It has also been suggested that turnover of dNTPs may be the signal for initiation of appropriate repair pathways (23). Previous studies have suggested that deaminated base recognition, a function unique to family-B archaeal replicative DNA polymerases (26), acts to proofread the template strand and prevent replication beyond uracil/hypoxanthine (10–14). Uracil can arise in DNA by deamination of cytosine in C:G base-pairs to give a pro-mutagenic U:G mispair, a process favoured by the hyperthermophilic environments of many archaea (27–29). Uracil is mistaken for thymine by most DNA polymerases and unless the U:G mismatch is repaired, replication results in 50% of the progeny inheriting a C:G → T:A transition mutation. Similarly, deamination of adenine to hypoxanthine results in A:T → G:C transitions following replication. Stalling of replication by the archaeal polymerase prevents such deaminated base-induced mutations and presumably also initiates DNA repair by an, as yet unknown pathway, that probably involves error-free recombination (30,31). This article shows that the 3′–5′ exonuclease activity of archaeal polymerases plays a role in preventing replication beyond uracil/hypoxanthine, further checking the template strand proofreading function of the deaminated base-binding pocket. During replication, stalling takes place when uracil/hypoxanthine is encountered at +4, largely switching off both the polymerase and exonuclease activities. Any further progression of the polymerase towards the deaminated base results in strong activation of the exonuclease activity, which degrades and shortens the extending primer (Figure 7). As a consequence, the separation between the deaminated base and the primer–template junction is restored, strongly decreasing the probability that the polymerase proceeds beyond uracil/hypoxanthine. The influence of the exonuclease activity is apparent from Figure 5; an exo− variant gives more full-length product in the presence of uracil than the wild type enzyme with a functional proofreading activity. Repeated polymerase/exonuclease cycles in response to deaminated bases is reminiscent of ‘idling’ described above. However, until now, triggering of exonuclease activity, essential for ‘idling’, has been dependent on the mismatched base-pair produced when a polymerase copies a damaged base. Such mismatches both stimulate exonuclease activity and are difficult to extend (3,23). The archaeal polymerase response to uracil/hypoxanthine does involve a damaged base, but not a base-pair mismatch as the deaminated base remains in the single-stranded region of the template. Rather, ‘idling’ in this instance flows from the specific capture of uracil/hypoxanthine by the N-terminal domain of the polymerase (Figure 7).
FUNDING
UK Biotechnology and Biological Sciences Research Council [grant no. BBS/B/0560 to B.A.C]. Funding for open access charge: UK Biotechnology and Biological Research Council (BBSRC).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Pauline Heslop for providing expert technical assistance.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Joyce CM. How DNA travels between the separate polymerase and 3′–5′ exonuclease sites of DNA polymerase I (Klenow fragment) J. Biol. Chem. 1989;264:10858–10866. [PubMed] [Google Scholar]
- 2.Kunkel TA. DNA replication fidelity. J. Biol. Chem. 1992;267:18251–18254. [PubMed] [Google Scholar]
- 3.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 4.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 5.Benkovic SJ, Valentine AM, Salinas S. Replisome-mediated DNA replication. Annu. Rev. Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 6.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 7.Lundberg KS, Shoemaker DD, Adams MWW, Short JM, Sorge JA, Mathur EJ. High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene. 1991;101:1–6. doi: 10.1016/0378-1119(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 8.Cline J, Braman JC, Hogrefe HH. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 1996;24:3546–3551. doi: 10.1093/nar/24.18.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopfner K-P, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl Acad. Sci. USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl Acad. Sci. USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 12.Shuttleworth G, Fogg MJ, Kurpiewski MR, Jen-Jacobson L, Connolly BA. Recognition of the pro-mutagenic base uracil by family B DNA polymerases from archaea. J. Mol. Biol. 2004;337:621–634. doi: 10.1016/j.jmb.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA. Uracil recognition in archaeal DNA polymerases captured by X-ray crystallography. J. Mol. Biol. 2008;381:529–539. doi: 10.1016/j.jmb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Connolly BA. Recognition of deaminated bases by archaeal family-B polymerases. Biochem. Soc. Trans. 2009;37:65–68. doi: 10.1042/BST0370065. [DOI] [PubMed] [Google Scholar]
- 15.Gill S, O’Neill R, Lewis RJ, Connolly BA. Interaction of the family-B DNA polymerase from the archaeon Pyrococcus furiosus with deaminated bases. J. Mol. Biol. 2007;372:855–863. doi: 10.1016/j.jmb.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Gruz P, Shimizu M, Pisani FM, DeFelice M, Kanke Y, Nohmi T. Processing of DNA lesions by archaeal DNA polymerases from Sulfolobus solfataricus. Nucleic Acids Res. 2003;31:4024–4030. doi: 10.1093/nar/gkg447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a Pol alpha family DNA polymerase. Cell. 2001;105:657–666. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 18.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 19.Emptage K, O’Neill R, Solovyova A, Connolly BA. Interplay between DNA polymerase and proliferating cell nuclear antigen switches off base excision repair of uracil and hypoxanthine during replication in archaea. J. Mol. Biol. 2008;383:762–771. doi: 10.1016/j.jmb.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Evans SJ, Fogg MJ, Mamone A, Davis M, Pearl LH, Connolly BA. Improving dideoxynucleotide-triphosphate utilisation by the hyper-thermophilic DNA polymerase from Pyrococcus furiosus. Nucleic Acids Res. 2000;28:1059–1066. doi: 10.1093/nar/28.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donlin MJ, Patel SS, Johnson KA. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- 22.Khare V, Eckert KA. The 3′–5′ exonuclease of T4 DNA polymerase removes premutagenic alkyl mispairs and contributes to futile cycling at 06 methylguanine lesions. J. Biol. Chem. 2001;276:24286–24292. doi: 10.1074/jbc.M011025200. [DOI] [PubMed] [Google Scholar]
- 23.Khare V, Eckert KA. The proofreading 3′–5′ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutation Res. 2002;510:45–54. doi: 10.1016/s0027-5107(02)00251-8. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz H, Shavitt O, Livneh Z. The role of exonucleolytic processing and polymerase-DNA association in vitro: significance for SOS targeted mutagenesis. J. Biol. Chem. 1988;263:18277–18285. [PubMed] [Google Scholar]
- 25.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PMJ. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wardle J, Burgers PMJ, Cann IKO, Darley K, Heslop P, Johansson E, Lin L, McGlynn P, Sanvoisin J, Stith CM, et al. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res. 2008;36:705–711. doi: 10.1093/nar/gkm1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;286:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 29.Schroeder GK, Wolfenden R. Rates of spontaneous disintegration of DNA and the rate enhancements produced by DNA glycosylases and deaminases. Biochemistry. 2007;46:13638–13647. doi: 10.1021/bi701480f. [DOI] [PubMed] [Google Scholar]
- 30.Baynton K, Fuchs RPP. Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci. 2000;25:74–79. doi: 10.1016/s0968-0004(99)01524-8. [DOI] [PubMed] [Google Scholar]
- 31.Cox MM. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 2001;35:53–82. doi: 10.1146/annurev.genet.35.102401.090016. [DOI] [PubMed] [Google Scholar]