Abstract
Mrt4 is a nucleolar component of the ribosome assembly machinery that shares notable similarity and competes for binding to the 25S rRNA GAR domain with the ribosomal protein P0. Here, we show that loss of function of either P0 or Mrt4 results in a deficit in 60S subunits, which is apparently due to impaired rRNA processing of 27S precursors. Mrt4, which shuttles between the nucleus and the cytoplasm, defines medium pre-60S particles. In contrast, P0 is absent from medium but present in late/cytoplasmic pre-60S complexes. The absence of Mrt4 notably increased the amount of P0 in nuclear Nop7–TAP complexes and causes P0 assembly to medium pre-60S particles. Upon P0 depletion, Mrt4 is relocated to the cytoplasm within aberrant 60S subunits. We conclude that Mrt4 controls the position and timing of P0 assembly. In turn, P0 is required for the release of Mrt4 and exchanges with this factor at the cytoplasm. Our results also suggest other P0 assembly alternatives.
INTRODUCTION
In all organisms, the biogenesis of ribosomes is a fundamental process mediated by trans-acting factors. Most of our knowledge concerning ribosome synthesis in eukaryotes derives from studies with Saccharomyces cerevisiae (1), where around 200 protein trans-acting factors and 75 small nucleolar RNAs (snoRNAs) have been identified (2–4). These factors, whose precise functions are still largely unknown, must assure speed, accuracy and directionality to the ribosome maturation process. Biogenesis of eukaryotic ribosomes involves the transcription, processing and modification of the pre-rRNAs in the nucleolus, the folding and the assembly of the pre-rRNA with the ribosomal proteins (r-proteins) and the export of the resulting pre-ribosomal particles to the cytoplasm, where the last assembly reactions take place (2–4). Pre-rRNA processing is a well-defined pathway and involves a complex series of sequential endo- and exo-nucleolytic reactions (Figure S1). However, the concomitant process of r-subunit assembly is still poorly understood. Although an outline of this process was described in the 1970s (5,6), a detailed characterization of the pre-ribosomal complexes emerged only recently due to the development of affinity purification methods and protein identification by mass spectrometry techniques (7–13). These studies have revealed several distinct, successive pre-ribosomal particles. The 90S particles contain the 35S pre-rRNA, many 40S r-subunit proteins and about 40 trans-acting factors (8,9,14,15). Endonucleolytic cleavage of 35S pre-rRNA at site A0–A2 (Figure S1) generates the early 43S and 66S pre-ribosomal particles. It appears that most of the factors associated with 90S particles are released after these cleavages (9–11). The early 43S particle is rapidly exported to the cytoplasm. Only a few factors and 40S r-proteins are needed to finish the maturation of 40S r-subunits (7,16). The early nucleolar pre-60S r-particles are the result of the association of ∼50 trans-acting factors and many 60S r-proteins with the 27SA2 pre-rRNA (10,11,17). The study of the RNA and protein composition of different purified pre-60S complexes is consistent with the presence of distinct intermediates that move from the nucleolus to the nucleoplasm and from there to the cytoplasm (7,11,18,19). These intermediates are termed, according to their position in the ribosome assembly pathway, early, medium, late and cytoplasmic pre-60S r-particles (3,12,13). Apparently, the complexity of the pre-60S r-particles decreases during their maturation and the export-competent particles have completed the pre-rRNA processing reactions (11,19). As for the pre-40S particles, last assembly reactions occur in the cytoplasm (3,20–23). From this, it must be assumed that (i) the trans-acting factors undergo cycles of association for function to and dissociation after function from the distinct pre-ribosomal particles and (ii) these particles undergo structural rearrangements during maturation. Insights into the approximate timing of association of some of these factors have been obtained by studying the composition of distinct purified pre-60S complexes from wild-type and mutant strains blocked at early, medium or late steps of ribosome maturation [(24,25) and references therein]. Evidence for structural rearrangements has also been recently obtained for both pre-40S and pre-60S r-particles (26,27).
In contrast to the trans-acting factors for ribosome biogenesis and excluding pioneer work (28), no specific studies on the course of the assembly of the r-proteins have been reported. This is due to the fact that r-proteins are common contaminants in purified complexes (29,30). Two recent reports have systematically approached the role of individual 40S r-proteins in pre-rRNA processing and their association with pre-ribosomal particles (16,31). However, equivalent analyses for the 60S r-proteins have not yet been reported. To date, only functional analyses on ribosome maturation have been reported for a few 60S r-proteins, such as Rpl3 (32), Rpl5 (33), Rpl10 (21,34,35), Rpl11 (36), Rpl25 (37) and Rpl33 (38).
We are interested in understanding the functions of the eukaryotic r-stalk. The stalk is a highly flexible protuberance of the 60S r-subunit essential for the interaction and function of a number of soluble translation factors (39). The eukaryotic stalk is composed of several phosphoproteins, named P proteins. In S. cerevisiae, the stalk is formed by two heterodimers of the acidic 12 kDa proteins (P1α-P2β and P1β-P2α) attached to the ribosome via the 32 kDa RNA binding protein P0. In contrast to the prokaryotic equivalent structure, the eukaryotic stalk undergoes a cyclic association/disassociation process during translation (40,41), which apparently generates various ribosome subpopulations with different stalk compositions. This ribosome heterogeneity seems to be on the basis of the proposed stalk translational regulatory capacity (42). In yeast, protein P0, which plays a central role in this structure, has been found associated with various pre-ribosomal complexes (9,11,17,43). However, the available data do not provide reliable information about its assembly. P0 has three distinct domains; an N-terminal one, which is responsible for binding to the GTPase-associated region (GAR) of 25S rRNA (44), a central one, which seems to contain at least two separated regions required to bind the P1α-P2β and P1β-P2α dimers (45,46) and a highly conserved C-terminal peptide, which is required for the protein activity in translation (47). Mrt4, a non-essential nucleolar protein likely required for 60S r-subunit biogenesis (10,48), is homologous to the N-terminal domain of P0 (49). Due to the significant degree of homology, it has been hypothesized that Mrt4 and P0 successively occupy the same rRNA binding site during ribosome synthesis, and indeed our results show that both proteins are mutually exclusive in r-particles (49). In this work, we have studied the effect of depletion of P0 and deletion of MRT4 on ribosome maturation. In addition, we have studied the dynamics of association and release of Mrt4 from pre-ribosomal particles and the relative position of a stable P0 assembly. Although other alternatives are possible, we propose a sequential association of Mrt4 and P0 with pre-60S r-particles. Mrt4 is a nucleo-cytoplasmic shuttling factor. When ribosome export is impaired, P0 remains cytoplasmic; therefore, the replacement of Mrt4 by P0 might take place mostly in the cytoplasm, being a prerequisite to recycle back Mrt4 to the nucleolus.
MATERIALS AND METHODS
Strains, plasmids and microbiological methods
The S. cerevisiae strains used in this study are listed in Table S1. Strains expressing TAP-tagged proteins were either purchased from Euroscarf or prepared by homologous recombination (50). PCR products were generated using the TAP module from plasmid pBS1479 as template (51) and selected pairs of oligonucleotides (see Table S2). When required, deletion of the MRT4 ORF was carried out as previously reported. In all cases, of the constructs were checked by PCR.
For in vivo depletion of P0, cells from W303dGP0 and W303dMGP0 were grown in 2% galactose medium (YPGal) at 30°C until mid-exponential phase (OD600 = 0.8), and then, transferred to 2% glucose medium (YPD) for the indicated times. As control, the wild-type W303-1B strain was used.
Information on the construction of the different plasmids used in this study can be found on the Supplementary Data section.
Sucrose gradient analyses
Polysome preparations were obtained from exponentially growing cells and studied by 7–50% sucrose gradient centrifugation exactly as described (52). Ten A260 of extracts was loaded in each gradient.
RNA analyses
RNA extraction, northern hybridization and primer extension analyses were carried out according to (53). RNA was extracted from samples corresponding to 10 OD600 units of exponentially growing cells. RNA samples to equal amounts of OD600 units (ca. 5 µg for W303-1B) were loaded in 1.2% agarose–formaldehyde gels (high-molecular mass RNAs) and 7% polyacrylamide–urea gels (low-molecular-mass RNAs) or used in primer extension reactions. Sequences of oligonucleotides used for RNA hybridization and primer extension analyses have been described previously [(32); see Figure S1A for their annealing location in the 35S pre-rRNA]. Phosphorimager analysis was performed in a FLA-5100 imaging system (Fujifilm) at the Biology Service (CITIUS) from the University of Seville.
Fluorescence microscopy
To test pre-ribosomal particle export, the appropriate strains were co-transformed with a DsRed-Nop1 construct and a second plasmid harbouring Rpl25-eGFP (34) or a Rps2-eGFP fusion protein (54). To study the subcellular localization of P0, plasmids YCplac111-RPP0-eGFP, pUG35-RPP0-yGFP and pU23-RPP0-yGFP were used. To study the subcellular localization of Mrt4, Nug1, Nsa1, Rix1 and Rix7, plasmids YCplac111-MRT4-eGFP, pRS315-NUG1-eGFP (19), YCplac111-NSA1-eGFP (55), pRS315-RIX1-eGFP and YCplac111-RIX7-eGFP were used, respectively. pRS315-NUG1-eGFP, YCplac111-NSA1-eGFP, pRS315-RIX1-eGFP and YCplac111-RIX7-eGFP were a generous gift from J. Bassler. Transformants were selected in minimal medium lacking the adequate requirements. Then, several transformants were grown to mid-log phase in selective liquid medium or subjected to in vivo depletion for different times. Strain Y2944, which harbours a genomic GFP-tagged Nop7 (11), was used to determine the subcellular localization of Nop7. When required, Hoechst 33342 (1 µg/ml final concentration) was added to the cultures during the last 30 min of growth. Acquisition was done in a Leica DMR fluorescence microscope. Images were acquired with a DC350F digital camera and its software and processed with Adobe Photoshop 7.0 for grey levels. Alternatively, an Axiovert 200 Zeiss microscope and a Coolsnap FX CCD camera were used.
Heterokaryon assays were done as described (56) with slight modifications; briefly, cells were grown in synthetic medium until OD600 of ∼0.8 at 30°C. Then, equal amount of cells expressing distinct GFP constructs and cells of the kar1-1 strain (EY93) were mixed. Mixes were concentrated by filtration onto 0.45 µm MF-Millipore membrane filters and then placed onto YPD plates. Mating was monitored by microscopy and after ∼1 h, when the first heterokaryons started to appear, the filters were placed on YPD plates containing 50 µg/ml cycloheximide and incubated for another hour. Then, cells were washed off the membranes and analysed by fluorescence microscopy as above.
Leptomycin B (LMB) experiments were carried out as described (57) with slight modifications; briefly, cells from AJY1539 and AJY1539dM strains were transformed with either pRS315-RPL25-eGFP or YCplac111-RPP0-eGFP and grown at 30°C in the appropriate selective minimal medium to an OD600 of ∼0.6. Then, cells from 2 ml culture were concentrated 2-fold in fresh medium and split. LMB (Tocris Biosciences) in 20% ethanol or 20% ethanol alone was added to the cultures at a final concentration of 200 ng/ml LMB–0.16% ethanol or 0.16% ethanol, respectively. Cultures were incubated for 1–3 h and then cells were inspected by fluorescence microscopy as above.
Antibodies and western blotting
Total yeast protein extracts were prepared and analysed by western blotting according to standard procedures. Ribosomal particles analysed in Figure 8A were isolated as described (49). Monoclonal 3BH5 antibodies, which are specific to the conserved C-terminal end of the stalk r-proteins (58), were used to identify P0. For detection of P0-C, polyclonal rabbit antibodies were prepared against a custom peptide comprising amino acids K8 to C28 of P0. Polyclonal rabbit anti-Mrt4 antibodies were previously described (49). Anti-Rpl1 polyclonal rabbit antibodies were a gift from F. Lacroute (59). Anti-GFP antibodies were purchased from Santa Cruz Biotechnology.
Figure 8.
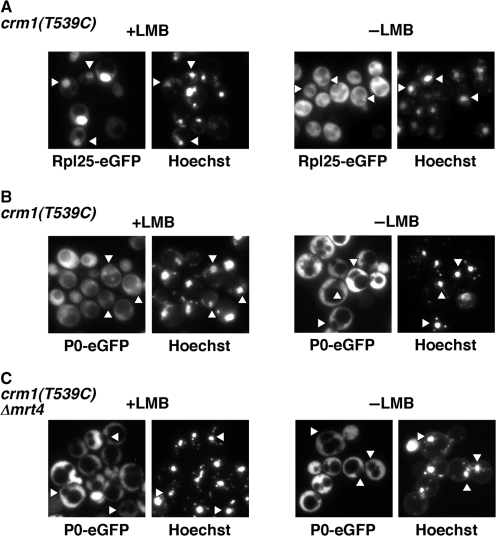
Location of P0-eGFP after inhibition of nuclear export of pre-ribosomes with Leptomycin B. (A) Rpl25-eGFP and (B) P0-eGFP were expressed in the LMB-sensitive AJY1539 strain, which harbours a wild-type MRT4 allele. (C) P0-eGFP was also express in AJY1539dM strain, which carries an mrt4 null allele. Cells were treated with 200 ng/ml LMB for 3 h (+LMB). Untreated cells were used in parallel (−LMB). The GFP signal was analysed by fluorescence microscopy. Triangles indicate the position of the nuclei.
Affinity purification of TAP-tagged proteins
TAP purifications were performed as exactly described (49) following a standard procedure (10,50). Purified complexes were analysed in Tris–glycine SDS 4–20% gradient polyacrylamide gels (Invitrogen). To normalised the amount of purified complex to be loaded in comparative studies, a sample was previously resolved in the same conditions and dyed with a silver or Sypro Ruby stain according to manufacturer's; instructions (Invitrogen).
In-gel protein digestion and sample preparation for mass spectrometry
Protein bands from Sypro Ruby-stained gels were used for mass spectrometric identification (CNB Proteomics Facility, Cantoblanco, Madrid).
MALDI peptide mass fingerprinting, MS/MS analysis and database searching
For MALDI-TOF/TOF analysis, samples were automatically acquired in an ABi 4800 MALDI TOF/TOF mass spectrometer (Applied Biosystems) in positive ion reflector mode (the ion acceleration voltage was 25 kV to MS acquisition and 1 kV to MS/MS) and the obtained spectra were stored into the ABi 4000 Series.
RESULTS
Depletion of P0 and deletion of Mrt4 lead to a deficiency in 60S r-subunits
As an approach to the phenotypic analysis of Mrt4 and P0, we used strains W303dM (Δmrt4), W303dGP0 (GAL::RPP0) and W303dMGP0 (Δmrt4 GAL::RPP0) (Table S1). W303dM is a mrt4 null strain, which has a slight slow-growth (sg) phenotype at 30°C (49). W303dGP0 is a conditional P0 null strain, which carries a genomic RPP0 ORF under the control of a GAL1 promoter (60). W303dMGP0 is a double Δmrt4 GAL::RPP0 mutant (49). In YPGal, there were no significant differences in the growth rate between GAL::RPP0 and an isogenic wild-type strain or between the double Δmrt4 GAL::RPP0 and the isogenic mrt4 null strain (data not shown). Nine to twelve hours after the transfer of the conditional P0 strains to YPD, the growth rate began to slow down and finally stopped. Concomitantly, there was a depletion of P0, as previously described (60).
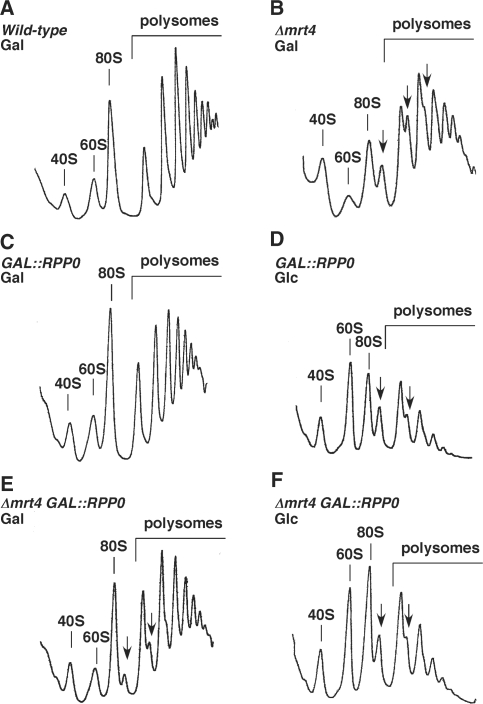
To determine the effects of the absence of Mrt4 and P0 in ribosome metabolism, we performed polysome profile analysis of the different strains. In agreement with a previous report (10), deletion of MRT4 is accompanied by a clear alteration on the polysome profile; the free 60S/40S ratio decreased and half-mer polysomes accumulated (Figure 1B). Upon P0 depletion by a 12 h shift to YPD, the Δmrt4 GAL::RPP0 strain showed an overall decrease in polysomes and an accumulation of half-mer polysomes (Figure 1F) similar to that reported for the GAL::RPP0 strain [(60) and Figure 1D]. Strikingly, a slight alleviation of the 60S r-subunit shortage was reproducibly observed for Δmrt4 GAL::RPP0 extracts obtained in YPGal compared to those for the mrt4 null strain (Figure 1B and E). Moreover, an increase rather than the expected decrease in the free 60S/40S ratio was detected for the P0-depleted strains after the shift to YPD (Figure 1D and F), which is probably due to the accumulation of arrested pre-60S r-particles (see Discussion section).
Figure 1.
Absence of Mrt4 and depletion of P0 results in a deficit in 60S r-subunits. Polysome profiles are shown for W303-1B (Wild-type) (A) and W303dM (Δmrt4) cells (B) grown in YPGal at 30°C, W303dGP0 (GAL::RPP0) and W303dMGP0 (Δmrt4 GAL::RPP0) cells grown in YPGal at 30°C (C and E, respectively) or shifted to YPD for 12 h (D and F, respectively). The peaks of free 40S and 60S r-subunits, 80S and polysomes are indicated. Half-mers are labelled by arrows.
Altogether, these results indicate that P0 depletion and absence of Mrt4 lead to a net deficit in active 60S r-subunits. Both proteins genetically interact since the polysome profile phenotype shown by the mrt4 null strain can be partially suppressed by increased dosage of P0 from the GAL promoter, which has been previously shown to slightly increase the amount of P0 in the GAL::RPP0 strain (60).
Both P0 and Mrt4 are required for normal pre-rRNA processing
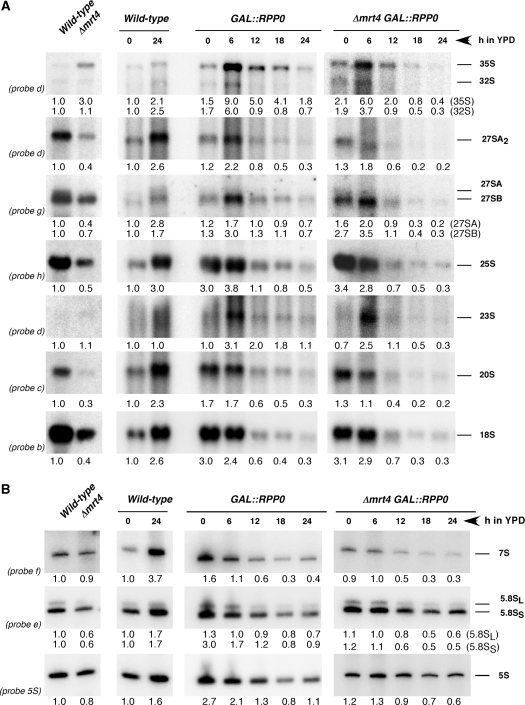
To characterize the basis of the 60S r-subunit deficit in the mutant strains, we analysed the effect of depletion of P0 and/or deletion of MRT4 on pre-rRNA processing. Total RNA was extracted from the wild-type and Δmrt4 strains grown in YPD at 30°C and from both GAL::RPP0 and Δmrt4 GAL::RPP0 strains during a time course of P0 depletion. Then, steady-state levels of pre- and mature rRNA were analysed by northern hybridization. Deletion of MRT4 resulted in a significant decrease in mature 18S and 25S rRNAs (Figure 2A) and mature 5.8S and 5S rRNAs (Figure 2B). This decrease was more drastic upon depletion of P0. A mild increase of the 35S/32S pre-rRNA ratio was observed following loss of Mrt4 or at early times of depletion of P0; this was accompanied by reduced levels of 27SA2 and 20S pre-rRNAs and the detection of the aberrant 23S pre-rRNA; however, pre-rRNAs are eventually turned over at late times of depletion of P0 (Figure 2A). In addition, levels of 27SB and 7S pre-rRNAs decreased in the absence of Mrt4 but especially upon P0 depletion (Figure 2A and B).
Figure 2.
Effects of the absence of Mrt4 and the depletion of P0 on steady-state levels of pre-rRNAs and mature rRNAs. W303-1B (Wild-type) and W303dM (Δmrt4) cells were grown in YPD at 30°C. W303-1B (Wild-type), W303dGP0 (GAL::RPP0) and W303dMGP0 (Δmrt4 GAL::RPP0) cells were grown in YPGal at 30°C, shifted to YPD and harvested at the indicated times; then, total RNA was extracted. (A) Northern analysis of high-molecular-mass pre- and rRNAs. (B) Northern analysis of low-molecular-mass pre- and rRNAs. Probes, between parentheses, are described in Supplementary Figure S1A. Signal intensities were measured by phosphorimager scanning; values (indicated below each lane) were normalised to those obtained for the wild-type strain grown either in YPD or YPGal, arbitrarily set at 1.0.
To discriminate between the 27SA2 and 27SA3 pre-rRNAs and distinguish between the 27SBL and 27SBS pre-rRNAs, we performed primer extension analyses. As shown in Figure S2, the quantities of the cDNAs terminating at sites A2, A3, B1L and B1S decreased significantly upon depletion of P0 and to a lesser extent upon MRT4 deletion.
Altogether, our data indicate that P0 depletion results in similar although more drastic pre-rRNA processing defects than deletion of MRT4.
Depletion of P0 impairs export of pre-60S r-particles from the nucleus to the cytoplasm
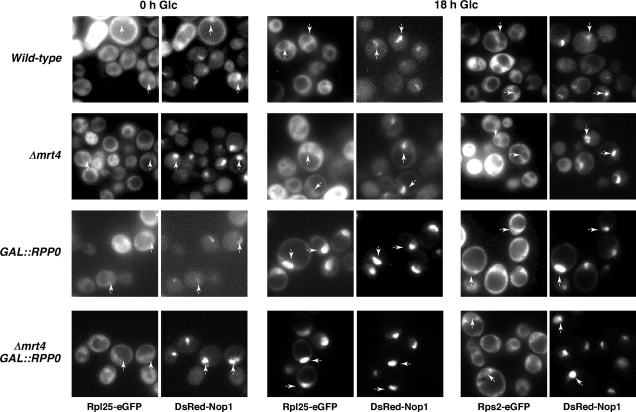
To determine whether the Mrt4 absence or the P0 depletion impairs nuclear export of pre-60S r-particles, we analysed the location of the 60S reporter Rpl25-eGFP (34) in the different strains. While Rpl25-eGFP was found, as expected for a r-protein, predominantly in the cytoplasm in all strains under permissive culture conditions, most of the cells from both GAL::RPP0 and Δmrt4 GAL::RPP0 strains exhibited an accumulation of the reporter in the nucleus, following a 12–18 h shift to a glucose medium (Figure 3). In some cells, the fluorescence signal for Rpl25-eGFP was restricted to the nucleolus, which was detected with the nucleolar marker DsRed-Nop1. In contrast, nuclear accumulation of Rpl25-eGFP was not observed in either the mrt4 null or the wild-type strain, under our experimental conditions. All these phenomena are specific for the large r-subunit, since no nuclear fluorescence accumulation was observed when we studied the location of the 40S r-subunit reporter Rps2-eGFP (54) either in galactose (data not shown) or in a glucose medium (Figure 3).
Figure 3.
Depletion of P0 leads to nuclear retention of the 60S r-subunit reporter Rpl25-eGFP. W303-1B (Wild-type), W303dM (Δmrt4), W303dGP0 (GAL::RPP0) and W303dMGP0 (Δmrt4 GAL::RPP0) cells expressing Rpl25-eGFP and DsRed-Nop1 were grown in SGal-Leu-Trp (0 h Glc) or shifted for 18 h to SD-Leu-Trp (18 h Glc). As a control, the same strains expressing Rps2-eGFP and DsRed-Nop1 were used. The GFP and DsRed signals were analysed by fluorescence microscopy. Arrows indicate the position of the nucleoli.
We conclude that both intra-nuclear and nucleo-cytoplasmic transport of pre-60S r-particles is blocked upon depletion of P0 but not after deletion of MRT4.
Mrt4 is associated with medium pre-60S r-particles
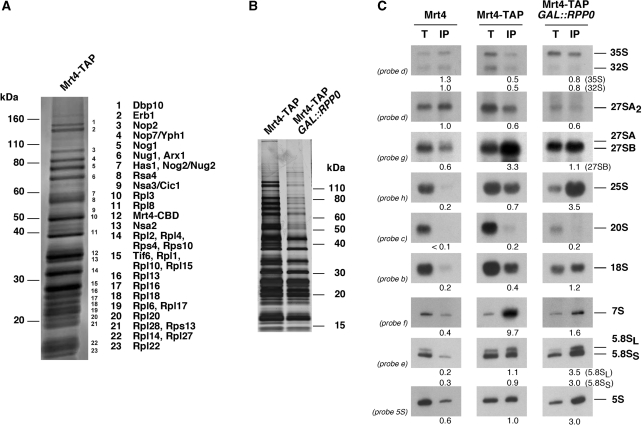
Since the interaction of Mrt4 and P0 with r-particles is mutually exclusive (49), we reasoned that defining the association of Mrt4 precisely with pre-ribosomal particles would help understand the position and timing of the P0 assembly. In sucrose gradients, Mrt4 is associated with high-molecular-mass particles of a size overlapping the 60S/80S peaks (49). To characterize these particles, we tagged Mrt4 with a C-terminal TAP cassette. After TAP purification, Mrt4 co-purifed with several proteins (Figure 4A). Mass spectrometry identification of the most abundant bands revealed nucle(ol)ar pre-60S r-assembly factors and r-proteins mostly from the 60S r-subunit. Among these proteins, we found Nop7, Erb1 and Nsa3, which are indicators of early pre-60S r-particles (10,11,61), Nug1 and Nog2, which are indicators of medium/late pre-60S r-particles (11,62) and Arx1, which is an indicator of a late/cytoplasmic pre-60S r-particle (11). To determine which pre-rRNA species are present in the purified particles, co-precipitated RNA was analysed by northern. As shown in Figure 4C, there was a significant enrichment for 7S pre-rRNA and to a lesser extent, for 27SB pre-rRNA in the purified particles. In addition, mature rRNAs were modestly co-precipitated over the background levels detected for those rRNAs purified from the untagged strain.
Figure 4.
Mrt4 is associated with medium pre-60S r-particles in wild-type cells but with almost mature 60S r-subunits upon depletion of P0. (A) The protein composition of the Mrt4–TAP complexes from extracts of W303dM pTAPC111-MRT4 cells (Mrt4–TAP) was analysed by 4–20% gradient SDS–PAGE and stained with Sypro Ruby. The indicated proteins were identified by mass spectrometry. (B) Equivalent amounts of Mrt4–TAP complexes from W303dM pTAPC111-MRT4 (Mrt4–TAP) and W303dMGP0 pTAPC111-MRT4 upon 12 h depletion of P0 (Mrt4–TAP GAL::RPP0) were compared. Purified complexes were analysed as above but gels were stained with silver. (C) Northern analysis of the pre- and mature rRNAs present within Mrt4–TAP complexes. RNA was extracted from cell extracts (lanes T) and affinity-purified samples (lanes IP) prepared from W303-1B (Mrt4), W303dM pTAPC111-MRT4 (Mrt4–TAP) or W303dMGP0 pTAPC111-MRT4 cells upon 12 h depletion of P0 (Mrt4–TAP GAL::RPP0). Probes are indicated. Signal intensity was measured by phosphorimager scanning; values (below each IP lane) refer to as the percentage of each pre-rRNA or mature rRNA recovered after purification.
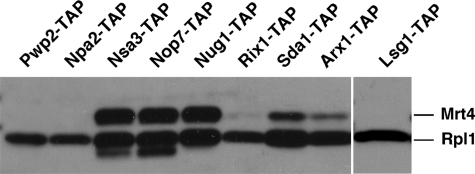
To further address the binding properties of Mrt4 to pre-ribosomal particles, we screened for the presence of Mrt4 in TAP purification of selected 90S (Pwp2/Utp1) (8,9,15) and factors (Npa2/Urb2, Nsa3/Cic1, Nop7/Yph1, Nug1, Rix1/Ipi2, Sda1, Arx1 and Kre35/Lsg1) known to purify early, medium, late and cytoplasmic pre-60S r-particles (10,11,19,63,64). As a result (Figure 5), Mrt4 was enriched in early (Nsa3–TAP and Nop7–TAP) and medium (Nug1–TAP) pre-60S r-particles while it was notably reduced in late (Sda1–TAP and Rix1–TAP) and late/cytoplasmic (Arx1–TAP) pre-60S complexes. Mrt4 was absent from 90S (Pwp2–TAP), the earliest pre-60S (Npa2–TAP) and cytoplasmic pre-60S r-particles (Lsg1–TAP).
Figure 5.
Presence of Mrt4 in pre-ribosomal particles. The TAP procedure was performed from wild-type cells expressing the TAP-tagged version of the indicated proteins. Equivalent amounts of the corresponding purified complexes were subjected to SDS–PAGE and western blotting with antibodies against Mrt4 and Rpl1.
Mrt4 shuttles between the nucleus and the cytoplasm
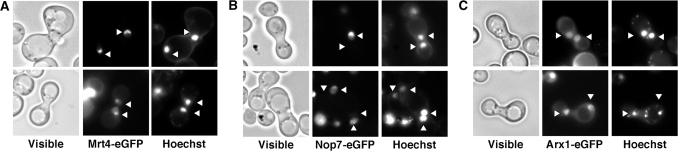
Mrt4 has been mostly localized in the nucleolus with a fainter nucleoplasmic staining at steady-state levels (48) (Figure S3A). To test whether Mrt4 shuttles between the nucleus and the cytoplasm, the Δmrt4 strain expressing Mrt4-eGFP was mated with a strain carrying the kar1-1 mutation, which prevents nuclear fusion following mating, leading to formation of heterokaryons. As a positive control, we used Arx1-eGFP, which is a late shuttling pre-60S r-assembly factor (56). As a negative control, we used the nucleolar Nop7-eGFP, which is expected to be retained in the nucleus (65). As shown in Figure 6, Mrt4-eGFP, just as Arx1-eGFP, was detected in both nuclei of heterokaryons while Nop7-eGFP was localized to only one of the nuclei. As an additional experiment, we co-expressed Mrt4-eGFP and DsRed-Nop1 in the same strain, which was also crossed with the kar1-1 strain. In this case, while Mrt4-eGFP was detected in both nuclei of heterokaryons, DsRed-Nop1 was only found in one of them (Figure S4).
Figure 6.
Mrt4-eGFP does shuttle from the nucleus to the cytoplasm in a heterokaryon assay. Strains expressing Mrt4-eGFP (A), Nop7-eGFP (B) and Arx1-eGFP (C) were mated with a kar1-1 strain as described in ‘Materials and Methods’ section. Heterokaryons, which were identified under brightfield illumination (visible), were analysed by fluorescence microscopy for the GFP signal. The nucleoplasm was visualized by Hoechst 33342 staining. Triangles point to the positions of the nucleoli.
Altogether, these data indicate that Mrt4 is a stable component of medium nucle(ol)ar pre-60S r-particles containing 7S pre-rRNAs. Mrt4 apparently associates with pre-60S r-particles at some point after the formation of 27SB pre-rRNA and dissociates from particles after nuclear export. Then, Mrt4 must be rapidly recycled back to the nucle(ol)us.
P0 associates with late pre-60S r-particles
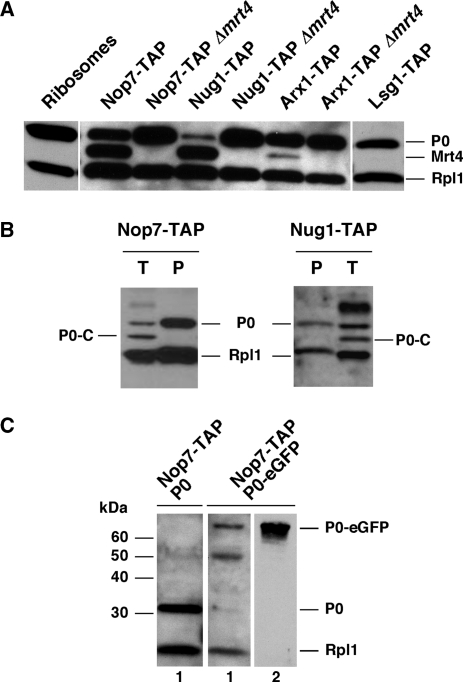
To address whether the binding of P0 to pre-60S r-particles is concomitant to dissociation of Mrt4, we analysed selected TAP-purified complexes by western blotting for the presence of P0. Taken into account the previous data (Figure 5), Nop7–TAP, Nug1–TAP and Arx1–TAP were selected as representative of the distinct phases of 60S r-subunit assembly. In addition, Lsg1–TAP and a preparation of purified ribosomes were included. As shown in Figure 7A, while the P0/Mrt4 band intensity ratio was roughly one in the Nop7–TAP complexes, the P0 signal was drastically reduced in the Nug1–TAP while the Mrt4 signal was reduced in the Arx1–TAP and absent in the Lsg1–TAP preparation.
Figure 7.
Assembly of P0 to pre-60S r-particles in wild-type and mrt4 null mutant cells. (A) The TAP procedure was performed with wild-type and mrt4 null cells expressing the TAP-tagged version of the indicated proteins. Equivalent amounts of the corresponding purified complexes were compared by SDS–PAGE and western blotting with antibodies against Mrt4, P0 and Rpl1. Band intensities were compared with those obtained for a ribosome preparation. (B) An equal amount of extracts from W303dMGP0 cells expressing, as a sole source of P0, a truncated P0 protein (P0-C) were mixed with extracts from W303dMNop7–TAP or dMNug1–TAP strains, which express a wild-type P0 and either Nop7–TAP or Nug1–TAP, respectively. Then, the Nop7–TAP and Nug1–TAP complexes were purified by the TAP procedure. Aliquots from the starting mix of extracts (T) and the corresponding purified complexes (P) were compared by western blotting with antibodies against P0 and Rpl1. (C) The TAP procedure was performed with W303dGP0Nop7–TAP cells expressing P0-eGFP (Nop7–TAP P0-eGFP) and W303dMNop7–TAP cells (Nop7–TAP P0) grown in YPD. Equivalent amounts of the corresponding purified complexes were subjected to western blotting with antibodies against P0 and Rpl1 (1) or the GFP protein (2).
Next, we tested whether the association of P0 with pre-60S r-particles was dependent on Mrt4. Thus, Nop7–TAP, Nug1–TAP and Arx1–TAP were also purified from mrt4 null cells and the presence of P0 again analysed by western blotting (Figure 7A). The amount of P0 found in the Arx1–TAP complexes was not affected by the absence of Mrt4; however, a significant increase was detected in the Nop7–TAP and the Nug1–TAP particles purified from mrt4 null cells. In both cases, the P0/Rpl1 ratio was similar to that of purified ribosomes (Figure 7A).
TAP purifications can be contaminated by r-proteins (29,30). Therefore, to exclude the possibility that association of P0 with the tested TAP complexes is the consequence of unspecific contamination during purification, we mixed equal amounts of cells from Δmrt4 cells expressing TAP-tagged or untagged versions of either Nop7 or Nug1. The untagged cells expressed as a sole source of functional P0 a truncated protein (P0-C) lacking the last 21 amino acids (47), easily distinguishable from the wild-type protein by SDS–PAGE. After purification from the mixed extracts, only the wild-type but not the truncated P0-C protein was found associated with the TAP purified particles (Figure 7B). Similar results were obtained using a His6-P0 as a contamination marker (data not shown). Thus, under the purification procedure assayed, the association of P0 with the purified particles was not a result of contamination.
We conclude that, in wild-type cells, P0 exchanges with Mrt4 at a very late stage during 60S r-subunit biogenesis, most specifically at the interface between Nug1 and Arx1 containing r-particles. However, P0 is also present in theoretically earlier pre-60S r-particles such as the ones containing Nop7 (but see Discussion section). The results obtained upon deletion of MRT4 indicate that Mrt4 is not strictly required for P0 assembly in agreement with its non-essential role. In the absence of competition by Mrt4, P0 can prematurely associate with the medium pre-60S r-particles.
P0 localizes in the cytoplasm after blocking nuclear export of pre-ribosomal particles
To address whether the Mrt4 to P0 exchange takes place in the nucleus or in the cytoplasm, we monitored localization of a fully functional P0-eGFP in a LMB sensitive strain (57). This sensitivity is due to the presence of the T539C mutation in the exportin Crm1/Xpo1. Treatment of the sensitive strain with LMB impairs export of pre-ribosomal particles and causes their accumulation in the nucleus (34). As shown in Figure 8A, the Rpl25-eGFP was localized in the cytoplasm in untreated cells but trapped in the nucleus after a treatment with LMB. In contrast, P0-eGFP, which is also found in the cytoplasm in untreated cells, remained cytoplasmic after a treatment with LMB (Figure 8B). This location is not altered by the deletion of MRT4 in the LMB sensitive strain (Figure 8C). Similar results were obtained using other P0-eGFP constructs described in ‘Materials and Methods’ section (data not shown). It must be noted that GFP tagging of P0 might not hinder its import to the nucleus as indicated by the association of P0-eGFP with the Nop7–TAP complexes (Figure 7C). Moreover, when cell fractionation was performed by centrifugation, P0-eGFP was exclusively present in the pellet but not in the S100 supernatant fraction even after the LMB treatment. Thus, P0 seems not to accumulate as a free protein in the cytoplasm upon pre-ribosomal particle export inhibition (data not shown).
Altogether, these data strongly suggest that, independently of the presence of Mrt4, P0 assembles in cytoplasmic pre-60S r-particles.
P0 is required for the release of Mrt4 from pre-60S r-particles
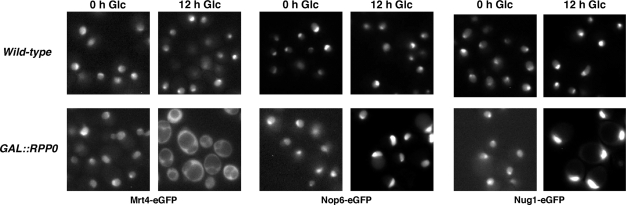
To address a possible role of P0 in the release of Mrt4 from pre-60S r-particles, we re-analysed the dynamics of 60S assembly upon P0 depletion. Thus, when Δmrt4 GAL::RPP0 YCplac111-MRT4-eGFP cells were depleted of P0, Mrt4-eGFP was strongly mislocalized to the cytoplasm (Figures 9 and S3B). No mislocation of other GFP-tagged pre-ribosomal markers, such as Nop6, Nop7, Nsa1, Rix1 and Rix7, was observed (Figure 9 and data not shown). Interestingly, a weak cytoplasmic signal was found for Nug1-eGFP upon P0 depletion (Figure 9). However, our data indicate that compromising the Nug1 function by the nug1-1, nug1-2 and nug1-ΔN1 alleles (gifts from J. Bassler) (19,66) does not result in cytoplasmic mislocation of Mrt4-eGFP (data not shown).
Figure 9.
Mrt4 is not properly recycled back to the nucleus and it is retained on cytoplasmic 60S r-particles upon depletion of P0. W303-1B (Wild-type) and W303dGP0 (GAL::RPP0) cells expressing Mrt4-eGFP, Nop6-eGFP or Nug1-eGFP were grown in SGal-Leu (0 h Glc) and shifted for 12 h to SD-Leu (12 h Glc). The subcellular location of these reporters was analysed by fluorescence microscopy.
To determine the distribution of this cytoplasmic Mrt4, we fractionated extracts from the GAL::RPP0 strain after a 12 h shift to YPD by sucrose gradient and fractions were subjected to western blotting for detection of Mrt4. We could not detect significant changes in the pattern of sedimentation of Mrt4 upon P0 depletion, which was still found in the 60S–80S region of gradients (data not shown). This result indicates that Mrt4 remains bound to high-molecular-mass complexes, most likely aberrant cytoplasmic pre-60S r-particles, upon P0 depletion. To confirm this hypothesis, Mrt4–TAP was purified from GAL::RPP0 cells upon depletion of P0. Analysis of the purified particles by SDS–PAGE revealed an important reduction of high-molecular-mass proteins, which we previously identified as pre-60S factors, and a relative enrichment in r-proteins (Figure 4B). Northern analysis revealed co-enrichment of mature rRNAs specially those from the 60S r-subunit (Figure 4C). Interestingly, Nop7–TAP and Arx1–TAP-tagged preparations from P0-depleted extracts of the GAL::RPP0 strain lack Mrt4 (Figure S5), suggesting that these two proteins have dissociated from the aberrant particles.
Altogether, our data indicate that Mrt4 does require P0 to be efficiently displaced from cytoplasmic pre-60S r-particles in order to recycle back to the nucleus.
DISCUSSION
In this work, we have addressed the functional characterisation in ribosome biogenesis of P0, the central component of the r-stalk, and its homologous r-protein-like Mrt4. The architecture of this essential r-domain has been extensively studied mainly in prokaryotes by distinct techniques, which evaluate the interactions between its components (39). The eukaryotic r-stalk structure is less stable than its prokaryotic counterpart and seems to modulate the cell translation activity through a mechanism that involves the exchange between ribosome-bound and free cytoplasmic stalk components (42). Unfortunately, there is little information available on the role of P0 and its prokaryotic counterpart L10 in ribosome biogenesis. In eukaryotes, a notable sequence homology exists between P0 and the non-essential ribosome synthesis factor Mrt4, which suggests a functional relationship between both proteins (49). In yeast, the absence of either P0 or Mrt4, which is lethal only in the first case, leads to a deficit of active 60S relative to 40S r-subunits [(10,60) and Figure 1]. In agreement with this information, we show herein that both proteins are required for the production of 25S and 5.8S rRNAs (Figure 2 and Figure S2); deletion of MRT4 and depletion of P0 affects pre-rRNA processing leading to a discrete accumulation of 35S and upon early times of P0 depletion also of aberrant 23S pre-rRNA. This is a general feature in many strains affected in 60S r-subunit biogenesis (1) that most likely occurs due to failure of recycling of trans-acting factors required for the early pre-rRNA cleavages at sites A0–A2 that improperly dissociate from defective pre-60S r-particles (13). Most importantly, the absence of Mrt4 and the depletion of P0 lead to reduced steady-state levels of all 27S and 7S pre-rRNAs and, therefore, of mature 25S and 5.8S rRNAs. In general, the pre-rRNA processing defects detected upon depletion of P0 were more drastic than those after the deletion of MRT4. In addition, depletion of P0 is epistatic over and synergistic with the absence of Mrt4, confirming the functional relationship between the two proteins. Since it is unlikely that either P0 or Mrt4 have a direct role in 27S pre-rRNA processing reactions, we assume that the depletion of P0 and the absence of Mrt4 lead to defective assembly of early pre-60S r-particles, which causes destabilisation and efficient degradation of the 27S pre-rRNAs and their products. More relevant concerning the final ribosome yield of the conditional systems tested is the fact that P0 depleted but not MRT4 deleted cells accumulated Rpl25-eGFP within the nucleus (Figure 3). Rpl25 has been suggested as assembling early during ribosome biogenesis (28). This result suggests a block in nucleo-cytoplasmic export of defective pre-60S r-particles that lack P0 and/or other factor limited upon P0 depletion but not of pre-60S r-particles assembled in the absence of Mrt4.
Mrt4 has been previously shown to localize to the nucleolus (48) and found to be associated with many pre-60S r-particles (10,11,19,24,25). Moreover, Mrt4–TAP complexes co-enriched defined pre-60S r-particles (Figure 4). Analysis of the RNA composition of these complexes indicates that Mrt4 might bind to pre-60S r-particles soon after formation of 27SB pre-rRNAs and apparently dissociate only when pre-rRNA processing reactions have been completed. At steady-state levels, Mrt4 appears to stably concentrate in medium/late pre-60S r-particles, which contains 7S pre-rRNA (Figures 4 and 5). The protein composition of the Mrt4–TAP complexes also reflects this fact and indeed most of the trans-acting factors and r-proteins identified have been previously listed as components of Nug1–TAP complexes, which are considered the representatives of nuclear medium pre-60S r-particles (11,13,19). On the other hand, our heterokaryon assay data strongly suggest that Mrt4 travels associated with those nuclear pre-60S r-particles that exit to the cytoplasm (Figure 6). There, Mrt4 dissociates from the particles and is efficiently reimported to the nucleus in order to maintain its steady-state nuclear distribution. Shuttling factors often require other proteins to be released from the cytoplasmic pre-60S r-particles and/or recycled back to the nucleus (20,22,23,57,67–70), amongst them, Drg1, which is an ATPase required for the release of Arx1, Rlp24, Nog1 and Tif6 (23), and Efl1/Ria1 and Lsg1, which are GTPases required for optimal dissociation of Tif6 and Nmd3, respectively (22,57). Little is known about Mrt4 releasing factors; our data suggest that the GTPase Nug1 does not seem to have this role since Mrt4 does not mislocalize upon loss of its function. Very recently, while this manuscript was being prepared, Johnson and co-workers have shown that the efficient release of Mrt4 from pre-60S r-particles requires the participation of the non-essential phosphatase Yvh1 that associates with late pre-60S r-particles (A.W. Johnson, personal communication).
When and how is P0 assembled? We have previously shown that P0 and Mrt4 interact in a mutually exclusive manner to the 25S rRNA GAR domain (49). Taking this into account, a simple model where Mrt4 and P0 successively occupy the GAR domain in pre-60S r-particles and mature 60S r-subunits, respectively, can be envisaged for the timing of P0 assembly. Since our data strongly suggest that Mrt4 dissociates from late, most likely cytoplasmic, pre-60S r-particles, the exchange of Mrt4 with P0 might take place mainly in the cytoplasm, which is agreement with the interpretation of LMB experiments. Thus, if P0 predominantly assembled in the nucleus, we would expect the export of P0-containing pre-60S r-particles to be Crm1-dependent in the LMB-sensitive strain. However, functional GFP-tagged P0 did not relocalize to the nucleus in the presence of LMB even in the absence of Mrt4 (Figure 8). Moreover, we show here that, in wild-type cells, P0 is practically absent from some Mrt4-containing medium pre-60S r-particles such as the Nug1–TAP complexes (Figure 7). These results are in agreement with those previously reported for Nog1-, Nog2- and Rlp24–TAP complexes (62), Nsa1-defined pre-60S r-particles (55) and Nsa3–TAP and Ssf1–TAP complexes (11,71), all of which lack P0 as tested using either mass-spectrometry or specific antibodies. On the contrary, P0 is clearly present in late complexes such as the ones obtained using Arx1–TAP as bait, which contain little Mrt4 and cytoplasmic complexes such as the Lsg1–TAP ones, which lack Mrt4 (Figure 7).
Our data also demonstrate that release of Mrt4 from pre-60S particles requires the presence of P0. This conclusion is based on the fact that Mrt4-eGFP mislocalized to the cytoplasm upon P0 depletion (Figure 9), where it is still associated with high-molecular-mass complexes, most likely inactive almost mature 60S r-subunits lacking P0 (Figure 4B and C). These complexes strongly accumulated increasing the free 60S peak in polysome profiles even in the absence of bound Mrt4 (Figure 1D and F). Our experimental approaches do not address whether Mrt4 is preferentially released from pre-60S r-particles via a direct displacement by P0 or via a trans-acting factor. As above mentioned, it has been recently shown that Yvh1 is required for removing Mrt4 from late pre-60S r-particles that are then susceptible to bind P0 in the cytoplasm (A.W. Johnson, personal communication).
In apparent contradiction with the previous data, which strongly support that an important part of P0 is assembled in the cytoplasm after an exchange with Mrt4, the Nop7–TAP purified complexes contain a significant amount of both Mrt4 and P0. The presence of both proteins in these particles reflect heterogeneous composition since it is known that Nop7–TAP complexes are a mixture of several early, medium and even late pre-60S intermediates (10), see also (11,55). Nevertheless, since Nop7 seems to be a nuclear protein, which is unable to shuttle between the nucleus and the cytoplasm (Figure 6), some P0 must assemble in the nucleus. Accordingly, the study of the P0 assembly dynamics in the mrt4 null mutant also leads to interesting conclusions being the most obvious one that P0 does not strictly require Mrt4 for association with pre-60S r-particles. Strikingly, in mrt4 null cells, the amount of P0 increases in both Nop7-and Nug1–TAP particles to proportions close to that of mature ribosomes (Figure 7), indicating that in the absence of Mrt4, the nuclear assembly of P0 is enhanced. Some of these particles might be subjected to surveillance and degradation, which provide an explanation for the net 60S r-subunit deficit in mrt4 null cells; however, since active ribosomes are still synthesized in these cells, this alternative Mrt4-independent P0 assembly pathway that may operate even in the presence of Mrt4 cannot be excluded.
Several arguments could help to understand the apparent discrepancy of the data supporting both a nuclear and a cytoplasmic assembly of P0: (i) the fraction of P0 that assemble in the nucleus could be minor versus that assembling in the cytoplasm and thus P0-eGFP should not be detected there after the LMB treatment, (ii) export of pre-60S r-particles containing P0 could be independent of Crm1, (iii) the GFP bait, although shown not to interfere with the function of P0 and its capability of association with nuclear Nop7–TAP complexes (Figure 7C), could influence the import of the P0-eGFP protein in the nucleus after the treatment with LMB, as earlier discussed (21). Further experiments are required to solve the questions raised; however, the possibility that ribosome biogenesis operates as a mesh of pathways rather than as a linear series of events has been previously suggested (12). This is an appealing possibility as a source of eukaryotic ribosome heterogeneity (72), which starts being considered on the bases of new ribosome-dependent translation regulatory mechanisms (42,73). The existence of alternative assembly pathways, at least for some r-components, might also explain some of the paradoxical paralogue-specific effects recently reported for duplicated genes encoding r-proteins (74). Figure S6 summarizes the conclusions of this study concerning the role of Mrt4 and the assembly of P0 in wild-type conditions, in the absence of Mrt4 or upon depletion of P0.
In Escherichia coli, L10 is a component of the RI50[1] complex, which is the first in vitro reconstitution intermediate of 50S r-subunits (75). In vivo, it seems that L10 assembles in the p150S complex, which is the earliest of the three pre-50S r-particles detected (76). Thus, the prokaryotic and eukaryotic scenarios seem to be different and it prompted us to speculate whether Mrt4 appeared during evolution to optimize ribosome assembly by preventing early P0 assembly. Our structural models indicate that P0 binds a little tighter than Mrt4 to the GAR domain (49); therefore, it is possible that the premature binding of P0 to early nuclear pre-ribosomal particles could impede its appropriate maturation by interfering in defined structural rearrangements within these particles. Additionally, replacement of an r-protein-like factor by its r-protein counterpart might provide directionality to the ribosome synthesis process throughout different subcellular regions (nucleolus, nucleoplasm, cytoplasm) and opportunities for quality control mechanisms. Mrt4-P0 is not the sole example of a paralogue pair comprised of a non-ribosomal factor and a r-protein. Factors Imp3, Rlp7, Rlp24 show significant homology to r-proteins Rps9, Rpl7 and Rpl24, respectively (24,77–79). It would be appealing to understand whether or not all these pairs have arisen during evolution to deal with similar situations during ribosome biogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science and Innovation (MICINN) and FEDER (BFU2007-60151 to J.d.l.C., BFU2007-64280 to M.R. and BFU2006-00365 to J.P.G.B.), the Andalusian Government (CVI-271, P07-CVI-02623 and P08-CVI-03508 to J.d.l.C.) and Ramón Areces Foundation (Institutional Grant to C.B.M.S.O.). Funding for open access charge: Ministerio de Ciencia e Innovacion, Spain.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank J.L. Woolford, Jr. for training M.R.-M. in his lab on the TAP method. We are indebted to the colleagues mentioned in the text for their gift of material used in this study. The authors are grateful to A. Fernández-Pevida for plasmid preparations. The authors thank A.W. Johnson for communicating results prior to publication. M.R.-M., J.J. G.-G. and R.F.-V. are recipients of FPI fellowships from MICINN.
REFERENCES
- 1.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 2.Kressler D, Linder P, de la Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 4.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner JR. The assembly of ribosomes in yeast. J. Biol. Chem. 1971;246:447–454. [PubMed] [Google Scholar]
- 6.Trapman J, Retèl J, Planta RJ. Ribosomal precursor particles from yeast. Exp. Cell Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- 7.Schäfer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 10.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 11.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 13.de la Cruz J, Kressler D, Linder P. In: Nucleolus. Olson MOJ, editor. Georgetown, Landes Bioscience/eurekah.com: Kluwer Academic; 2004. pp. 258–285. [Google Scholar]
- 14.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 15.Pérez-Fernández J, Roman A, de Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell Biol. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 19.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 20.Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West M, Hedges JB, Chen A, Johnson AW. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell Biol. 2005;25:3802–3813. doi: 10.1128/MCB.25.9.3802-3813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 23.Pertschy B, Saveanu C, Zisser G, Lebreton A, Tengg M, Jacquier A, Liebminger E, Nobis B, Kappel L, van der Klei I, et al. Cytoplasmic recycling of 60S preribosomal factors depends on the AAA protein Drg1. Mol. Cell Biol. 2007;27:6581–6592. doi: 10.1128/MCB.00668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saveanu C, Namane A, Gleizes PE, Lebreton A, Rousselle JC, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lebreton A, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M, Saveanu C. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008;36:4988–4999. doi: 10.1093/nar/gkn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schäfer T, Maco B, Petfalski E, Tollervey D, Bottcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 27.Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol. Cell. 2004;15:295–301. doi: 10.1016/j.molcel.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 28.Kruiswijk T, Planta RJ, Krop JM. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 29.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 30.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 31.Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 32.Rosado IV, Kressler D, de la Cruz J. Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis. Nucleic Acids Res. 2007;35:4203–4213. doi: 10.1093/nar/gkm388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshmukh M, Stark J, Yeh LC, Lee JC, Woolford JL., Jr Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J. Biol. Chem. 1995;270:30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- 34.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60S ribosomal subunit depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisinger DP, Dick FA, Trumpower BL. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moritz M, Pulaski BA, Woolford J.L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol. Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Beekvelt CA, de Graaff-Vincent M, Faber AW, van't; Riet J, Venema J, Raué HA. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:5001–5008. doi: 10.1093/nar/29.24.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martín-Marcos P, Hinnebusch AG, Tamame M. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell Biol. 2007;27:5968–5985. doi: 10.1128/MCB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 40.Remacha M, Jiménez-Díaz A, Santos C, Briones E, Zambrano R, Rodríguez Gabriel MA, Guarinos E, Ballesta JPG. Proteins P1, P2, and P0, components of the eukaryotic ribosome stalk. New structural and functional aspects. Biochem. Cell Biol. 1995;73:959–968. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]
- 41.Zinker S, Warner JR. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J. Biol. Chem. 1976;251:1799–1807. [PubMed] [Google Scholar]
- 42.Ballesta JPG, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Progr. Nucleic Acid Res. Mol. Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- 43.Horsey EW, Jakovljevic J, Miles TD, Harnpicharnchai P, Woolford J.L., Jr Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos C, Ballesta JPG. Characterization of the 26S rRNA-binding domain in Saccharomyces cerevisiae ribosomal stalk phosphoprotein P0. Mol. Microbiol. 2005;58:217–226. doi: 10.1111/j.1365-2958.2005.04816.x. [DOI] [PubMed] [Google Scholar]
- 45.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 46.Pérez-Fernández J, Remacha M, Ballesta JPG. The acidic protein binding site is partially hidden in the free Saccharomyces cerevisiae ribosomal stalk protein P0. Biochemistry. 2005;44:5532–5540. doi: 10.1021/bi047332r. [DOI] [PubMed] [Google Scholar]
- 47.Santos C, Ballesta JPG. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J. Biol. Chem. 1995;270:20608–20614. doi: 10.1074/jbc.270.35.20608. [DOI] [PubMed] [Google Scholar]
- 48.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'S;hea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 49.Rodríguez-Mateos M, Abia D, García-Gómez JJ, Morreale A, de la Cruz J, Santos C, Remacha M, Ballesta JPG. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009;37:3514–3521. doi: 10.1093/nar/gkp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 51.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 52.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venema J, Planta RJ, Raué HA. In: Protein synthesis: Methods and protocols. Martin R, editor. Vol. 77. Totowa, NJ: Humana Press; 1998. pp. 257–270. [Google Scholar]
- 54.Milkereit P, Strauss D, Bassler J, Gadal O, Kuhn H, Schutz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc-complex specifically involved in the formation and nuclear export of ribosomal 40S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 55.Kressler D, Roser D, Pertschy B, Hurt E. The AAA ATPase Rix7 powers progression of ribosome biogenesis by stripping Nsa1 from pre-60S particles. J. Cell Biol. 2008;181:935–944. doi: 10.1083/jcb.200801181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belaya K, Tollervey D, Kos M. FLIPing heterokaryons to analyze nucleo-cytoplasmic shuttling of yeast proteins. RNA. 2006;12:921–930. doi: 10.1261/rna.2301806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hedges J, West M, Johnson AW. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilella MD, Remacha M, Ortiz BL, Mendez E, Ballesta JP. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44' have different functional roles. Eur. J. Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 59.Petitjean A, Bonneaud N, Lacroute F. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archeabacterial L1 ribsomal protein. Mol. Cell Biol. 1995;15:5071–5081. doi: 10.1128/mcb.15.9.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos C, Ballesta JPG. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Biol. Chem. 1994;269:15689–15696. [PubMed] [Google Scholar]
- 61.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford J.L., Jr Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kallstrom G, Hedges J, Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosado IV, Dez C, Lebaron S, Caizergues-Ferrer M, Henry Y, de la Cruz J. Characterization of Saccharomyces cerevisiae Npa2p (Urb2p) reveals a low-molecular-mass complex containing Dbp6p, Npa1p (Urb1p), Nop8p, and Rsa3p involved in early steps of 60S ribosomal subunit biogenesis. Mol. Cell Biol. 2006;27:1207–1221. doi: 10.1128/MCB.01523-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford J.L., Jr Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002;8:150–165. doi: 10.1017/s1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassler J, Kallas M, Hurt E. The NUG1 GTPase reveals and N-terminal RNA-binding domain that is essential for association with 60 S pre-ribosomal particles. J. Biol. Chem. 2006;281:24737–24744. doi: 10.1074/jbc.M604261200. [DOI] [PubMed] [Google Scholar]
- 67.Hung NJ, Johnson AW. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. RNA. 2007;13:1570–1581. doi: 10.1261/rna.585007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nature Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 70.Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA. The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA. 2007;104:1558–1563. doi: 10.1073/pnas.0610704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fatica A, Cronshaw AD, Dlakic M, Tollervey D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 72.Ramagopal S. Are eukaryotic ribosomes heterogeneous? Affirmations on the horizon. Biochem. Cell Biol. 1992;70:269–272. doi: 10.1139/o92-042. [DOI] [PubMed] [Google Scholar]
- 73.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc. Natl Acad. Sci. USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nierhaus KH, Bordasch K, Homann HE. Ribosomal proteins. XLIII. In vivo assembly of Escherichia coli ribosomal proteins. J. Mol. Biol. 1973;74:587–597. doi: 10.1016/0022-2836(73)90049-1. [DOI] [PubMed] [Google Scholar]
- 77.Gadal O, Strauss D, Petfalski E, Gleizes PE, Gas N, Tollervey D, Hurt E. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002;157:941–951. doi: 10.1083/jcb.200111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dunbar DA, Dragon F, Lee SJ, Baserga SJ. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl Acad. Sci. USA. 2000;97:13027–13032. doi: 10.1073/pnas.97.24.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee SJ, Baserga SJ. Imp3p and Imp4p; two specific components of the U3 small nucleolar ribonucleoprotein that are essential for 18S rRNA processing. Mol. Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.