Abstract
Important regions of rRNA are rich in nucleotide modifications that can have strong effects on ribosome biogenesis and translation efficiency. Here, we examine the influence of pseudouridylation and 2′-O-methylation on translation accuracy in yeast, by deleting the corresponding guide snoRNAs. The regions analyzed were: the decoding centre (eight modifications), and two intersubunit bridge domains—the A-site finger and Helix 69 (six and five modifications). Results show that a number of modifications influence accuracy with effects ranging from 0.3- to 2.4-fold of wild-type activity. Blocking subsets of modifications, especially from the decoding region, impairs stop codon termination and reading frame maintenance. Unexpectedly, several Helix 69 mutants possess ribosomes with increased fidelity. Consistent with strong positional and synergistic effects is the finding that single deletions can have a more pronounced phenotype than multiple deficiencies in the same region. Altogether, the results demonstrate that rRNA modifications have significant roles in translation accuracy.
INTRODUCTION
Decoding genetic information with high accuracy is critical for all organisms. While loss of fidelity compromises the functions of the protein products, there is also a high energy cost, related to additional synthesis and turnover activities. Indeed, translation of mRNA into proteins is the most energy consuming pathway in the cell and it is thus of prime importance that the mechanisms involved are sufficiently precise to keep errors at a low, tolerable level. However, it has long been known that decoding accuracy is not maximal, but instead optimal, apparently due to the high energy demand of achieving perfect accuracy (1,2). During the last several years, solving ribosome structures at atomic level resolution has greatly increased our knowledge of the events involved in decoding and provided valuable clues to understanding the structural basis of decoding accuracy. Nonetheless, there is still much to learn about the precise mechanism of decoding, not least of which are the processes that allow the ribosome to maintain correct reading frame and stop at the termination codon (3,4). In this context studies have addressed the possibility that nucleotide modifications in rRNA may influence translation accuracy (5–7).
Modified nucleotides are present in ribosomes in all three kingdoms of life, although with different complexity and created by different synthesis strategies. Two major types of modification dominate: 2′-O-methylation (Nm) and conversion of uridine to pseudouridine (Ψ). In eukaryotes, cytoplasmic rRNA modifications are primarily formed by small nucleolar RNA (snoRNA) protein complexes (snoRNPs). Whereas most snoRNPs create modifications in rRNA, a few are required for pre-rRNA cleavages. In Archaea, homologues of snoRNPs (sRNPs) form Nm and Ψ in both rRNA and tRNA, revealing that the mechanisms are ancient and conserved (8,9). In contrast, modification of rRNA in eubacteria is carried out by site-specific enzymes without RNA co-factors.
The snoRNPs are classified into two groupings, known as the box C/D and box H/ACA families, based on short, conserved sequences in the snoRNA components. Most C/D snoRNPs form Nm modifications and most H/ACA snoRNPs mediate Ψ formation. In each family, site specificity is provided by a unique guide snoRNA and catalysis by a common snoRNP protein. The mechanisms by which those modifications occur were first described in yeast (10–12). The C/D boxes occur at the 5′- and 3′-ends of the snoRNAs and in many cases variant boxes also occur internally. Methylation is targeted by a single, long guide sequence adjacent to the box D/D’ elements. This family of snoRNPs contains four common core proteins: Nop1p (fibrillarin, the methyltransferase), Snu13p, Nop56p and Nop58p. The canonical H/ACA snoRNAs possess two extended hairpin structures and these contain an internal bulge with two short guide sequences that select uridines to be isomerized. The H and ACA boxes occur adjacent to these structures, respectively. The H/ACA snoRNPs also contain four distinct core proteins: Cbf5p (dyskerin, the pseudouridine synthase), Gar1p, Nhp2p and Nop10p (13,14). Relevant to the present study, modifications made by snoRNPs can be blocked selectively by genetically deleting the appropriate guide snoRNAs, which blocks function of the cognate snoRNP.
A striking feature of the modified nucleotides in rRNA is that they are highly concentrated in functionally important regions of the ribosome (15,16). This correlation gave rise to the widely accepted view that modifications have active roles in the translation process, through resulting structural alterations. Indeed, both Nm and Ψ modifications have the potential to stabilize RNA folding domains, as shown by NMR solution studies (17–19). Methylation of 2′-OH sites endows a nucleotide with greater hydrophobicity, protects against nucleolytic attack and stabilizes helices, and thus can benefit inter- or intra-molecular interactions (20). Pseudouridine exerts a subtle, but significant rigidifying influence on the sugar–phosphate backbone and enhances base stacking. In addition, Ψ provides an additional donor site for hydrogen-bond formation, which can stabilize RNA–RNA or RNA–protein interactions (21). Because of their enrichment in the inner core of the ribosome and on subunit surfaces, analysis of the effects of rRNA modifications is an essential aim for ribosome research.
Evidence that the Nm and Ψ modifications in the ribosome are important at the global level came from examining effects of blocking their formation in yeast, with point mutations in the catalytic snoRNP proteins. The most effective mutations caused severely impaired growth rates, indicating that the modifications may not be needed for survival, but, are essential for robust growth (22,23). Presently, it appears that all yeast snoRNAs that target Ψ and Nm modifications to rRNA have been deleted on an individual basis, with no strong effect on growth. In a few cases, deleting single guide snoRNAs has caused observable changes in growth (slight), amino-acid incorporation rate or ribosome structure (24–27). Similarly, deleting bacterial modifying enzymes for rRNA has generally resulted in little or no affect, with a few exceptions, such as RlmL (28). Effects of depleting multiple modifications systematically from important regions of the ribosome have also been examined, and in some cases revealed substantial defects in growth rate, ribosome production and translation rate efficiency (5,7,25,29,30). These findings provide further evidence that fully functional ribosomes require many modifications.
The aim of the present study was to test collections of yeast mutants defective in rRNA modifications, for the ability to support translational accuracy. The functional regions examined are: the decoding center (DC) in the small subunit, and the A-site finger (ASF) and Helix 69 (H69) domains in the large subunit. Translation accuracy was quantified using a set of reporter constructs based on sequences that direct high level of programmed translational errors. Strains with altered accuracy were also tested for resistance to a set of ribosome-based antibiotics. The results demonstrate that modifications in these regions can modulate ribosome accuracy in different ways and also provide insights into the potential mechanisms involved.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains lacking single or multiple snoRNA genes were constructed by sequential gene disruption in strain YS602 MATα, ade2-101, his3-11, 15, trp1-901, ura3-52, leu2-3,112 and are described elsewhere (5,29,30). Strains are described in Supplementary Table S1. Cells were grown in rich medium (YPD) or in synthetic minimal media according to standard procedures. SnoRNA descriptions can be found at http://people.biochem.umass.edu/sfournier/fournierlab/.
Quantification of translation accuracy
Strains were transformed with a set of dual reporter plasmids, carrying an upstream lacZ and a downstream firefly luciferase reading frame separated either by an in-frame stop codon in the Tobacco Mosaic Virus (TMV) readthrough context (CCA GCA GGA ACA CAA TAG/TGA/TAA CAA TTA CAG TGG), the –1 frameshift infectious bronchitis virus (IBV) sequence (TATTTAAACGGGTACGGGGTATCAGTCAGCTCGGCTGGTACCCCTTGCAAAGCGAGCCTCA), the +1 frameshift Ever Shorter Telomeres (EST3) (CAAATACTTAGTTGAGTTTTCC) and Ornithine decarboxylase AntiZyme (OAZ1) (TGGTGCGCGTGACATCCCTCTA) sequences or the control sequence (CCA GCA GGA ACA CAA CAG CAA TTA CAG TGG). β-Galactosidase activity and the luciferase activity were estimated as already described (31). The firefly luciferase activity reflects the translation error level, while the β-Galactosidase activity serves as an internal normalization control. Error efficiency was expressed as the percent of luciferase/β-Galactosidase activity ratio compared to an in-frame fusion construct. The fold wild type (WT) value corresponded to the mutant versus WT efficiency ratio. Statistical significance was determined using the Mann–Whitney test (XL STAT 2007 software).
Antibiotic sensitivity test
Strains were grown in YPD to an OD600 of 1. Ten-fold dilutions were done in YPD from 1 to 10−5 and dilutions 10−2 to 10−5 were spotted on YPD plates containing the indicated antibiotics. The following concentrations were used: G418 (50 µg/ml), neomycin B (5 mg/ml), kanamycin A (5 mg/ml), doxycycline (700 µg/ml). Plates were incubated at 30°C for 3 days.
Computational analysis of ribosome structure
The crystal structure of the Escherichia coli ribosome was visualized and analyzed using PYMOL DeLano (http://pymol.org/). The PDB files (PDB ID: 2J00 and 2J01) were downloaded from the Protein Data Bank (http://www.rcsb.org) and have been modified to have A-site tRNA added by hand fitting.
RESULTS
The frequencies of translational misincorporation and stop codon readthrough are in the range of 10−3 to 10−4, while more deleterious frameshifting errors occur even less often (10−5). Although very low levels of translation errors can be detected in vivo, the signal/noise ratio of such quantification is low and can mask significant variations, especially decreases in error rate. Programmed translational errors include several related phenomena, known collectively as recoding, which end with a local alteration of the normal rules of decoding (32,33). The mechanisms at play generally involve only normal components of the translation machinery, and a number of studies have demonstrated the role of both mRNA sequences and higher structures in stimulating errors. These sequences provide valuable tools to monitor the efficiency of translational accuracy in various conditions, and significant variations in programmed errors can easily be quantified using reporter constructs (31,34,35). Here, we used a dual reporter system that relies on the relative levels of luciferase versus β-Galactosidase activities when encoded in the same mRNA, but separated by a sequence that induces a high error level. Accuracy of the elongation step of translation was systematically examined here using sequences known to promote either −1 (IBV) or +1 (EST3) frameshifting. For the termination step, a readthrough sequence derived from the TMV that contains the UAG stop codon in-frame was used. When required, the +1 OAZ frameshifting sequence and the UAA and UGA stop codons in the TMV context were tested (see ‘Materials and Methods’ section).
The mutants examined for each ribosome region are depleted of one or more modifications, caused by genetic deletion of the corresponding guide snoRNAs. The modifications affected are described in Figure 1 and Table 1. The test strains for the three regions (ASF, DC and H69) have been described previously. Results from these studies have been integrated in the tables. Error rates (readthrough and frameshifting) displayed by each mutant strain were compared to those obtained in the WT, and a mutant/WT factor was determined. At least five independent experiments (up to 20) were performed. The significance of variation was evaluated using a non-parametric statistical test to calculate a P-value (Mann–Whitney).
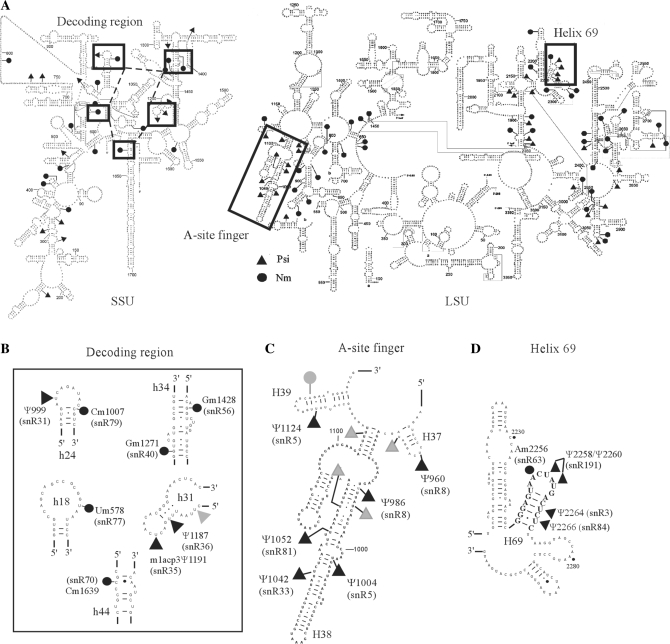
Figure 1.
Modification map of S. cerevisiae rRNA. (A) Secondary structure of the small subunit (SSU) and large subunit (LSU) rRNAs with Nm and Ψ modifications. The modification-rich regions featured in this study are boxed. (B) Modifications in the decoding region in the SSU. The partial sequences of five stem–loop regions implicated in decoding are shown. Modifications are identified with yeast rRNA position numbers, and names of the corresponding guide snoRNAs are given in parentheses. A gray triangle indicates a modification in helix 31 that is not implicated in decoding. (C) Modifications in the ASF region in the LSU. Two flanking helices examined are also shown. Gray dots and triangles indicate modifications not examined in this study. (D) Modifications in H69 in the LSU. Flanking stem–loop structures are also given.
Table 1.
Modifications in the three functional regions examined
| Position | snoRNA | Helix | Conservation of sites | Association or function |
|---|---|---|---|---|
| A-site finger | ||||
| Ψ960 | snR8 | helix 37 | Ψ1664 (human) | |
| Ψ986 | snR8 | helix 38 | ||
| Ψ1004 | snR5 | helix 38 | Ψ1731 (human), Ψ1013 (A. thaliana) | |
| Ψ1042 | snR33 | helix 38 | Ψ1769 (human) | |
| Ψ1052 | snR81a | helix 38 | Ψ1779 (human) | |
| Ψ1124 | snR5 | helix 39 | Ψ1849 (human) | |
| Decoding center | ||||
| Um578 | snR77 | helix 18 | Um627 (human) Um580 (A. thaliana) | A-region |
| Ψ999 | snR31 | helix 24 | Ψ1056 (human) Ψ1000 (A. thaliana) | 3 nt next IF3 Association with 50S E-region |
| Cm1007 | snR79 | helix 24 | Next E-site tRNA | |
| Ψ1187 | snR36 | helix 31 | Ψ1244 (human) | Aa region |
| M1acp3Ψ1191 | snR35 | helix 31 | Gm966 (E. coli) Ψ1248 (human) | P-site tRNA Protein synthesis |
| Gm1271 | snR40b | helix 34 | Gm1328 (human) Gm 1272 (A. thaliana) | Downstream cross-links to mRNA A-region |
| Gm1428 | snR56 | helix 34 | Gm1490 (human) Gm 1431 (A. thaliana) | 2 nt next cross-links site to mRNA Aa region |
| Cm1639 | snR70 | helix 44 | Cm1402 (E. coli) Cm1703 (human) Cm 1641 (A. thaliana) | Downstream cross-links to mRNA P-region |
| Helix 69 | ||||
| Am2256 | snR63 | helix 69 | Am3749 (human) | A-site tRNA D-stem Contact H44 (DC) |
| Ψ2258 | snR191 | helix 69 | m3Ψ1915 (E. coli) Ψ3741 (human) Ψ2248 (A. thaliana) | A-site tRNA D-stem Contact H44 (DC) Translational fidelity Contact RF3 Translation termination |
| Ψ2260 | snR191 | helix 69 | Ψ1917 (E. coli) Ψ3743 (human) Ψ2250 (A. thaliana) | Translation termination |
| Ψ2264 | snR3c | helix 69 | Ψ3747 (human) | |
| Ψ2266 | snR84 | helix 69 | Ψ3749 (human) | P-site tRNA D-stem |
asnR81 also guides position Ψ42 in U2 snRNA.
bsnR40 also guides position Um898 in 25 S rRNA.
csnR3 also guides Ψ2129 in helix 64 and Ψ2133 in helix 65.
A-site finger
The ASF is formed by a long helix (Helix 38 or H38) in folding domain II of the 25 S rRNA, and is conserved in all three kingdoms of life. The ASF is part of the B1a intersubunit bridge between the large and small subunits. It protrudes into the intersubunit space above the tRNA at the ribosomal A site and makes contact with the A-site tRNA through the tip region of H38, and with 5 S rRNA through a segment in the upper portion of the ASF. In E. coli the ASF also establishes contact with protein S13 (S15 for Saccharomyces cerevisiae). H38 in S. cerevisiae contains seven Ψs and three other Ψs occur in short helices that flank H38. The neighboring helices (H37 and H39) fold together with the basal portion of H38 and may be in position to influence the behavior of the ASF.
A total of six modifications were analyzed in the ASF region, four in the middle portion of H38 and two from the flanking helices (Figure 1C). We note that the bacterial ASF lacks modifications, whereas eukaryotic ASF helices have several Ψs and three of these are conserved in mammals as well as yeast. The deletions examined here include the cluster of these conserved modifications (Table 1). Three Ψs in the ASF were not included as the corresponding snoRNAs also direct modifications to three other rRNA positions outside of the ASF, which would complicate data interpretation. Thus, the mutants studied only lack Ψs in the ASF region itself or in combination with one or two Ψs in the adjacent helices. There were five mutants deleted variously for one, two and all six modifications.
No significant effects were observed with mutants deleted of one or two of the six Ψs, on readthrough (UAG stop codon) and −1 frameshifting levels (Table 2, columns 2–5). Small effects were seen for +1 frameshifting for two mutants: one lacking a single modification (Ψ1042; Table 2, column 2), and the other lacking two modifications (Ψ960 and Ψ986; Table 2, column 5); here, slight, but significant decreases in error rate were detected (0.8-fold WT). These results indicate that none of the six ASF modifications tested has a strong effect on translation accuracy. A strain deleted of all six modifications exhibited no change in the capacity to maintain correct reading frame (+1 and −1) or to recognize the UAG stop codon (Table 2, column 6). In previous studies, mutations in the distal portion of the ASF were shown to affect the accuracy of translation. Shortening the E. coli ASF caused an increase (∼1.44-fold) of +1 frameshifting (36) while in S. cerevisiae, a point mutation led to a decrease (∼0.65-fold) in suppression of a UAA nonsense codon (37). These data prompted us to test the readthrough level using UGA and UAA targets with the multiply depleted test strain; UGA is the least efficient stop codon in eukaryotes. A significant increase in UGA suppression level was revealed (1.4-fold, P < 0.037). In contrast, readthrough of the UAA stop codon was equivalent to the WT level (P = 0.148). We also tested a sequence derived from the S. cerevisiae OAZ gene, which drives +1 frameshifting. A UGA termination codon is part of the conserved OAZ frameshift site and is the main stimulating element in S. cerevisiae (38). The frameshift level was affected, showing a significant decrease (0.7-fold, P < 0.0001). This result confirms the protective effect of this Ψ cluster on UGA readthrough by an independent way.
Table 2.
Translational accuracy of A-site finger mutants
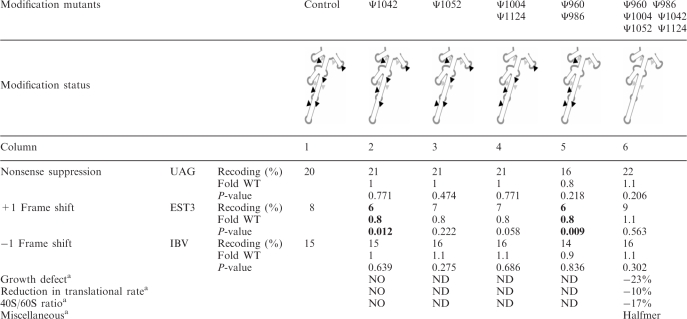 |
Statistically significant values are in bold.
aData from (29). ND: Not Determined; NO: Not Obvious.
In summary, loss of individual modifications in the ASF and flanking helices slightly modifies the maintenance of translational reading frame. No effect is observed when using a combination of all six Ψ deletions. This situation causes a new phenotype, namely, elevation of UGA nonsense codon readthrough that is correlated with an impact on the OAZ +1 frameshift level.
Decoding center
The DC, located in the ribosomal small subunit, includes the A and P sites of codon–anticodon interactions and thus, has major influences on translation accuracy. It was therefore of great interest to test the functional relevance of rRNA modifications in this region. Data from structural studies, chemical protection, cross-linking and genetic analysis in E. coli suggest that the DC includes portions of helices 24, 31, 34, 44 and the 530 loop of 16 S rRNA. The homologous regions in yeast are presumed to be functionally equivalent. In E. coli, the G530 loop and A1492 and A1493 of 16 S rRNA play a central role in control of codon–anticodon pairing accuracy, through the proofreading activity of the ribosome (39). The functional DC is not fully delimited and includes rRNA segments that are separated in the primary structure, but are close to one another in crystal structures. The DC region in yeast contains eight modified nucleotides: five Nm and three Ψ; one Ψ undergoes additional modifications to form 1-methyl-3-(3-amino-3-carboxypropyl) Ψ(m1acp3Ψ) (Figure 1B). Seven of these modifications occur in mammalian cells and two occur in E. coli (Table 1). In yeast, four of the eight DC modifications are in the A-region. Two are located on the tRNA-entry side of the A-site tRNA and we will refer to these as A-region modifications (Um578 and Gm1271). The other two are located above tRNA in the A site and we will refer to these as Aa-region modifications (A site above) (Ψ1187 and Gm1428). Of the four remaining modifications, two are in the P-region (m1acp3Ψ1191 and Cm1639), and two are in the E-region (Ψ999 and Cm1007).
Analyses of translation accuracy were carried out using a set of 14 deletion mutants, lacking individual modifications (four mutants) or different combinations of modifications (10 mutants) (Table 3) (30). Two strains that lack modifications in the P-region or a combination of the Aa- and P-regions exhibited a significant change in termination accuracy, which was readily apparent using the UAG stop codon (1.3–1.5-fold of WT level; Table 3, columns 3 and 9). Both mutants lack Cm1639 in the P-region: one lacks this Nm alone, the other also lacks m1acp3Ψ1191 (P-region) and Gm1428 (Aa-region). Interestingly, three other combinations of Cm1639 deletions did not affect termination, including combinations with Gm1428 or with Ψ1187 and m1acp3Ψ1191, all in the Aa- and P-regions (Table 3, columns 8, 14 and 15). This striking observation indicates that the effect depends not only on a particular modification, but also on the specific pattern of deletions and residual modifications, suggesting strong synergistic relationships between modifications. Altogether, the deletion results show that translation termination has low sensitivity to loss of modifications in the DC region.
Table 3.
Translational accuracy of decoding region mutants
 |
#Processing of 20 S to 18 S delayed. Statistically significant values are in bold.
aData from (30). ND: Not Determined; NO: Not Obvious.
In contrast, moderate to strong frameshifting effects were detected for 11 of the 14 strains examined (for either +1 or −1 contexts). As for readthrough, deleting the P-region Cm1639 had an impact on +1 frameshifting alone or in four combinations with other Aa- and P-region deletions. The +1 frameshift activity was 1.7-fold that of WT for loss of the Cm1639 modification alone (Table 3, column 3) and varied from 1.8- to 2.4-fold for the other mutant strains, which included two, three and four deletions from the Aa- and P-regions (Table 3; columns 8, 9, 14 and 15). Blocking the two E-region modifications reduced +1 frameshift activity (0.6-fold; Table 3, column 6). The values for four other mutants with three deletions each (Table 3, columns 10–13) are not statistically different from those obtained with the various single deletions (P-values ranging from 0.244 to 0.579). These last strains included two mutants deleted of both A-region modifications and one Aa-region modification; one mutant deleted of one A-region, one Aa-region and one P-region modifications; and one mutant deleted of two modifications in the Aa-region and one modification in the P-region.
On examining effects on −1 frameshifting, we observed that loss of three individual modifications alone from the Aa- and P-regions increased frameshift activity (Ψ1187; Table 3, column 5, Gm1428 column 2 and Cm1639 column 3, respectively). Notably, loss of Ψ1187 from the Aa-region led to a 1.2-fold increase in −1 frameshifting, but to a decrease when combined with deletion of a P-region modification (m1acp3Ψ1191; Table 3, column 7) or with two A-region modifications (Um578 and Gm1271; Table 3, column 10) (0.5-fold). Increased −1 frameshift activity was also observed for two strains deleted of a mix of Aa- and P-region modifications, as follows: one Aa- and one P-region modification (Table 3, column 8), one Aa- and two P-region modifications (Table 3, column 9). No effect was detected for a strain deleted of the two E-region modifications. Similarly, no effect was detected for four multiply deleted strains lacking other combinations of A-, Aa- and P-region modifications (Table 3, columns 11–14). Behavior of one strain lacking four modifications was difficult to interpret due to internal variations of error level that resulted in a P-value of 0.110 (Table 3, column 15).
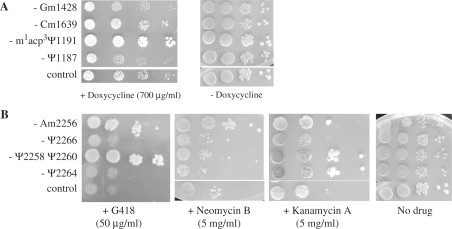
The rRNA site that undergoes hypermodification in yeast (m1acp3Ψ1191) is also modified at the corresponding site in E. coli, yielding mG966 in the 970 loop of 16 S rRNA. Interestingly, the 970 loop is involved in tetracycline binding (40,41) and a mutation at position 966 confers resistance to the antibiotic. These relationships prompted us to test the sensitivity of the m1acp3Ψ1191 deletion strain to doxycycline, a tetracycline analog. Strikingly, this strain showed substantially increased resistance to doxycycline. This finding suggests that tetracycline and related analogs target the same position in both organisms (Figure 2A).
Figure 2.
Sensitivity of rRNA modification mutants to translational inhibitors. WT and mutant strains lacking individual modification were spotted as 10-fold dilutions on YPD and YPD containing the indicated antibiotic media. Cells were incubated for 3 days at 30°C. (A) Loss of m1acp3Ψ1191 modification in the DC leads to doxycycline resistance. (B) Loss of modification in H69 induces aminoglycoside resistance.
In summary, modifications in the Aa-, P- and E-regions have significant roles in maintaining translation accuracy, whereas the two A-region modifications have only minor influence. Both Aa-region and P-region modifications are involved, as blocking these individually affects either stop codon readthrough or +1 and −1 frameshifting. Fidelity is also altered with deletion of multiple Aa- and P- region modifications in different combinations. Effects on both readthrough and +1/−1 frameshifting were seen for two strains deleted of at least the P-region modification Cm1639. Finally, blocking the two E-region modifications together only affects +1 frameshift activity. Taken together, the data indicate that Aa- and P-region modifications have greater influence on maintaining normal accuracy than A-region modifications.
Helix 69
H69 is a short stem–loop structure of 19 nucleotides located in domain IV of 25 S rRNA. It is a component of the intersubunit bridge B2a, along with portions of helix 44 of 18 S rRNA. In addition to its important role in subunit joining, it has been established in E. coli that the H69 loop and stem contact the D-stem of tRNAs in the A site and P site, respectively, suggesting a role(s) in translation accuracy. Interactions between H69 and various translational factors have also been described for E. coli. In particular, relationships between H69 and RF2 and RF3 were established by cryoelectron microscopy experiments, linking H69 with termination of protein synthesis (42–44). Earlier evidence of these relationships came from the observation that blocking formation of E. coli H69 modifications Ψ1911, Ψ1915 and Ψ1917, by inactivating the pseudouridine synthase RluD, impaired translation termination; readthrough activity increased by up to 14-fold for the UGA codon (6). Further evidence of the importance of H69 comes from our findings that yeast lacking assorted H69 modifications can be defective in growth, subunit joining and translation rate (5). Mutants from that collection are featured in the present study.
The relatively small H69 domain in S. cerevisiae is rich in modifications, with two Ψs in the stem (Ψ2264, Ψ2266) and two Ψs plus one Nm in the loop (Am2256, Ψ2258, Ψ2260) (Figure 1D). All these modifications are conserved in humans. The Ψ2258 modification (Ψ1915 in E. coli), is of particular interest evolutionarily, since it is conserved in bacterial variants with the minimal genomes described thus far (45). These five modifications are guided by four snoRNAs, with one snoRNA targeting two sites (Table 1). We tested six strains deleted for individual modifications and in three combinations of two to four deletions.
Remarkably, the results point to the involvement of at least four and possibly all five modifications in translation accuracy for both the elongation and termination phases (Table 4). Readthrough was affected for each stop codon tested with the four single deletion strains. The general trend was a decrease of readthrough (between 0.3- and 0.7-fold). The mutant lacking Ψ2258 and Ψ2260 (Table 4, column 4) was an exception, showing a small increase for the UAG stop codon (1.2-fold). Frameshift efficiency was affected to a small, but significant extent (0.4- to 0.7-fold WT) for three mutants lacking one modification and the mutants lacking two to three modifications. Two strains were affected in both +1 and −1 frameshifting (lacking either Ψ2266 alone or lacking Ψ2258 and Ψ2260; Table 4, columns 3 and 4), but strains lacking Ψ2264 or Am2256 alone (Table 4, columns 5 and 2) displayed a normal level of +1 or −1 frameshifting, respectively. Strikingly, loss of Ψ2266 or both Ψ2258 and Ψ2260 altered nonsense suppression and reading frame maintenance, both forward and backward.
Table 4.
Translational accuracy of Helix 69 mutants
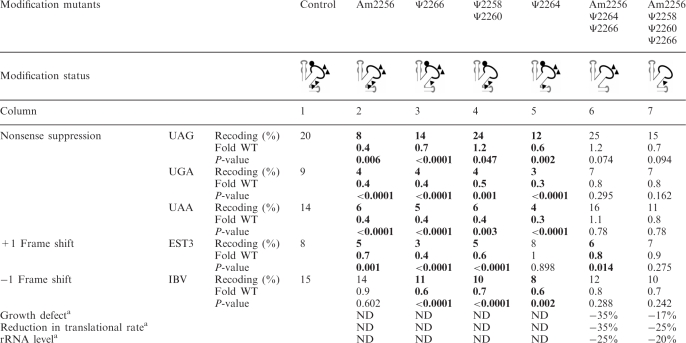 |
Statistically significant values are in bold.
aData from (5). ND: Not Determined; NO: Not Obvious.
For strains lacking three or four modifications (Table 4, columns 6 and 7), a small decrease in accuracy was observed for both readthrough and frameshifting, although the differences were not statistically significant. For stop codon recognition and −1 frameshifting, strains deleted of three or four modifications behaved similarly to WT controls. However, the strain deleted of three modifications displayed a small decrease of +1 frameshift levels (0.8-fold WT) (Table 4). We did not test a strain deleted for all five modifications, due to a strong growth impairment that did not allow precise quantification of translation accuracy (5).
Sensitivity to aminoglycoside antibiotics was evaluated as well. These drugs interfere with translocation of tRNA from the A site to the P site and induce translational misreading. Three mutants lacking one or two modifications show increased resistance to either one or two of the three drugs, in strain-specific fashion (Figure 2B). Deleting Ψ2264 (in the stem) caused increased resistance to kanamycin, but not to G418 or neomycin, whereas deleting Ψ2258 and Ψ2260 (both in the loop) yielded increased resistance to G418 and kanamycin, but not to neomycin. The differences presumably reflect different binding patterns for the drugs, as evidenced by different positional effects of modification on binding.
In summary, three single deletions and one double deletion of H69 modifications altered both elongation and termination accuracy. Interestingly, loss of three or four modifications has only a slight effect, consistent with some of the combinatorial effects observed for the DC and ASF. Three mutants showing increased translation accuracy also exhibit increased resistance to aminoglycoside antibiotics, suggesting a correlation between translation fidelity and ribosome structural changes.
DISCUSSION
The precise role of rRNA modifications on the function of the ribosome remains to be assessed. Earlier depletion analyses for clusters of modifications in the yeast ribosome revealed a range of effects, variously, for growth, ribosome biogenesis and translation efficiency (5,25,29,30). In the main, the modifications created by snoRNPs improve the fitness of the eukaryotic ribosome at a number of levels, including subunit formation and stability, polysome formation, translation rate and ribosome structure. In the present article, we made use of a collection of modification-depleted strains to systematically explore the role of rRNA modifications on translational accuracy.
The major finding of this work is that several modifications in the two intersubunit bridges and DC affect translation accuracy. These alterations are specific since strains exhibiting large effects on translational rate, ribosome status or cell growth are not affected for accuracy and conversely. In addition, each strain displays a given pattern with either readthrough or frameshifting or both being affected. Unexpectedly, loss of certain modifications also generated hyperaccurate ribosomes. This is reminiscent of the E. coli Um2552 modification that is a negative modulator of translational accuracy (46). These lower readthrough levels observed with single deletions in H69 (Table 4) correlate with higher resistance to aminoglycosides. Similarly increased frameshift and readthrough activities have been reported for loss of Ψs in the peptidyltransferase centre, accompanied by increased sensitivity to the antibiotics anisomycin and sparsomycin (7). These observations point to the fact that accuracy has a defined cost in terms of protein synthesis, it might thus be beneficial to the cell to balance accuracy with speed and energy consumption.
Altogether, reading frame maintenance was the property most often perturbed in the mutants we analyzed. In particular, +1 frameshifting was affected for the three regions and varied for six of the nine strains deleted for single modifications and for nine of seventeen strains with multiple modifications. This situation might be related to the mechanism of +1 frameshifting, which is easily subject to modulation through codon/anticodon interactions. Notably, several multiply-deleted mutants are less affected than mutants with single deletions. This finding is reminiscent of earlier effects seen for the ASF region. This study showed that the specific distribution pattern of deleted and residual modifications is important, and that mutants with the same number of deletions, but different patterns can exhibit different effects on growth, polysome formation and translation rate (29). The present results indicate that modification effects on ribosome accuracy can also be combinatorial in nature, with the absence of one modification being able to counteract or amplify the absence of another(s).
Apart from these important general conclusions, tentative molecular interpretations of some effects can also be proposed, exploiting the high degree of conservation between bacterial and eukaryotic ribosomes, especially in the catalytic domains (47,48).
A-site finger
The seven Ψs in the mid- and lower portions of the ASF helix can be predicted to reduce its conformational flexibility. This damping effect could be even greater for the long lever-like upper portion of the ASF region (Figure S1) (29). Similarly, the three Ψs in the H37 and H39 helices, which interact with the basal portion of H38, could influence the activity of the entire three-helix structure. Thus, variations in the modification pattern could have different consequences on translation fidelity. We observed that loss of six Ψ modifications induces nonsense suppression of the UGA stop codon. During termination, the release factor eRF1 is in competition with near cognate tRNAs that can establish unconventional base pairing with the stop codon. Nucleotides in the upper portion of H38 are in close proximity to the D-loop of the A-site tRNA and are thought to influence tRNA movement (49). In E. coli a tRNAtrp mutation in the D-loop (position 24) was shown to affect rejection of this near cognate tRNA, leading to increased UGA suppressor activity (50). Moreover, a G1020C mutation in yeast H38 was proposed to favor discrimination between the tRNA D-loop and the analogous topological region of eRF1 (37). Thus, loss of H38 modifications might alter the discrimination between a near cognate tRNA and eRF1, and thereby favor nonsense suppression. A reduction in +1 ribosomal frameshifting was also observed in two ASF mutant strains, including the mutant deleted of Ψ1042 which is the nearest modification to the tip of H38. This is reminiscent of effects reported for deletion of the tip of H38 in E. coli (36). The efficiency of +1 frameshifting relies on the presence of either a stop codon (OAZ) or a hungry codon (EST3) at the A site. The ribosome pauses at these codons with a resulting shift in reading frame. If ASF modifications alter the A-site environment, frameshifting could be altered as well.
Decoding center
Due to the wide variety of interactions that could be influenced by modifications in the DC, a global explanation of the effects is difficult to develop. Thus, our interpretation focuses on examining the position of each modification linked to an effect, with the aim of obtaining clues about its role.
The absence of Ψ1187 and Gm1428, in the Aa-region, individually led to increases in −1 frameshifting (1.2-fold). Although we cannot explain the precise basis of the effect that blocking these modifications has on −1 frameshifting, it is tempting to propose that the modifications strengthen the codon/anticodon interaction and thereby, decrease the probability of simultaneous slippage of the P- and A-site tRNAs.
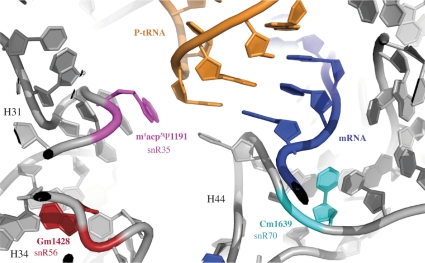
The m1acp3Ψ1191 modification includes an aminocarboxypropyl side chain, which bears an electronic charge and is expected to have high steric clash (Figure 3; depicted in magenta). This nucleotide is close to wobble base 34 of P-site tRNA. The wobble position is frequently modified, which extends the decoding capacity of a tRNA to additional codons (51). Disrupting m1acp3Ψ1191 formation led to an increase in the +1 frameshifting level (1.3-fold). Therefore, the observed phenotypic effects might be influenced not only by combinatorial losses of modifications in rRNA, but changes in tRNA modification as well.
Figure 3.
Two modifications in the DC are in close proximity with tRNA and mRNA at the P site. 3D representation of the rRNA bases of interest mapped onto E. coli P-region. Modified nucleotides and the corresponding guide snoRNAs are shown in various colors, mRNA is shown in blue and P-tRNA is shown in orange.
Blocking Nm of DC nucleotide C1639, alone or in combination with the three DC modifications just discussed above, resulted in decreases in translation accuracy for all three types of recoding targets examined. Among the DC modifications tested, loss of this methylation produced the greatest number of pleiotropic effects. This modified nucleotide (Figure 3; with Cm1639 depicted in cyan) is in close proximity with the third base of the mRNA codon at the P site, a position that is crucial for translational accuracy. Interestingly, results from a high-resolution growth phenotyping analysis of yeast lacking different methylation guide snoRNAs, implicated the Cm1639 modification with a wide range of environmental perturbations (52). Thus, this modification could have an important role in modulating ribosomal activity during environmental stress. The fact that the corresponding positions in E. coli and human rRNAs are also modified, with the unusual m4Cm1402 and Cm1703, respectively, provides additional evidence of a beneficial effect. In the context of translational termination and reading frame maintenance, loss of this 2′-O-methyl modification has a negative impact on ribosome function.
Helix 69
The most notable deletion effects for this region are substantial increases in translation accuracy, for both elongation and termination. Also interesting, the improved accuracy correlates well with increases in resistance to G418, kanamycin A and neomycin B. These aminoglycoside drugs are known to undermine translation fidelity by binding to specific nucleotides in H44 (SSU) and H69 (LSU) (53). This induces misincorporation through a conformational change that mimics binding of cognate tRNA. The reduced sensitivity seen with the aminoglycoside antibiotics could reflect structure changes in H69 that are transmitted to H44 and affect drug binding. The increase in termination efficiency might result from increased discrimination against a near-cognate tRNA, which may be involved with the increase in aminoglycoside resistance.
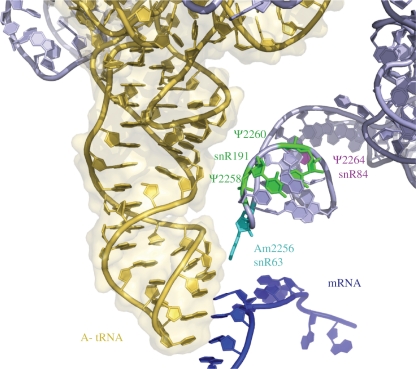
Two modifications that appear to be in good position to influence the rigidity of H69 through increased base stacking are Ψ2264 and Ψ2266. Consistent with a structural change that may be relevant, blocking formation of Ψ2264 (Figure 4; depicted in pink) was shown earlier to increase the chemical reactivity of the H69 nucleotide A2252 with dimethylsulfate (5). The neighboring Ψ2266, is shown in crystal structures to contact the sugar–phosphate backbones of nucleotides 25 and 26 of P-site tRNA (49), adding stabilizing influence during tRNA–ribosome interactions.
Figure 4.
Modifications within H69. 3D representation of the rRNA nucleotides of interest mapped onto E. coli H69. Modified nucleotides and the corresponding guide snoRNAs are shown in color, mRNA is shown in blue and A-site tRNA is shown in yellow.
The two Ψs near the tip region of H69, Ψ2258 and Ψ2260, (Figure 4; depicted in green) occur in functionally promising positions and are also located near the Nm modification in this domain (Am2256; Figure 4; depicted in light blue). These two Ψs are conserved in E. coli (Ψ1915 and Ψ1917) and inactivation of the RluD pseudouridine synthase responsible for their formation leads to a growth defect (6). Moreover, mutational analysis of E. coli H69 positions 1912 to 1918 identified variants with increased +1 or −1 frameshifting and UGA and UAG readthrough, including a Ψ1915A mutation that caused, variously, ∼2- to 5-fold increase in these frameshifting and readthrough activities (54).
In the deduced 3D model of yeast ribosome modifications, Am2256 (Figure 4) is predicted to be 2.77 Å from base-37 of the A-site tRNA, which is a highly modified purine that plays a crucial role in the decoding ability of tRNA. We speculate that the four Ψs in H69 have a stabilizing effect on the structure and this could help modulate orientation of Am2256 which is blocked in the 3′-endo conformation, toward position-37 of the tRNA located in the A site. Another possibility is that modified nucleotides in H69 might contact the D-loop of the tRNA and improve accuracy in this way.
In conclusion, we have demonstrated that deleting Ψ and Nm modifications from three functionally important regions of the yeast ribosome modulates ribosome accuracy at the level of both reading frame maintenance and translation termination. The ribosome regions are in close proximity with tRNA and many of the key changes observed are proposed to reflect modification effects on codon/anticodon interactions. Since tRNA functions are highly modulated by modifications, further examination of their effects on accuracy is warranted. Similarly, undertaking studies of potential relationships between rRNA and tRNA modifications seems promising too, with the prospect of revealing important new aspects of control and additional new roles for modified nucleotides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale pour la Recherche [grant ANR-06-BLAN-0391-01 to J.P.R.], the Association pour la Recherche sur le Cancer [grant 3849 to J.P.R.], the Association Française contre les Myopathies [grant 12652 to J.P.R.] and the US National Institutes of Health [grant Gm19351 to M.J.F.]. Funding for open access charge: ANR-06-BLAN-0391-01.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Olivier Namy for providing the 3D pictures of the ribosome. They also thank Henri Grosjean and Wayne Decatur for stimulating discussions.
REFERENCES
- 1.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 2.Kirkwood TBL, Rosenberger RF, Galas DJ. Accuracy in Molecular Processes. London: Chapman and Hall; 1986. [Google Scholar]
- 3.Petry S, Weixlbaumer A, Ramakrishnan V. The termination of translation. Curr. Opinion Struc. Biol. 2008;18:70–77. doi: 10.1016/j.sbi.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 4.von der Haar T, Tuite MF. Regulated translational bypass of stop codons in yeast. Trends Microbiol. 2007;15:78–86. doi: 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Ejby M, Sorensen MA, Pedersen S. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl Acad. Sci. USA. 2007;104:19410–19415. doi: 10.1073/pnas.0706558104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baxter-Roshek JL, Petrov AN, Dinman JD. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE. 2007;2:e174. doi: 10.1371/journal.pone.0000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 9.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Exp. 2002;10:17–39. [PMC free article] [PubMed] [Google Scholar]
- 10.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 11.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 12.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 13.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reichow SL, Hamma T, Ferre-D'A;mare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brimacombe R, Mitchell P, Osswald M, Stade K, Bochkariov D. Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 1993;7:161–167. doi: 10.1096/fasebj.7.1.8422963. [DOI] [PubMed] [Google Scholar]
- 16.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 17.Rife JP, Moore PB. The structure of a methylated tetraloop in 16S ribosomal RNA. Structure. 1998;6:747–756. doi: 10.1016/s0969-2126(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 18.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabello-Villegas J, Nikonowicz EP. Solution structure of psi32-modified anticodon stem-loop of Escherichia coli tRNAPhe. Nucleic Acids Res. 2005;33:6961–6971. doi: 10.1093/nar/gki1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2'-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 21.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 22.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 24.Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol. Cell. 2004;16:663–669. doi: 10.1016/j.molcel.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 25.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 26.Bonnerot C, Pintard L, Lutfalla G. Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol. Cell. 2003;12:1309–1315. doi: 10.1016/s1097-2765(03)00435-0. [DOI] [PubMed] [Google Scholar]
- 27.Badis G, Fromont-Racine M, Jacquier A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9:771–779. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sergiev PV, Bogdanov AA, Dontsova OA. Ribosomal RNA guanine-(N2)-methyltransferases and their targets. Nucleic Acids Res. 2007;35:2295–2301. doi: 10.1093/nar/gkm104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J. Biol. Chem. 2008;283:26026–26036. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 33.Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- 34.Namy O, Hatin I, Stahl G, Liu H, Barnay S, Bidou L, Rousset JP. Gene overexpression as a tool for identifying new trans-acting factors involved in translation termination in Saccharomyces cerevisiae. Genetics. 2002;161:585–594. doi: 10.1093/genetics/161.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 37.Rakauskaite R, Dinman JD. An arc of unpaired “hinge bases” facilitates information exchange among functional centers of the ribosome. Mol. Cell Biol. 2006;26:8992–9002. doi: 10.1128/MCB.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov IP, Gesteland RF, Atkins JF. Evolutionary specialization of recoding: frameshifting in the expression of S. cerevisiae antizyme mRNA is via an atypical antizyme shift site but is still +1. RNA. 2006;12:332–337. doi: 10.1261/rna.2245906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noller HF. Biochemical characterization of the ribosomal decoding site. Biochimie. 2006;88:935–941. doi: 10.1016/j.biochi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 40.Brodersen DE, Clemons W.M., Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 41.Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C, et al. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 2001;20:1829–1839. doi: 10.1093/emboj/20.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 43.Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- 44.Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322:953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Crecy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life. 2007;59:634–658. doi: 10.1080/15216540701604632. [DOI] [PubMed] [Google Scholar]
- 46.Widerak M, Kern R, Malki A, Richarme G. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene. 2005;347:109–114. doi: 10.1016/j.gene.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 47.Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 48.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008;36:D178–D183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 50.Smith D, Yarus M. Transfer RNA structure and coding specificity. I. Evidence that a D-arm mutation reduces tRNA dissociation from the ribosome. J. Mol. Biol. 1989;206:489–501. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- 51.Agris PF, Vendeix FA, Graham WD. tRNA's; wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 52.Esguerra J, Warringer J, Blomberg A. Functional importance of individual rRNA 2′-O-ribose methylations revealed by high-resolution phenotyping. RNA (New York, NY) 2008;14:649–656. doi: 10.1261/rna.845808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struc. Mol. Biol. 2007;14:727–732. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 54.Hirabayashi N, Sato NS, Suzuki T. Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J. Biol. Chem. 2006;281:17203–17211. doi: 10.1074/jbc.M511728200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.