Abstract
To unravel regulatory networks of genes functioning during embryonic development, information on in situ gene expression is required. Enormous amounts of such data are available in literature, where each paper reports on a limited number of genes and developmental stages. The best way to make these data accessible is via spatio-temporal gene expression atlases. Eleven atlases, describing developing vertebrates and covering at least 100 genes, were reviewed. This review focuses on: (i) the used anatomical framework, (ii) the handling of input data and (iii) the retrieval of information. Our aim is to provide insights into both the possibilities of the atlases, as well as to describe what more than a decade of developmental gene expression atlases can teach us about the requirements of the design of the ‘ideal atlas’. This review shows that most ingredients needed to develop the ideal atlas are already applied to some extent in at least one of the discussed atlases. A review of these atlases shows that the ideal atlas should be based on a spatial framework, i.e. a series of 3D reference models, which is anatomically annotated using an ontology with sufficient resolution, both for relations as well as for anatomical terms.
INTRODUCTION
Information regarding the level and the location of the expression of genes is required to unravel the function of those genes during embryonic development. A wealth of information on gene expression levels in different organs and developmental stages in several species has been obtained with microarray, and more recently next-generation sequencing, studies. These data are collated and made available by the major databases, ArrayExpress (1) and NCBI GEO (2). The recently launched Gene Expression Atlas (http://www.ebi.ac.uk/gxa) aims to make these data accessible to non-expert biologists; the data are retrieved from ArrayExpress, and enriched through curation and statistical analysis.
These microarray data are mostly based on organ and tissue samples containing different cell types. The observed differential expression can be used to identify candidate genes related to different conditions or states of the harvested tissue samples. However, to test hypotheses on regulatory interactions of the identified genes on the cellular level, in situ gene expression information is required. Gene products, mRNA as well as proteins, have been visualized in whole-mount stained tissue samples and histological sections to determine the pattern of gene expression in the organ or tissue of interest. Enormous amounts of such in situ data are available in literature, where each paper reports on a limited number of genes, developmental stages and species.
Microarray data, giving the expression level of a large number of genes in a limited number of tissues per experiment have been collected in large scale databases. A similar joining of data on in situ gene expression is hampered by the larger variety of techniques employed to generate these data. Automation of the techniques used to determine in situ expression in the last decade enabled the start of large scale in situ visualization projects. This resulted in wealth of data on in situ gene expression of large number of genes from different species and developmental stages and exacerbated the problems in retrieving information from literature.
To remedy this situation, several initiatives were started during the last decade to make these data accessible via spatio-temporal gene expression atlases. We define a gene expression atlas as a database containing anatomically annotated in situ gene expression information. In other words, such a gene expression atlas describes gene expression within anatomically defined structures. These expression patterns can be based on the in situ visualization of the expression levels of mRNAs, proteins or transgenic reporters.
Note that, the microarray-based gene expression databases do not match our definition of a spatio-temporal atlas. However, via the gene identifier the gene expression levels determined with microarray studies can be linked to the in situ gene expression information contained in these atlases. Such a link is for instance implemented in GXD.
We selected all gene expression atlases that fit our definition, restricting ourselves to atlases describing developing vertebrates and covering at least 100 genes. To the best of our knowledge, 11 atlases (Table 1) meet these criteria. These 11 atlases were reviewed to illustrate the different approaches used to build developmental gene expression atlases.
Table 1.
Atlas overview
| Name | Full project name and website | Species | References |
|---|---|---|---|
| GEISHA | Gene expression in situ hybridization analysis http://geisha.arizona.edu/geisha/ | Chicken | (3,4,31) |
| MEPD | The Medaka Gene Expression Pattern Database http://ani.embl.de:8080/mepd/ | Medaka | (6,30) |
| EMAGE | Edinburgh Mouse Gene Expression database www.emouseatlas.ora/emage/ | Mouse | (7–13) |
| GenePaint.org | GenePaint.org www.genepaint.org | Mouse | (14,15) |
| GENSATa | Gene Expression Nervous System Atlasa www.ncbi.nlm.nih.gov/projects/gensat/ | Mouse | (16) |
| GUDMAP | Genito Urinary Development Molecular Anatomy Project www.qudmap.org/index.html | Mouse | (17,18) |
| GXD | Gene Expression Database www.informatics.jax.org/expression.shtml | Mouse | (7,19–21,32) |
| EURExpress | EURExpress www.eurexpress.org/ee/intro.html | Mouse | |
| EuReGeneDb | European Renal Genome Project www.euregene.org/euregenedb/pages/db home.html | Mouse | (17) |
| XGEbase | European Renal Genome Project www.euregene.org/xgebase/pages/entry page.html | Xenopus | (22,23) |
| ZFIN | The Zebrafish Model Organism Database http://zfin.org | Zebrafish | (5) |
Basic information on the developmental gene expression atlases.
aAlso known as BGEM.
Just over a decade ago, shortly after the fruition of research techniques to visualize gene expression patterns, the first developmental gene expression atlases started to emerge. Already in 1994, the available gene expression data from different modalities accumulated rapidly and the question was raised how to acquire, manage, analyse, interpret and disseminate these data (7). The need was expressed to answer both simple queries, such as when and where a particular gene is active, and more complex queries that require a deeper understanding of developmental processes (7). To enable such queries, a project was presented to develop a database to gather and retrieve information from diverse resources.
A few years later this project resulted in the developmental gene expression atlases GXD and EMAGE (Table 1). The cooperation between these atlases has the ultimate goal to create an integrated resource of mouse developmental gene expression information (7). In 1997, a prototype of a database was presented in which gene expression data from different sources was combined and anatomically annotated: the GXD (19). At the same time, the underlying database of EMAGE was described in detail. Spatial mapping of in situ hybridization (ISH) images to morphological reference models was then announced (8).
Publications on developmental gene expression atlases containing data from high throughput ISH projects appeared a few years later. In 2000, it was shown that whole-mount ISH techniques could be adapted to perform high-throughput gene expression analyses on mouse (25) and medaka (26). In 2003 and 2004, similar high-throughput techniques resulted in mouse atlases like GENSAT, covering the nervous system at different developmental and post-natal stages (16) and GenePaint.org, covering the whole embryo at a single stage of development (14) (Table 2).
Table 2.
Atlas contents
| Name | Mutants | Ages/Developmental stages | Nr Genesa | Annotation |
|---|---|---|---|---|
| GEISHA | Yes | HHb 2–27 | 1025 | A |
| MEPD | No | Iwamatsu stages 15–44 | 1227 | O |
| EMAGE | Yes | TS 1–28 | 2533 (2488c,10 012d) | O,S |
| GenePaint.org | No | E10.5, E14.5 (whole embryo), E15.5 (Head), P7, P57 (Brain) | 1296 (16 412d) | A |
| GENSAT | No | E10.5, 15.5, P7 and Adult | 3525 | A |
| GUDMAP | No | TSe 17–24, 26–28 | 2836 | O |
| GXD | Yes | TSe 1–26 and Adult | 8786 | O |
| EURExpress | No | TSe 23 (E14.5) | 14 900 (18 697d) | O |
| EuReGeneDb | No | TSe 23–28 | 406 | O |
| XGEbase | No | NFf 20–40 | 210 | O |
| ZFIN | Yes | All 44 stages | 10 501 | O |
Specific information on the developmental gene expression atlases. Annotation is coded as anatomical annotation using a controlled vocabulary (A), using an ontology (O) and using a spatial framework (S).
aTextual annotated.
bHamburger and Hamilton.
cSpatial annotated.
dIncluding genes within not annotated images.
eTheiler Stage.
fNeidhart and Faber.
In this review we will focus on: (i) the anatomical framework used by, (ii) the handling of the input of data in, and (iii) the retrieval of information from the atlases. Our aim is to provide insights both into the possibilities of the atlases, as well as to describe what more than a decade of developmental atlases can teach us about the requirements of the design of the ‘ideal atlas’.
RESULTS
Framework
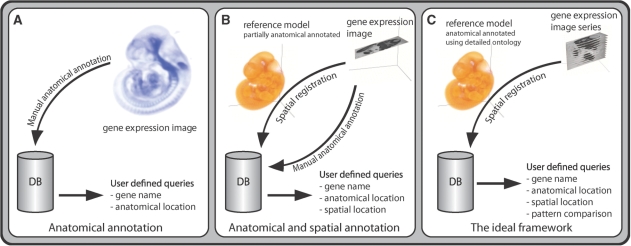
To enable comparison of gene expression data, information regarding these data has to be stored in such a way that they can be retrieved. In all atlases this is done using frameworks that always include a list of anatomical terms to specify the location at which an expressed gene is observed. Most often, in this annotation, the categories used to define the expression type are ‘scattered’, ‘regional’ and ‘ubiquitous’, whereas the expression strength is usually categorized as ‘negative’, ‘unknown’, ‘weak’, ‘moderate’ or ‘strong’. In addition to this ‘anatomical annotation’, gene expression patterns can be spatially mapped into a 2D or 3D reference model, which we will refer to as ‘spatial annotation’ (Figure 1).
Figure 1.
Frameworks. Panels A and B show the different frameworks used in the discussed atlases. All atlases except for EMAGE use a purely text-based annotation (A). In EMAGE the gene expression images are mapped into reference models (B). The ideal framework is shown in (C). Basically, this framework is used by the Allen Brain Atlas (except for the detailed ontology). The approach involves the mapping of sets of in situ gene expression images to a spatial framework. Enabling automated textual annotation and analysis based on the spatial location of the expression of a gene.
Anatomical annotation
The complexity of the vocabularies used for anatomical annotation ranges from an unsorted list of terms to complete ontologies. An ontology can be defined as a computer-interpretable description of a knowledge domain. Definitions, such as classes and relations, are used to associate the entities within such a knowledge domain (27). The most basic relation used in ontologies is the subsumption relation ‘is a’. In the domain of a developmental gene expression atlas, important additional relations include ‘part of’, ‘develops from’ and ‘gives rise to’ which can link anatomical structures within and between developmental stages. The ontologies used in EMAGE, GXD, ZFIN, XGEbase and MEPD are available via the Open Biomedical Ontologies website (http://www.obofoundry.org/). Within the OBO foundry ongoing efforts are made to design ontologies in such a way that they are interoperable and logically well formed. This includes the standardization of the used relations (28).
EMAGE developed a widely used anatomical ontology covering Theiler stages (29) (TS) 1 to 26 in mouse development and ‘part of’ relationships within each stage (9). This ontology was adopted by EURExpress and extended by the GXD with its own vocabulary for adult mice (21) which also includes the ‘part of’ relationship. EuReGeneDb and GUDMAP further extended this ontology to a higher resolution for the genito-urinary tract (17).
ZFIN developed its own zebrafish anatomical ontology which covers structures of each of the 44 developmental stages of the zebrafish, arranged by functional systems (5). Within these systems there are 2210 different embryological and anatomical terms defined with ‘part of’, ‘develops from’, ‘start stage’ and ‘end stage’ relations (http://www.zfin.org). The developers of MEPD created a similar ontology containing 4173 terms for the 44 developmental stages of medaka, which follows the terminology used for zebrafish, mouse and Drosophila when possible (30). The relations used in their ontology are ‘part of’, ‘is a’ and ‘develops from’. The ‘develops from’ relation is only used to link developmental stages and is not used at the organ level.
In a combined anatomical and genetics approach, XGEbase developed a schematic model of the stage NF 35/36 Xenopus pronephric nephron based on whole mount in situ hybridization, using 91 pronephric markers. These marker genes were projected onto the model to define segments (22). Anatomical terms were used to name those segments and to annotate the gene expression patterns within the pronephric nephron (23). For non-renal tissues the expression patterns were annotated using the Xenopus Anatomy Ontology, which includes the relations ‘part of’, ‘develops from’, ‘preceded by’, ‘start stage’ and ‘end stage’. The ‘preceded by’ relation was used only to link the different developmental stages and not at the organ level.
Eight out of 11 atlases use an ontology in which relationships are defined. The other three; GENSAT, GEISHA and GenePaint.org are based on vocabularies without specified relations. On the website of GENSAT an unstructured list of nervous system structures and cell types are available. GenePaint.org uses a controlled vocabulary for the E14.5 embryo (15). This vocabulary contains 96 structures of which 70 are unique ‘leaves’ meaning that they are not further divided into substructures; most of the terms used are suggested by the Mouse Atlas Project (10,14). Similarly, since a comprehensive chicken anatomical ontology is not available (3), the GEISHA team generated a controlled anatomical vocabulary based on a mouse atlas (4).
Spatial annotation
Gene expression patterns can also be annotated by their spatial location in addition to the annotation using anatomical terms. This is done by mapping the gene expression images into a common reference space which is defined by reference models representing the embryonic morphology at the different stages of development. Currently, the only developmental gene expression atlas using spatial annotation is EMAGE. This spatial annotation is done by using reference models which cover Theiler Stages (TS) 7–20. Of those reference models, TS 7–14 and 20 are anatomically annotated (11). The reference models are 3D reconstructions made from images of stained histological sections from either wax-embedded or plastic-embedded specimens. The acquired digital images have a cellular resolution of 4x4x7 and 2x2x2 microns per voxel, respectively (12). These reference models are used to spatially annotate images of sectioned and whole-mount stained embryos, using the 3D space or a 2D projection of the reference models, respectively. Recently, reference models ranging from TS 15 to TS 19 have been linked to each other with anchor points. These anchor point are used to represent morphological positions that are roughly similar between pairs of reference models.
The other atlases do not apply spatial annotation. However, MEPD is developing a 3D framework which will be used as a 2D and 3D expression annotation tool (30). GenePaint.org, EuReGeneDb and XGEbase provide series of anatomically annotated reference image(s) of the whole embryo, the kidney and the pronephric nephron, respectively. In EuReGeneDb and XGEbase the different structures in these images are interactively linked to the vocabulary, facilitating searches for the genes expressed in those structures.
Input of expression information
The discussed atlases started either as (part of) a project of one or more laboratories to make their own gene expression data available for the outside world in a practical way (MEPD, GenePaint.org, GENSAT, GUDMAP, EURExpress, EureGeneDB and XGEBase) or as community recourse (GEISHA, EMAGE, GXD and ZFIN) with the aim to collect and make available all kinds of gene expression information. The latter atlases and MEPD accept data from external parties or they extract information from literature (Table 3). The most important aspect of input of gene expression information is the anatomical annotation of the gene expression pattern.
Table 3.
Data input
| Name | Curation by expert | Accepts external data | Includes literature | Invites suggestions |
|---|---|---|---|---|
| GEISHA | y | na | y | y |
| MEPD | – | y | n | – |
| EMAGE | y | y | y | – |
| GenePaint.org | yb | n | n | y |
| GENSAT | – | n | n | – |
| GUDMAP | y | n | n | y |
| GXD | y | y | y | – |
| EURExpress | – | n | n | – |
| EuReGeneDb | – | n | n | – |
| XGEbase | – | n | n | – |
| ZFIN | y | y | y | – |
Information on data input. Input modalities are coded as yes (y), no (n) and unknown (–). The ‘invites suggestions’ column indicates whether the atlas will consider suggestions on which genes, or probe sequences, to include in the atlas.
aA submission webpage is under construction.
bIndepently confirmed by a second annotator.
Entries
Five atlases, GEISHA, EMAGE, GXD, GenePaint.org, ZFIN and GUDMAP, mention that specialized curators or annotators are involved in the entry of new images (Table 3) (5,13,15,18,20,31). In Genepaint.org anatomical annotations are even independently confirmed by a second expert (15). In EMAGE the anatomical annotation of the section can be automatically generated when spatial mapping is performed into the 3D space of an anatomically annotated reference model (11). The partners of the GUDMAP consortium monthly submit their data to the editorial office, which curates these data and makes them available within the database (18).
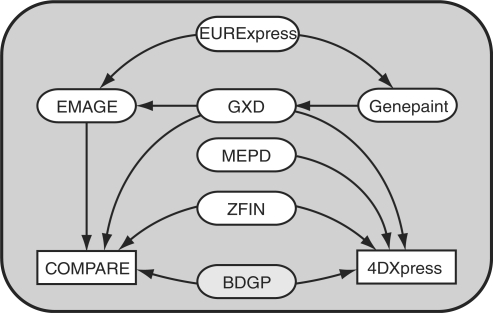
The atlases EMAGE, GXD, MEPD and ZFIN accept data from other than directly related parties (Table 3) (5,6,13,32). In EMAGE submitted data becomes publicly available only when the original paper containing these data is published (13). GEISHA, EMAGE, GXD and ZFIN incorporate information from literature in their databases by manually annotating the published images (Table 3). As shown in Figure 2 data are also exchanged between atlases.
Figure 2.
Data exchange. The arrows indicate the direction of data exchange between the different reviewed vertebrate atlases (rounded white cells). Only atlases that exchange data are included. The BDGP gene expression atlas (grey cell) holds expression data for Drosophila (http://www.fruitfly.org/cgi-bin/ex/insitu.pl). The rectangular cells are websites that combine the expression information of more than one species by importing data from the discussed atlases.
To facilitate contribution of data by external users, several atlases developed publically available software. EMAGE uses MAPaint software for spatial mapping. For EURExpress the application FIATAS (Fast Image AnnoTAtion Software) is used to anatomically annotate the expression patterns. GXD developed the application GEN (Gene Expression Notebook) to store information concerning the gene expression data and submit them to the GXD (20).
Data quality
For the users of an atlas it is important to find information on the quality of the data they retrieve. Atlases based on first party material, have protocols for data creation. The GENSAT website provides protocols for the generation of reporter mice and for the applied histological procedures. Similarly, all standard operating procedures are available on the GUDMAP website. The GEISHA website gives the protocols for their high throughput project. For XGEbase the information on how experiments were performed is described in a publication (23).
GenePaint.org does not provide procedures but states that: ‘At all stages of data production, quality control steps are implemented. This includes sequence validation of templates, tissue quality assessment and inclusion of positive and negative controls for each ISH experiment’ (14). EURExpress indicates to use the same procedures as GenePaint.org.
For literature-curated and external party submissions the atlases use other quality checks. All data in GUDMAP is curated by their editorial office (18). ZFIN uses experts for curation of literature data (5). Unpublished data is carefully curated by GEISHA, before it is published on the website (31).
EMAGE gives specific information on data quality for every image on its website; curators scored both the quality of the images as well as the morphological similarity between the data specimen and the 3D reference model after spatial mapping; a three level ranking system (‘good’, ‘moderate’ and ‘poor’) is used for both parameters (13).
It is not clear from the available information what kind of quality controls the GXD, EuReGeneDb and MEPD have implemented.
Information retrieval
For a user of an atlas it is important to know what kind of information can be found, how complete the database is, and which search options are available. In other words: can I ask the question I have and is the answer to it available?
Database contents
As indicators of database completeness the number of genes or developmental stages contained in every atlas can be used. These indicators give only a rough indication because the number of structures annotated and the number of developmental stages varie per gene. As shown in Table 2, both the number of genes and the covered developmental stages vary widely between the atlases. Note that some atlases contain, besides annotated data, also data which have not yet been annotated. ZFIN harbours the largest collection of genes while the full spectrum of developmental stages is covered: for over 10 000 different genes an anatomical annotation is available.
Search possibilities
The usefulness of a gene expression atlas depends strongly on the way in which data can be retrieved. An overview of the search possibilities is given in Table 4. All atlases offer the basic browse and query options on anatomical structure, gene and developmental stage. In most atlases gene names are extended with, or even linked to, GenBank, Ensembl, NCBI or Gene Ontology entries. Genepaint.org, EURExpress and GXD also enable the search for a probe sequence, which through a BLAST search, enables a search for database entries based on homologous sequences. Similarly GUDMAP and GXD offer a search on gene function through a link with Gene Ontology. Several search items are available in a subset of atlases: in GXD and ZFIN the user can search for literature, in ZFIN, GUDMAP, GXD and GENSAT the type of assay used to determine the expression pattern can be queried and gene expression strength and type are search options in MEPD and GENSAT.
Table 4.
Search possibilities
| Name | Gene | Dev stage | Location | Assay type | Expression type | Pattern similarity | Gene homologs/ paralogs | Sequence | Genotype | SQL acces | Gene function | Author | Spatial location |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEISHA | + | + | + | − | − | − | − | − | − | − | − | + | − |
| MEPD | + | + | + | − | +a | − | +b | + | − | − | − | − | − |
| EMAP/EMAGE | + | + | + | − | − | + | − | − | + | + | − | − | + |
| GenePaint.org | + | + | + | − | − | − | +b | + | −c | − | − | − | − |
| GENSAT | + | + | +d | + | + | − | − | − | − | − | − | − | − |
| GUDMAP | + | + | + | + | + | − | − | − | − | − | +e | − | − |
| GXD | + | + | + | + | − | − | + | + | + | + | +e | + | +f |
| EURExpress | + | − | + | − | − | − | − | + | − | − | − | − | − |
| EuReGeneDb | + | − | + | − | − | − | − | − | − | − | − | − | +f |
| XGEbase | + | + | + | − | − | − | − | − | − | − | − | − | +f |
| ZFIN | + | + | + | + | − | − | − | − | + | − | − | + | − |
The search possibilities offered by the atlases are coded as present (+) or absent (−).
aStrength and pattern.
bVia sequence/BLAST search.
cOnly on strain.
dAlso celltype.
eVia GO annotation.
fVia annotated image.
EMAGE offers the very interesting and unique option to search for genes with similar expression patterns, which results in hierarchical trees of genes. EMAGE also enables spatial queries which can be composed by painting an arbitrary region, either onto a lateral whole mount view or into the 3D space of a reference model (11). Recently, these searches are extended with the possibility to search simultaneously within different developmental stages.
Implementation of the search functions varies per atlas. Most atlases provide search options by text queries and/or by browsing a hierarchical tree of anatomical terms. To help the user, the text queries can be composed by using a query generator disguised as a form on the web interface or by clicking on an anatomical location in a model image. The latter is implemented in XGEbase and EureGeneDb.
The ultimate form of search freedom is direct (SQL) access to the underlying database, which is provided by some atlases on request (GXD, EMAGE). This form of data access is only an option for expert users, because it requires programming skills and knowledge of the database architecture.
Related work
Some databases that did not meet our inclusion criteria contain aspects that are valuable for the future extensions of gene expression atlases of vertebrate development, which will be discussed below.
The Berkeley Drosophila Transcription Network Project (BDTNP) created a point-cloud reference model representing the nuclei that make up a Drosophila embryo during the 50 min before gastrulation (33). This reference model is based on confocal images with cells labelled for two genes: even-skipped (eve) and fushi tarazu (ftz). The expression patterns of these genes serve to register other embryos to the reference model. By staining for one of the reference genes, and a gene-of-interest with an unknown expression pattern the gene expression pattern can be mapped to the reference model. This can be done at a cellular resolution, because at this stage of development the Drosophila embryo consists of a constant number of cells (34).
The Allen Brain atlas uses a fully anatomically annotated 3D reference model of the adult mouse brain which serves as spatial framework to map images of gene expression patterns (24). For each defined area within this reference framework statistics such as the average expression intensity and number of expressing cells are determined (24). The developers of the Allen Brain atlas are currently developing the ‘Developing Mouse Brain Atlas’ (http://developingmouse.brain-map.org/). When available, this will be an atlas that offers extensive possibilities as it will be based on the framework of the Allen Brain atlas.
COMPARE (35) and 4DXpress (36) are databases that combine the expression information of more than one species by importing gene expression data from the gene expression atlases that were already set up for these model organisms (Figure 2). Both databases include expression information of the mouse, zebrafish and Drosophila; 4DXpress also includes information from medaka (Figure 2). Both databases link between species using gene homologs and orthologs. 4DXpress also provides links between developmental stages, though at a limited resolution (36).
DISCUSSION
This inventory of gene expression atlases reveals that a number of atlas projects is thriving and is offering data on the in situ expression patterns of an increasing number of genes and developmental stages for all commonly studied species. However, despite the availability of this wealth of information, the number of references to these atlases in papers on the genetic regulation of development is limited, indicating an underutilization by developmental biologists. For example, an extensive search (without time constraint) for references to GenePaint.org resulted in only 37 articles that presented biological results. In contrast with this, the search: ‘gene expression’ AND ‘mouse’ AND ‘development’ resulted in over 4000 hits for articles published in 2008 alone. It may be argued that people tend to cite the original research papers, instead of the databases. However, in the case of GenePaint.org, only original data is included in the atlas, forcing the researcher to cite the database itself. Moreover, micro-array databases are frequently cited; for instance NCBI GEO is already cited over 5000 times (37).
It seems that most papers that show images to illustrate the localization of the expression of the described genes ignore the available in situ atlases as source for such images. One reason might be that in situ experiments are relatively easy and inexpensive to replicate. Although no conclusive information is available on the reasons for this disregard, the above inventory may point to some ways to improve the visibility of in situ atlases in literature, and by that fully exploit the potential of the atlases. These aspects mainly concern the framework used to annotate the expression patterns, the relations between the anatomical terms used in the annotation, and, to a lesser extent, the third party contributions to the data content, the data quality and the information retrieval.
Frameworks
To facilitate the use of in situ gene expression atlases for non-morphologists, the best framework combines an anatomical and a spatial annotation. There are three major drawbacks of the use of only an anatomical annotation; (i) manual annotation is very laborious, (ii) the exact borders of developing structures are often controversial or poorly defined (17) and (iii) the delineation of gene expression patterns does not necessarily agree with borders of known anatomical structures (8,38). However, although anatomical annotation may be far from perfect, it is hard to imagine how one can efficiently communicate about gene expression without some kind of anatomical annotation.
The laboriousness of anatomical annotation can be tackled by the use of annotated 3D reference models similar to the spatial frameworks implemented in EMAGE (12). Of course the 3D anatomical annotation of reference models is also a laborious task. However, this process has to be done only once after which the anatomical annotation of gene expression images can be instantly and automatically derived from their position in the annotated spatial reference model.
Image registration techniques that automate this spatial mapping process would make the spatial mapping even more practically applicable. There is still a lot of data available in the EMAGE database which is not yet mapped, reflecting the current practical problem of spatial mapping. In the BDTNP atlas, mapping is partially automated (33) using marker genes to facilitate the fitting into the reference model (34). The big challenge of this automated approach is to overcome the significant biological variation in embryo morphology. Especially, the temporal variation in organ development hampers the mapping of sections to 3D reference models of whole embryos. Mapping gene expression data from mutant embryos will pose additional challenges because their morphology can be very different from that of wild-type embryos.
There are of course several other problems that can make it more difficult to obtain a successful mapping. First, development is a continuous process but there will necessarily be only a limited number of time points available within a series of 3D reference models. Additionally, there will be technical variation between tissue samples, such as differences in background staining, tissue shrinkage and deformation. The last three problems will be more prominent for atlases that include data from different external resources. Moreover, techniques to register 2D images into 3D models which are needed to map arbitrary single sections into 3D reference models are still in their infancy.
We currently try to circumvent the biological variation in embryo morphology by focussing on gene expression in a specific organ instead of the whole embryo (39). However, a danger when focussing on an individual organ is that the area studied can be too narrow to cover the full development of that organ. For example, the main source for the growth of the early embryonic heart tube is the addition of cells from the pericardial mesoderm, which is not part of the heart (40). Therefore gene expression in tissues surrounding the developing organ should be included when all genes involved in its development are to be considered.
The additional complication with anatomical annotation is the controversy that often surrounds anatomical nomenclature. These controversies are often longstanding, deep and very complex, and range from completely different names for the same structure to structures with completely different names and borders. An example of such a controversy in heart research is the designation of the sinus venosus, of which the very existence is even discussed in birds and mammals (41–43). Such controversies are not limited to heart development as Little stated that ‘in research in kidney development common terms are in use without clear histological boundaries’ (17). These controversies can be, at least partially, solved by using annotated reference models. Obviously, it will be very difficult to obtain clear descriptions of gene expression patterns when ambiguous anatomical terms are used. Since the meaning of terms used in anatomically annotated reference models will be clarified by their 3D spatial depiction in the models, it will be evident from the reference models what the defined borders of named structures are, even for controversial structures or borders. This will remove most of the ambiguity in the terms used in anatomical annotations.
A final complicating aspect of using anatomical annotation is that gene expression patterns do not respect borders of anatomical structures. This problem can be addressed by combining the anatomical annotation of a spatial framework with an annotation based on the expression profiles of a limited set of genes with known expression domains. To describe the ‘gene of interest’ more accurately, the anatomical annotation can thus be extended with a biologically more solid basis. In this ‘genetic annotation’ approach, the names of the resulting compartments are partially based on traditional anatomical names and are further specified with the combined gene expression domains. This approach is used in XGEbase, where gene expression domains of marker genes are used to distinguish different segments in the pronephric nephron (22).
Ontologies
Ideally, an ontology with sufficient resolution for anatomical annotation is used. Basically, this resolution has two aspects. First, atlases differ with respect to the detail of the terminology used to describe the anatomical structures. In general one can say that atlases focusing on a specific organ (system) typically have a more detailed annotation than those covering whole embryos. Secondly, most atlases have a rather limited resolution with respect to the number and detail of the encoded relationships between structures. The use of ontologies with sufficient resolution has two major advantages to the use of a more simple annotation; (i) an ontology with a sufficient resolution on anatomical terms enables easier handling of variation of gene expression within anatomical structures, while (ii) sufficient relational resolution can reveal information not explicitly included in the atlases. The power of the atlases can be even further enhanced when the information within the atlases is combined, by establishing links between them via their ontologies.
A high anatomical resolution gives fewer problems with non-overlapping gene expression domains within a structure. This is because genes do not respect conventional borders and may be differently expressed at different locations within the same anatomical structure. With a higher resolution such differences can be better discriminated and described. Of course, this increase in anatomical resolution is limited by the biological variation in embryonic development.
A high relational resolution can encode valuable information for developmental biology and it can facilitate the automatic derivation of a potential large body of implicit knowledge contained in an atlas. To this end, ontologies should, next to the commonly implemented ‘is a’ and ‘part of’ relations, be extended with the use of ‘develops from’ and ‘gives rise to’ relations which can link anatomical structures within and between developmental stages at a (sub) organ level.
The ‘is a’ and ‘part of’ relations already offer important functionality used by the query systems of most of the atlases. When one wants to search for genes involved in artery development it would be a hassle to have to perform a search for every individual artery to find out which genes are expressed in arteries. The ‘is a’ relation enables the query system to automatically derive all arteries, and therefore enables the user to search for all genes expressed within all arteries at once. Similarly, when one wants to study genes involved in kidney development, one does not want to query for each part of the kidney separately to find all genes expressed in the kidney. With the ‘part of’ relation one could simply query the kidney and obtain results from all parts of the kidney.
Extension with the use of ‘develops from’ and ‘gives rise to’ relations can even give possibilities similar to the spatial links in EMAGE. Relations between structures and stages can then reveal relations between genes which can be the basis for new hypothesis on regulatory networks in time and space. The ‘gives rise to’ relationship also makes it possible to link gene expression in a structure at a certain stage in development to structures that do not exist. This would enable the identification of genes thus far unknown to be involved in the regulation of the development of these structures.
Links between atlases can complete the information within them, and knowledge on different species can be combined. To easily establish these links, compatible vocabularies or ontologies should be used. Currently a substantial part of the mouse atlases are compatible; the GXD uses the ontology of EMAGE whereas GUDMAP and EuReGeneDb use a higher resolution extension of the same ontology. However, there are still a large number of different vocabularies used in the other discussed atlases.
Though ontologies exist for each species, these ontologies have not yet been extensively explored across species. Currently such relations are being established within 4DXpress by mapping between the developmental stages of Drosophila, medeka, zebrafish and mouse (36). The resolution of this initial mapping is rather low, because the number of developmental stages in this mapping is limited to eight (36). As a result, the most important stages in organ development are pooled into only three stages: gastrulation, neurulation and organogenesis. The low temporal resolution of this link between species makes meaningful comparisons between gene expression patterns during these phases of development very difficult. To avoid the reduction of classes (in this case developmental stages) resulting from the use of one common ontology, one could consider to map the individual species ontologies onto each other or a more abstract ontology of a class of species such as vertebrates.
Mapping species that are closely related makes it possible to maintain a high resolution which is not possible when the species are evolutionary further apart. It is obvious that mapping between two fish species such as zebrafish and medaka could be performed with a higher resolution than the mapping of the ontologies of Drosophila and mouse. However, any cross-species mapping of ontologies will be a great challenge. The controversies that already surround the anatomical nomenclature within one species, will feature more prominently when different species are to be combined. Moreover, organ development may not be synchronous even within the same class of species, and in the worst case some structures might not even be present in every species to be mapped.
Handling input to preserve data quality
An essential prerequisite of gene expression atlases is that sufficient information is provided to judge the quality of the data. Two important aspects of the quality of the data deserve attention: (i) the way the expression patterns of genes are collected, and (ii) how and by whom the anatomical annotation is performed. It has added value when all data are collected using the same quality standards.
In most atlases the quality of first party data is validated. However, in the decision to include data from external parties, one has to weigh the number of included genes and/or development stages against the possible loss of data quality. Because of the diversity of ways to visualize in situ gene expression, the quality of external data varies by definition. When these data are included into the atlas by literature curation the data quality in the atlas can still be controlled although the curator is dependent on the information given in the source papers. When data are directly added by an external researcher data quality may no longer be guaranteed.
This uncertainty about the quality of the data in in situ gene expression atlases may be one of the reasons for their low citation rate. However, when the external submissions include standardized information on the performed experiments this would allow others to judge the quality of the presented data. For micro-array repositories, the MIAME standard describes the minimum information required to ensure that the microarray data can be easily interpreted and that their analysis can be independently verified (44). Recently a similar specification for in situ experiments was developed: the minimum information specification for in situ hybridization and immunohistochemistry [MISFISHIE (45)], The GXD, EMAGE and ZFIN databases are already able to archive the information in this way (45). Although, collecting all information is probably not a realistic goal for all data entries through literature curation, atlases should at least ensure that the user can judge whether the appropriate internal and external positive and negative controls have been used.
Another item that needs to be controlled to preserve data quality is the anatomical annotation of third party data. Anatomical annotation can be controlled by mapping the input images onto a common 3D reference model. An additional benefit of mapping onto a reference model would be that it allows direct comparison of different entries of the same gene. For example, in the BDTNP, the expression of each gene is measured in several embryos, mapped onto a reference model and averaged, resulting in more reliable data (33).
Although data which are not technically and anatomically standardized are not suited to form the basis for final conclusions, they can still be used to formulate hypotheses. Therefore, in the query results a distinction should be made between data of known and unknown quality in the atlas.
Information retrieval
Retrieval of the information from the databases is crucial for the users of an atlas. Apart from the standard search possibilities, some atlases already provide more advanced options such as searches on: genes having similar expression patterns, gene homologs and paralogs, sequence and genotype. However, to fully exploit the information contained in the atlases, atlases should enable the search for associations between genes. Such possibilities now become available within EMAGE, where genes with a similar spatial expression patterns have been clustered at a specified time point (13).
An inverse experiment is performed for the developing chicken limb where spatial domains where found based on the expression pattern of 7 different genes (46). Similarly, data on 1030 genes contained in GenePaint.org were clustered based on their anatomical annotation revealing genes involved in a regulatory network in the brain (15). This shows that new functions of genes can be discovered when atlases offer queries based on computational analysis such as clustering of genes with similar (spatial) expression patterns.
Equally important as adding new genes is that the atlases can be complemented with data of lineage, proliferation and other important cell biological processes. Together these morphological integrated expression and developmental data provide new insights into the understanding of morphogenetic processes.
Finally, to encourage the use of gene expression atlases and to make efficient use of the continuous inclusion of new data, researchers should be able to store their specified queries and be alerted when new information is available. Such an alert system can be similar to those offered by for instance Pubmed and Scopus.
CONCLUSION
More than a decade of developmental gene expression atlases has led to a number of thriving atlases that contain a wealth of publicly available information. However, in our opinion, the ideal atlas does not yet exist, which could be an explanation for the limited number of biomedical papers citing atlases.
This review shows that most ingredients needed to develop the ideal atlas are already applied to some extent in at least one of the discussed atlases. The ideal atlas should be based on a spatial framework, i.e. a series of 3D reference models, that are anatomically annotated using an ontology with sufficient resolution, both for relations as well as for anatomical terms. Relations should at least be used to link (sub) organs through time and to define ‘part of’ relations. The resolution of the anatomical terms should be high enough to cope with variation of gene expression within anatomical structures. To define structures with poorly described or disputed borders, genetic annotation should be used to extend the ontology. Anatomical annotation of expression data can be automated when these data are mapped into these reference models. Moreover, collecting all gene expression data into a common 3D reference model allows the study of gene interactions using computational analysis.
To fully exploit the stored data, atlases, including atlases of different species, need to be linked to each other. The required gene orthologies are already available. One of the main challenges for the future will be the development of a mapping between (anatomical) ontologies of different species.
When an atlas uses an ontology with sufficient spatial and temporal resolution, implicit knowledge within the atlas can be disclosed. As shown by experiments in several atlases, analysis of the data contained in the atlases can reveal new spatial domains or identify genes involved in a regulatory network. The implementation of these analysis options would convert the atlas from just an image library into a hypothesis-generating and -testing system.
FUNDING
EU seventh framework program as part of the project CHeartED [HEALTH-F2-2008-223040]. Funding for open access charge: Academic Medical Centre, Amsterdam.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Arie Hasman and Maurice van den Hoff for their valuable suggestions after critically reading the manuscript and Alexandre Soufan for discussions about the 3D gene expression analysis. The embryos in Figure 1 were reconstructed from high resolution image stacks provided by Timothy Mohun.
REFERENCES
- 1.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, Holloway E, Kolesnykov N, Lilja P, Lukk M, et al. ArrayExpress – a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35:D747–D750. doi: 10.1093/nar/gkl995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell GW, Yatskievych TA, Antin PB. GEISHA, a whole-mount in situ hybridization gene expression screen in chicken embryos. Dev. Dyn. 2004;229:677–687. doi: 10.1002/dvdy.10503. [DOI] [PubMed] [Google Scholar]
- 4.Antin PB, Kaur S, Stanislaw S, Davey S, Konieczka JH, Yatskievych TA, Darnell DK. Gallus expression in situ hybridization analysis: a chicken embryo gene expression database. Poult. Sci. 2007;86:1472–1477. doi: 10.1093/ps/86.7.1472. [DOI] [PubMed] [Google Scholar]
- 5.Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrich T, Ramialison M, Quiring R, Wittbrodt B, Furutani-Seiki M, Wittbrodt J, Kondoh H. MEPD: a Medaka gene expression pattern database. Nucleic Acids Res. 2003;31:72–74. doi: 10.1093/nar/gkg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ringwald M, Baldock RA, Bard JBL, Kaufman MH, Eppig JT, Richardson JE, Nadeau JH, Davidson DR. A database for mouse development. Science. 1994;265:2033–2034. doi: 10.1126/science.8091224. [DOI] [PubMed] [Google Scholar]
- 8.Davidson DR, Bard JBL, Brune R, Burger A, Dubreuil C, Hill W, Kaufman MH, Quinn J, Stark M, Baldock R. The mouse atlas and graphical gene-expression database. Semin. Cell Dev. Biol. 1997;8:509–517. doi: 10.1006/scdb.1997.0174. [DOI] [PubMed] [Google Scholar]
- 9.Burger A, Davidson DR, Baldock RA. Formalization of mouse embryo anatomy. Bioinformatics. 2004;20:259–267. doi: 10.1093/bioinformatics/btg400. [DOI] [PubMed] [Google Scholar]
- 10.Bard JBL, Kaufman MH, Dubreuil C, Brune RM, Burger A, Baldock RA, Davidson DR. An internet-accessible database of mouse developmental anatomy based on a systematic nomenclature. Mech. Dev. 1998;74:111–120. doi: 10.1016/s0925-4773(98)00069-0. [DOI] [PubMed] [Google Scholar]
- 11.Christiansen JH, Yang Y, Venkataraman S, Richardson L, Stevenson P, Burton N, Baldock RA, Davidson DR. EMAGE: a spatial database of gene expression patterns during mouse embryo development. Nucleic Acids Res. 2006;34:D637–D641. doi: 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldock RA, Bard JBL, Burger A, Burton N, Christiansen J, Feng G, Hill B, Houghton D, Kaufman MH, Rao J, et al. EMAP and EMAGE: a framework for understanding spatially organized data. Neuroinformatics. 2003;1:309–325. doi: 10.1385/NI:1:4:309. [DOI] [PubMed] [Google Scholar]
- 13.Venkataraman S, Stevenson P, Yang Y, Richardson L, Burton N, Perry TP, Smith P, Baldock RA, Davidson DR, Christiansen JH. EMAGE – Edinburgh Mouse Atlas of Gene Expression: 2008 update. Nucleic Acids Res. 2008;36:D860–D865. doi: 10.1093/nar/gkm938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visel A, Thaller C, Eichele G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32:D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visel A, Carson J, Oldekamp J, Warnecke M, Jakubcakuva V, Zhou X, Shaw CA, varez-Bolado G, Eichele G. Regulatory pathways analysis by high-throughput in situ hybridization. PLoS Genetics. 2007;3:1867–1883. doi: 10.1371/journal.pgen.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 17.Little MH, Brennan J, Georgas K, Davies JA, Davidson DR, Baldock RA, Beverdam A, Bertram JF, Capel B, Chiu HS, et al. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns. 2007;7:680–699. doi: 10.1016/j.modgep.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, et al. GUDMAP: the genitor-urinary developmental molecular anatomy project. J. Am. Soc. Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 19.Ringwald M, Davis GL, Smith AG, Trepanier LE, Begley DA, Richardson JE, Eppig JT. The mouse gene expression database GXD. Semin. Cell Dev. Biol. 1997;8:489–497. doi: 10.1006/scdb.1997.0177. [DOI] [PubMed] [Google Scholar]
- 20.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse gene expression database (GXD): 2007 update. Nucleic Acids Res. 2007;35:D618–D623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayamizu TF, Mangan M, Corradi JP, Kadin JA, Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reggiani L, Raciti D, Airik R, Kispert A, Brandli AW. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007;21:2358–2370. doi: 10.1101/gad.450707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raciti D, Reggiani L, Geffers L, Jiang Q, Bacchion F, Subrizi AE, Clements D, Tindal C, Davidson DR, Kaissling B, et al. Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 2008;9:R84. doi: 10.1186/gb-2008-9-5-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 25.Neidhardt L, Gasca S, Wertz K, Obermayr F, Worpenberg S, Lehrach H, Herrmann BG. Large-scale screen for genes controlling mammalian embryogenesis, using high-throughput gene expression analysis in mouse embryos. Mech. Dev. 2000;98:77–94. doi: 10.1016/s0925-4773(00)00453-6. [DOI] [PubMed] [Google Scholar]
- 26.Henrich T, Wittbrodt J. An in situ hybridization screen for the rapid isolation of differentially expressed genes. Dev. Genes Evol. 2000;210:28–33. doi: 10.1007/pl00008185. [DOI] [PubMed] [Google Scholar]
- 27.Gruber TR. Toward principles for the design of ontologies used for knowledge sharing. Intl. J. Human - Computer Studies. 1995;43:907–928. [Google Scholar]
- 28.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theiler K. The House Mouse, Development and Normal Stages from Fertilization to 4 Weeks of Age. Berlin: Springer-Verlag; 1972. [Google Scholar]
- 30.Henrich T, Ramialison M, Wittbrodt B, Assouline B, Bourrat F, Berger A, Himmelbauer H, Sasaki T, Shimizu N, Westerfield M, et al. MEPD: a resource for medaka gene expression patterns. Bioinformatics. 2005;21:3195–3197. doi: 10.1093/bioinformatics/bti478. [DOI] [PubMed] [Google Scholar]
- 31.Darnell DK, Kaur S, Stanislaw S, Davey S, Konieczka JH, Yatskievych TA, Antin PB. GEISHA: an in situ hybridization gene expression resource for the chicken embryo. Cytogenet. Genome Res. 2007;117:30–35. doi: 10.1159/000103162. [DOI] [PubMed] [Google Scholar]
- 32.Ringwald M, Eppig JT, Kadin JA, Richardson JE. GXD: a Gene Expression Database for the laboratory mouse: current status and recent enhancements. The Gene Expresison Database group. Nucleic Acids Res. 2000;28:115–119. doi: 10.1093/nar/28.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fowlkes CC, Hendriks CL, Keranen SV, Weber GH, Rubel O, Huang MY, Chatoor S, DePace AH, Simirenko L, Henriquez C, et al. A quantitative spatio-temporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 34.Luengo Hendriks CL, Keranen SVE, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: data acquisition pipeline. Genome Biol. 2006;7:R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salgado D, Gimenez G, Coulier F, Marcelle C. COMPARE, a multi-organism system for cross-species data comparison and transfer of information. Bioinformatics. 2008;24:447–449. doi: 10.1093/bioinformatics/btm599. [DOI] [PubMed] [Google Scholar]
- 36.Haudry Y, Berube H, Letunic I, Weeber PD, Gagneur J, Girardot C, Kapushesky M, Arendt D, Bork P, Brazma A, et al. 4DXpress: a database for cross-species expression pattern comparisons. Nucleic Acids Res. 2008;36:D847–D853. doi: 10.1093/nar/gkm797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savolainen SM, Foley JF, Elmore SA. Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol. Pathol. 2009;37:395–414. doi: 10.1177/0192623309335060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verbeek FJ, Lawson KA, Bard JB. Developmental bioinformatics: linking genetic data to virtual embryos. Int. J Dev. Biol. 1999;43:761–771. [PubMed] [Google Scholar]
- 39.de Boer BA, Ruijter JM, Voorbaak FPJM. Towards the automatic registration of histological sections into a 3D reference model. In: Dastani MM, de Jong E, editors. BNAIC'0;7. Proceedings of the 19th Belgian-Dutch Conference on Artificial Intelligence, Utrecht: 5–6 November 2007. Vol. X. 2007. pp. 41–48. [Google Scholar]
- 40.van den Berg G, Abu-Issa R, de Boer BA, Hutson MR, de Boer PA, Soufan AT, Ruijter JM, Kirby ML, van den Hoff MJ, Moorman AF. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ. Res. 2009;104:179–188. doi: 10.1161/CIRCRESAHA.108.185843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soufan AT, van den Hoff MJB, Ruijter JM, de Boer PAJ, Hagoort J, Webb S, Anderson RH, Moorman AFM. Reconstruction of the patterns of gene expression in the developing mouse heart reveals an architectural arrangement that facilitates the understanding of atrial malformations and arrhythmias. Circ. Res. 2004;95:1207–1215. doi: 10.1161/01.RES.0000150852.04747.e1. [DOI] [PubMed] [Google Scholar]
- 42.Tasaka H, Krug EL, Markwald RR. Origin of the pulmonary venous orifice in the mouse and its relation to the morphogenesis of the sinus venosus, extracardiac mesenchyme (spina vestibuli), and atrium. Anat. Rec. 1996;246:107–113. doi: 10.1002/(SICI)1097-0185(199609)246:1<107::AID-AR12>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 43.Webb S, Brown NA, Anderson RH, Richardson MK. Relationship in the chick of the developing pulmonary vein to the embryonic systemic venous sinus. Anat. Rec. 2000;259:67–75. doi: 10.1002/(SICI)1097-0185(20000501)259:1<67::AID-AR8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 44.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 45.Deutsch EW, Ball CA, Berman JJ, Bova GS, Brazma A, Bumgarner RE, Campbell D, Causton HC, Christiansen JH, Daian F, et al. Minimum information specification for in situ hybridization and immune-histochemistry experiments (MISFISHIE) Nat. Biotechnol. 2008;26:305–312. doi: 10.1038/nbt1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher ME, Clelland AK, Bain A, Baldock RA, Murphy P, Downie H, Tickle C, Davidson DR, Buckland RA. Integrating technologies for comparing 3D gene expression domains in the developing chick limb. Dev. Biol. 2008;317:13–23. doi: 10.1016/j.ydbio.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]