Abstract
Gene expression is regulated by combinations of transcription factors, which can be mapped to regulatory elements on a genome-wide scale using ChIP experiments. In a previous ChIP-chip study of USF1 and USF2 we found evidence also of binding of GABP, FOXA2 and HNF4a within the enriched regions. Here, we have applied ChIP-seq for these transcription factors and identified 3064 peaks of enrichment for GABP, 7266 for FOXA2 and 18783 for HNF4a. Distal elements with USF2 signal was frequently bound also by HNF4a and FOXA2. GABP peaks were found at transcription start sites, whereas 94% of FOXA2 and 90% of HNF4a peaks were located at other positions. We developed a method to accurately define TFBS within peaks, and found the predicted sites to have an elevated conservation level compared to peak centers; however the majority of bindings were not evolutionary conserved. An interaction between HNF4a and GABP was seen at TSS, with one-third of the HNF4a positive promoters being bound also by GABP, and this interaction was verified by co-immunoprecipitations.
INTRODUCTION
In each cell type the expression of genes is regulated by the action of a large number of transcription factors (TFs), but so far we have only a rudimentary knowledge of the location of the regulatory elements. Some are located upstream of genes but it is also well known that enhancers, silencers, locus control regions and boundary elements are frequent in the genome and can regulate the transcription of genes over large distances. It is also becoming clear that most genes have alternative promoters (1). Sequences that take part in gene regulation are characterized by open chromatin, and recent studies in CD4+ T-cells (2) have identified 95 000 such sites by mapping the sensitivity for DNaseI digestion. This finding is also supported by analyses in 1% of the genome which indicate that most types of cells have in the order of 100 000 sites of open chromatin (3). Which proteins that binds to these regulatory units is virtually unknown but can now be determined genome-wide in a systematic way in vivo using chromatin immunoprecipitation and high-resolution arrays (ChIP-chip) or direct sequencing of enriched fragments (ChIP-seq) (4–6).
In a recent genome-wide ChIP-chip study, we identified the binding sites for the TFs USF1 and USF2 (7) in HepG2 liver cells. In most cases USF1 and USF2 bind together at transcription start sites (TSS) but one striking finding was that sequences bound only by USF2 were mostly at distal positions and contained motif sequences for the hepatocytic nuclear factors HNF4a and FOXA2 (HNF3b). Furthermore, the recognition sequence for GABP (also called nuclear respiratory factor 2, NRF-2) was found to be overrepresented at TSS bound by the USFs. The nuclear receptor HNF4a is a major regulator of the hepatocytic phenotype and regulates genes involved in the control of lipid homeostasis (8). Mutations in HNF4a can cause maturity onset diabetes of the young (MODY1) (9), and single nucleotide polymorphisms (SNPs) in its promoter have been associated to type II diabetes (T2D) (10–12). FOXA2 has the ability to function as a pioneering factor during development by opening up compacted chromatin. It is also important for the normal function of several cell types, including the liver where it regulates the expression of genes involved in gluconeogenesis. To get a better understanding of which genes these factors regulates and to characterize the USF2-HNF distal regulatory regions we used ChIP-seq in HepG2 cells.
One inherent advantage of ChIP-seq over array based methods is the high resolution achieved by sequencing the ends of intact immunoprecipitated fragments, where ideally the random shearing around the bases bound by the TF can lead to true base pair resolution. However, in practice it is often not possible to define the exact binding sites from the aligned fragments if for example multiple binding sites for the TF are located close together or if too few fragment ends are sequenced. We developed a de novo motif finding algorithm which uses the expected enrichment of transcription factor binding sites (TFBS) in peak centers (5,13) in order to independently identify the most overrepresented motifs and thus predict the bases bound by the TFs.
We found a large overlap between the GABP and HNF4a peaks at TSS, indicating an interaction between these two TFs. We further investigated this using co-immunoprecipitations and found that these TFs are indeed in the same complexes within the cells. We also suggest that annotating the genome with ChIP-seq will help identify potential regulatory SNPs from genome-wide association studies (GWAS), since bindings of HNF4a and FOXA2 was found close to several SNPs associated to metabolic disorders.
MATERIALS AND METHODS
ChIP and sequencing
Chromatin immunoprecipitaation was performed essentially as described before (7) on 107–108 cells per reaction using antibodies sc-6554, sc-6556 and sc-13442 (Santa Cruz Biotechnology), see Supplementary Methods section for details. Sequencing was done on an Illumina 1G instrument according to the manufacturer’s protocol. For IgG and the first FOXA2 replicate libraries with insert sizes 50–150 and 250–450 bp was made, all other libraries had inserts of 100–300 bp. Antibodies for ChIP-PCR verifications were sc-101102 for NRF-1 and for GABP the monoclonal antibody sc-28312 was used. Sequences are available through MIAMExpress (http://www.ebi.ac.uk, E-MTAB-115).
Alignment and peak finding
The Eland software (Illumina) was used repeatedly to align reads with up to two mismatches against the hg18 reference genome, starting at 32 bases and shortening un-aligned reads by three bases at the 3′-end until the remaining alignments were 23 bases long. After evaluation of enrichment in aligned reads of different length we selected only reads aligning at 29 or 32 bases for the final analysis in order to minimize the number of false positive peaks introduced by misalignments. All aligned positions that were present in both the IgG and TF datasets were removed, as were all alignments within 1 Mb of the centromeric gaps. Some sequence libraries were found to have an unexpectedly high percentage of reads aligning to the same positions, indicating PCR artifacts or too deep sequencing for the library, thus for each library we collapsed reads on the same position and strand. Extended fragments of varying lengths were created by matching reads on opposing strands by identifying the nearest unmatched forward read facing each sorted reverse read, requiring a minimum of 100 bases between matched reads. If no unmatched read was found within the size limits in the library the read was allowed to extend to the maximum size in the sequencing library.
Peaks of enrichment was defined as groups of reads for which the fragments overlapped. The maximum overlap was used to define peak height, with the peak center taken as the midpoint for positions with maximum overlap. Each peak was given a mismatch score based on the average number of mismatches in the alignments contributing to the peak maximum, with the addition of one mismatch for each truncation step. Peaks were annotated with repeats by matching to the RepeatMasker table from the UCSC genome browser (14). SNPs within peaks were identified from the dbSNP database and the sequence around these positions was extracted from the aligned reads. Based on comparison with IgG peaks we removed all peaks that were either in satellite or rRNA repeats or had a mismatch score above 2.
To calculate overlaps between datasets (Table 1) we calculate the number of peaks in each dataset located within 500 bp of an entry in the other datasets. UCSC genes were used to define TSS positions, and CAGE-tag clusters were downloaded from the RIKEN database (15).
Table 1.
Number of significant peaks and overlaps between data sets
| FOXA2 | HNF4a | GABP | TSS | USF1 n = 2420 | USF2 n = 1314 | USF2-homo n = 240 | FOXA1 n = 7881 | FoxA2 n = 8677 | |
|---|---|---|---|---|---|---|---|---|---|
| FOXA2 | 7266 | 3564 | 125 | 454 | 183 | 208 | 72 | 1549 | 573 |
| HNF4a | 3546 | 18 783 | 690 | 1873 | 457 | 431 | 123 | 1051 | 880 |
| GABP | 125 | 699 | 3064 | 2609 | 162 | 125 | 17 | 31 | 108 |
Quantitative PCR validations
To define a cut-off for positive peaks we first calculate an FDR level of 10−4 based on a similar approach as used by Robertson et al. (13) (Supplementary Data), and then for each factor the enrichment of ∼20 regions with ChIP-seq signal was tested by qPCR using SYBR Green. We randomly picked peaks to cover the range of peak heights for each TF, including peaks below the FDR-based cut-off level and additionally included regions either unique or common for the different TFs. New ChIP material was prepared and all reactions were performed in triplicates. A quantitative value was calculated from a standard curve of input dilutions. As a background, four to six regions without ChIP-seq signal were run, and the quantitative values for each region was divided by the mean +2.5 SD of the negative regions. Fold enrichments ≥1.5 were considered significant.
Motif finding and conservation of TFBS
Each k-mer located within 150 bp of peak centers was given a score based on its frequency, average distance from peak center and enrichment in a window of 100 bases centered over the peak, with positions 100–150 bp on both sides of peak centers as a background. Repeats were selected against by only considering the outermost location of each k-mer in each sequence. Motifs were constructed from k-mers with at least 3-fold enrichment, and for each enriched k-mer a motif score was calculated by taking the sum of all scores for k-mers with one or two mismatches. For FOXA2 and GABP we used 8-mers with one mismatch, and for HNF4a we allowed two mismatches in 9-mers to better capture both halves of its motif. The top scoring motif was selected and all peaks with a match within 100 bp were removed, and the procedure was repeated until <5% of the peaks had been removed. Motifs were visualized and compared to the Transfac and Jaspar databases using STAMP (16). For additional motif discovery in masked sequences a two-fold enrichment of 8-mers was used allowing one mismatch from the core motif sequence. The conservation scores for the predicted TFBS and peak centers were collected from the UCSC phastCons28placMammal dataset for the 500 highest and lowest peaks with a TFBS located within 50 bp of peak center.
Gene Ontology and expression analysis
The distribution of Gene Ontology (GO)-annotations for protein coding genes (PCG) annotated to different peak fractions were compared to the distribution of GO-annotations for all the genes from the PCG-categories. The two-sided p-value for each GO-term was calculated using a Fishers’ exact test. The p-values were corrected for multiple hypotheses testing using Benjamin and Yekutieli (17) correction. Expression data from (18) and from GSE10021 was downloaded from Gene Expression Omnibus (NCBI) to compare expression levels for bound genes and for different cell types. A distance of 1 kb was used to associate bindings with expression and GO. All GO results are available in Supplementary file 2 and at http://www.anst.uu.se/stenr451/goseq/goseq/.
Co-immunoprecipitations
Nuclear extracts from HepG2 was incubated with TF antibodies or IgG and the immunoprecipitates were analyzed by western blot (details in Supplementary Methods section).
RESULTS
Alignment errors can affect ChIP-seq datasets
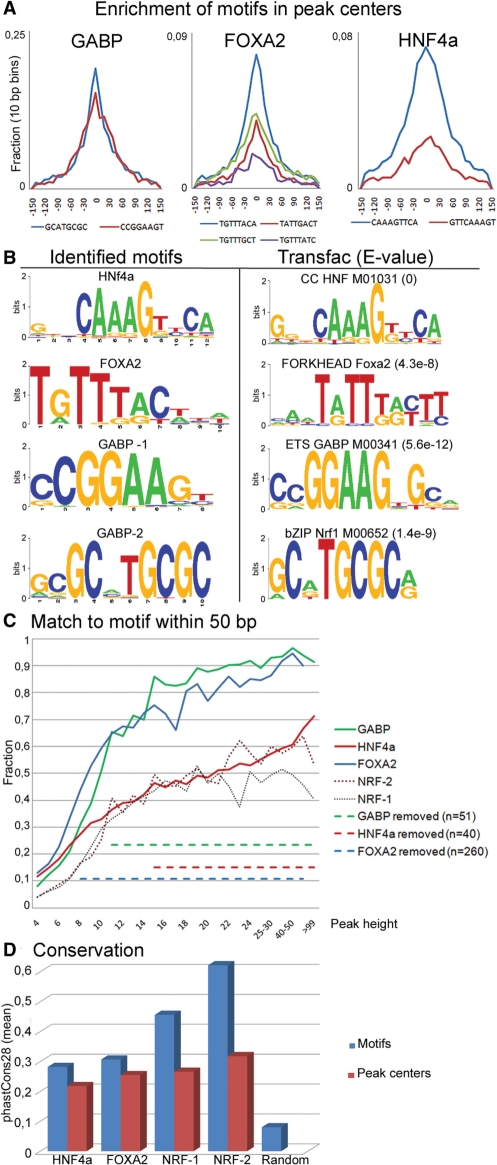
We used massively parallel sequencing to obtain ChIP-seq reads from chromatin immunoprecipitated with antibodies directed against HNF4a, FOXA2 and GABP and IgG. To maximize the number of usable reads we tested both reads with high and low quality scores for enrichment, and found that reads falling below the quality thresholds on the instrument were often correctly aligned at known binding sites. The number of uniquely aligned reads could further be increased substantially after truncation of the 3′-ends. However, we also found that the shorter alignments were more likely to be falsely placed over certain types of repeats and can thus increase the noise in ChIP-seq datasets (Supplementary Figure S1). We therefore required at most two mismatches to the reference in the first 29 bases of a read, which gave 3–7 million alignments for the different transcription factors. Although the negative control (IgG) gave fewer alignments, enrichment of the IgG alignments were seen in many locations. Most of these alignments were found to have a high number of mismatches to the reference and often to be located to satellite and rRNA repeats. We filtered the TF datasets based on this information and found that the removed peaks were not enriched for the TF motifs (Figure 1C, removed peaks).
Figure 1.
(A) Enrichment of the identified motifs in a 300 bp interval over peak centers, with the core sequences for each motif shown below the plot. (B) Sequence logos for the identified motifs and for the most similar motif from the Transfac database. (C) Fraction of peaks with at least one motif sequence within 50 bp of the center at different peak heights. Dotted lines indicate average motif content in removed peaks, starting at the significance threshold for each factor. (D) All identified motifs are significantly enriched for conserved bases compared to the centers of the same peaks.
Since the aligned reads represents the ends of enriched sonicated DNA, the TFBS are expected to be centered between reads from the forward and reverse strands. We found that using a fixed read extension to create virtual ChIP fragments, as was done in some of the first ChIP-seq studies (13), can lead to a shift in peak maxima if two binding sites are present at a short distance while other methods that scan for changes in strand preferences of reads (19) are prone to give too many peak calls in such situations (Supplementary Figure S2). We used the strand separation of read to create shorter fragments in peak centers by simulating paired-end reads within the size range defined by the sequencing libraries. This was found to give a better separation of close peaks and an estimation of peak height (overlaps) that better reflects the actual number of reads from each peak in simulated datasets.
To define a set of significantly enriched peaks we first calculated an FDR level of 10−4 at five overlaps for GABP, seven for FOXA2 and eight for HNF4a. Next, we tested enrichment around the cut-off using quantitative PCR and found a good correlation between enrichment and peak height, but several peaks for GABP and HNF4a close to the initial cut-off level were not enriched in the qPCR (Supplementary Figure S1D). Based on these results we required a minimum overlap of 11 for GABP, eight for FOXA2 and 15 for HNF4a in order to get lower false positive rates. This gave 3064 peaks for GABP, 7266 for FOXA2 and 18783 for HNF4a (Table 1). The datasets are presented as genome-wide overlap profiles to be visualized in the UCSC genome browser (14) (Supplementary Dataset 1).
De novo motif discovery in ChIP-seq datasets
A ChIP-seq experiment often leads to the identification of a very high number of enriched regions, and given the high resolution each TF target sequence can be expected to be enriched in peak centers. It is further possible that unspecific binding of the antibody or binding of a TF together with different partners could lead to enrichment of different motifs in subsets of peaks. We developed an algorithm to identify motifs which are independently enriched in peak centers, and validated our method by re-analyzing published ChIP-seq datasets from Valouev et al. (20). For the serum response factor SRF, we identified both motifs highly similar to the SRF motif as well as the ETS.1/GABP motif (Supplementary Figure S3), in concordance with the reported results from the de novo motif finding program MEME. For GABP we analyzed a larger dataset and found the expected motif in more than 12 000 peaks, which is almost twice the number of reported peaks in the original article. This shows that the addition of motif discovery to peak detection will allow identification of true binding sites also in sequences with low enrichment. Additionally, we found the method applicable also to lower resolution ChIP-chip data from the USF study (Supplementary Figure S3C and D).
We identified several variants of the expected motifs for FOXA2 and HNF4a (Supplementary Figure S4), and the combined motifs were found to be highly similar to the established motifs in the Transfac database (Figure 1B). Although HNF4a and FOXA2 in many cases are enriched in the same regions we were able to identify their individual binding sites, as exemplified in Figure 2C for the HNF1a promoter where exact matches to two previously reported (21) HNF4a sites and one FOXA2 site was found. Surprisingly, for GABP we could identify two equally enriched motifs, one that matches the expected motif for GABP/NRF-2 and one motif for the related factor NRF-1 (22). Although a large number of peaks contain both the NRF-1 and NRF-2 motif, the NRF-1 sites had a very tight distribution around peak centers and were present also in peaks without the NRF-2 motif (Figure 1A and B). Thus, this motif is likely representing the bound sequences in a subset of the GABP peaks rather than being identified by co-localization with NRF-2 motifs. We therefore divided the GABP peaks into two groups, NRF-1 and NRF-2, for further analysis.
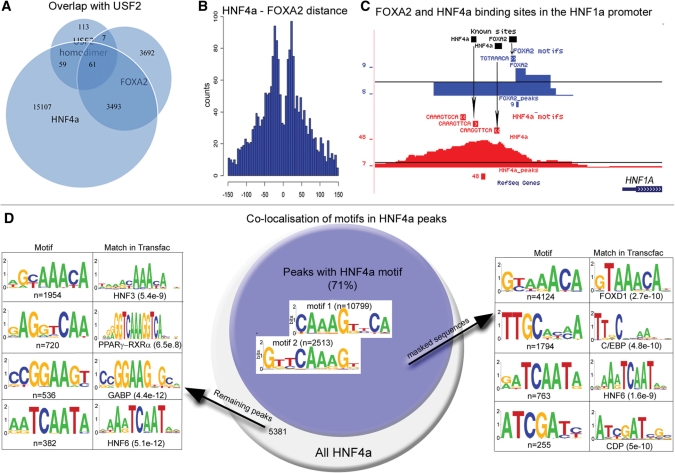
Figure 2.
Motifs in HNF4a and FOXA2 peaks. (A) Overlaps between HNF4a, FOXA2 and USF2 homodimers. (B) Distance between HNF4a and FOXA2 motifs in overlapping peaks. (C) The close location of FOXA2 and HNF4a sites are exemplified with identified motifs in the HNF1a promoter, with arrows indicating the positions of three sites taken from the literature. (D) Identification of co-localized motifs in HNF4a peaks. Peaks with a match within 100 bp to any of the two 9-bp motifs (blue circle, numbers indicate the number of peaks with the motifs) were reanalyzed after motif masking and found to contain HNF6, C/EBP, CDP and forkhead motifs (right panel). Reanalysis of the 5381 peaks with no match to the HNF4a using less stringent settings revealed motifs for HNF3, HNF6 and GABP (left panel).
Co-identified motifs in peaks correspond to in vivo binding
For HNF4a a relatively large number of peaks (29%) remained without a match to the motif (Figure 2D). We further analyzed these sequences using more relaxed settings and found the majority of these peaks to contain variants of the forkhead motif and also that subsets of peaks were enriched for HNF6 and GABP motifs. PPARγ-RXRα motifs were also identified; however, these sequences are highly similar to the HNF4a motif and could be direct targets of HNF4a. FOXA2 and HNF6 are both known to bind together with HNF4a which indicates that this subset of peaks is either due to indirect binding or binding to sequences that are more divergent from the consensus or less well centered in peaks. Additionally, we searched for motif in peaks where the identified HNF4a sequences had been masked and found both the FOXA and HNF6 motifs to be enriched close to the HNF4a sites, and further identified motifs for C/EBP and CDP which are also known to interact with HNFs (23,24). Almost 50% of the HNF4a regions with the FOXA2 motifs were overlapping with peaks in the FOXA2 dataset, compared to 22–26% for the HNF6 and RXR motifs and 20% of all HNF4a peaks. For the GABP motifs in HNF4a peaks, 63% were also found in the GABP ChIP-seq dataset. We therefore conclude that motifs identified in this way are likely to correspond to binding in vivo. Analysis of masked FOXA2 sequences revealed HNF4a and C/EBP motifs, and GABP motifs were found to co-localize with SP1 and AP-2 motifs (Supplementary Figure S5).
We also matched a collection of 66 known binding sequences for HNF4a from human studies (25) to our peaks and out of these, 50 were present at least once in the peak centers and 34 were present in the list of predicted binding sites. Interestingly, the sequences that were not common in the peaks had a lower information content (Supplementary Figure S4D), indicating that many of these sequences are not direct HNF4a targets in vivo. We also found that some known binding sites were present in peaks below our cut-off which led us to test to what extent lower signals could be due to direct binding of the TF. To do this, we searched for the most enriched sequences at each peak height below the cut-off and were able to identify the target sequences already in peaks with as few as three to six overlapping reads for the different factors. Although enrichments are low in these peaks (Figure 1C) the high total number of peaks at these heights means that the full repertoire of binding sites in HepG2 cells are larger than the number of significant peaks presented here. However, it is possible that bindings to motifs below our threshold are transient and therefore not as likely to give rise to biologically meaningful interactions.
Some binding sites are evolutionary conserved
We then asked whether the sequences bound by the transcription factors were shared with other species, thereby suggesting evolutionary constraint in transcription factor positioning in the genome. We analyzed the conservation scores for the predicted TFBS as well as the overlap between FOXA2 binding sites identified by ChIP-seq in human and mice. The TFBS for all factors were significantly more conserved than the corresponding peak centers, indicating that the identified motif positions to a large extent are true binding sites (Figure 1D). Additionally, we found a significant increase in conservation for motifs in the lowest scoring peaks (not shown), indicating that also many of the peaks close to our cut-off level also have an evolutionary conserved function. However, the majority of predicted binding sites for HNF4a and FOXA2 had low conservation scores. To further characterize the level of conservation between species we compared the FOXA2 binding sites to those presented by Wederell et al. in a recent ChIP-seq study of FoxA2 binding sites in mouse livers (26). This dataset was obtained using the same antibody and peaks were found to contain the same motifs in similar numbers, thus these datasets should be well suited to study evolutionary differences in the FOXA2 binding patterns. We used the UCSC liftOver program to convert coordinates from mouse to human and found 577 FOXA2 and 860 HNF4a sites to overlap with the converted FoxA2 peaks (Table 1). The signal for FOXA2 in HepG2 was significantly higher for the sites where an orthologous signal was found in mouse, indicating that higher peaks are more likely to have a functional role which has been conserved between the species. One example of such a binding is found upstream of the HNF4a gene, where both experiments identified the same TGTTGAC target sequence (Supplementary Figure S6). Next we used ChIP-seq results from a different mouse strain (27) to define a set of high-confidence sites and further restricted this dataset by only include sites for which the same motif sequence was found also at the converted location in the human genome. Although a larger proportion of these sites were common between human and mouse, the majority (two-third) were not, showing that there is a large variation in binding sites between the species.
Most FOXA2 and HNF4a peaks are far from TSSs
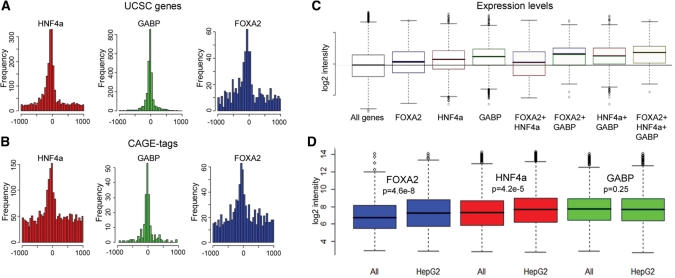
We found that only six and ten percent of all FOXA2 and HNF4a peaks respectively were located within 500 bp of a TSS (UCSC genes, Table 1). This is in line with what was found in the ENCODE regions in an earlier study (28). However, the increased resolution and coverage achieved by ChIP-seq allowed us to identify that both factors are enriched at positions −200 to +50 bp from the TSS (Supplementary Figure 3A). To investigate if the distal sites are in part due to binding to less well characterized TSS we mapped the peaks that were not found at a known TSS to the RIKEN CAGE-tag dataset. This revealed a similar pattern with an additional 255 peaks for FOXA2 and 651 for HNF4a located within +50 to −200 bp of a CAGE-tag cluster indicating that some novel transcripts or TSSs are regulated by these factors. To further characterize the distribution of binding sites, we considered bindings associated to genes if it were in a region from −10 to +1 kb of the gene body. Even with this broad definition only 60% of the peaks for the TFs were associated to genes, with 33% of all unique gene symbols associated with HNF4a binding and 16% with FOXA2. We have previously seen a difference in motif content for HNF4a binding sites in promoters (28) and in this study we found not only that fewer peaks at TSSs contain the motif but also that the average height for the peaks at TSS are lower, contrary to what is found for the other factors in the study (Supplementary Figure S7D). This is caused by the peaks without motifs, which indicates an indirect binding to these sites. We found that almost half of the FOXA2 peaks were co-localizing with HNF4a, often at a very close distance and with both motifs present (Figure 2B) and 20% of the FOXA2 peaks were close to sites bound by FOXA1 in MCF-7 cells (29), indicating that a large proportion of binding sites are shared between different FOXA proteins and used in different cell types.
HNF4a and FOXA2 but not GABP binds genes with liver-specific expression
To investigate if the TFs regulate the activity of nearby genes we matched binding sites within 1 kb of a TSS to the respective gene expression level in HepG2 (18). All factors were found to be associated with elevated gene expression, and genes with more than one factor at the TSS generally have a higher expression level (Figure 3C). To test if the bound genes have a liver-specific expression pattern we also compared the expression levels for the bound genes in 16 different cell lines (Figure 3D). Genes bound by HNF4a and FOXA2 have a significantly higher expression in HepG2 compared to in non-liver derived cell lines, and the average peak heights were higher for these TFs at genes with elevated expression in HepG2. GABP on the other hand did not show any significant difference in expression levels in different cell types. We also found the association with expression to be strongest for peaks close to the TSS (Supplementary Figure S8).
Figure 3.
Distribtion of peaks around TSSs and expression levels of bound genes. (A) All factors are enriched upstream of known TSS (UCSC genes) and a similar pattern (B) is seen around CAGE-tags for peaks that were not at a UCSC gene. (C) Genes bound by FOXA2, HNF4a and GABP have a higher expression than the average gene in HepG2, and genes where all factors are bound have the highest expression levels (P << 0.01 for all combinations). Horizontal line shows the median of all genes on the array. (D) Expression levels differ in HepG2 compared to non-liver derived cell lines for genes bound by HNF4a and FOXA2 but not for GABP.
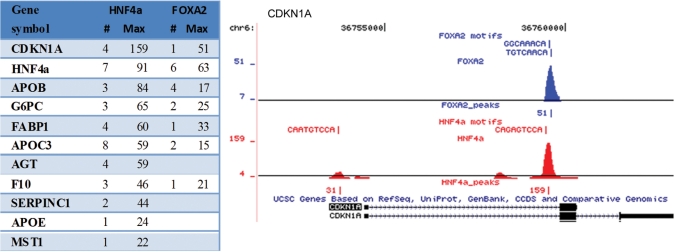
In a recent study, RNAi against HNF4a was shown to decrease the expression level of several genes (30). We observed HNF4a signal within 10 kb of all the reported genes, often with multiple sites throughout the gene body (Figure 4). A prominent example of this is the CDKN1A gene, where we could verify one of the reported binding sites in the CDKN1A promoter, but interestingly the strongest HNF4a signal in this locus was seen close to the last exon, in a region bound also by FOXA2.
Figure 4.
HNF4a and FOXA2 peaks in genes down-regulated by RNAi against HNF4a. The table shows the number of peaks and maximum overlaps within 10 kb of target genes. In most cases several binding sites with high enrichment were seen. The profile over the CDKNA1 locus illustrates that in many cases the binding sites are intragenic.
We extracted the GO (31) annotations for genes that were within 1 kb of a ChIP-seq signal and tested the distribution of these against all GO-annotations in all protein coding genes. GABP peaks were divided into two groups with either the NRF-2 or the NRF-1 motif, and the GABP/FOXA2, GABP/HNF4a and FOXA2/HNF4a combinations were tested. These results indicate that FOXA2 is binding close to genes involved in cellular organization and biogenesis, various metabolic processes and signal transduction. For peaks within 500 bp of TSS genes involved in response to stress and DNA repair were overrepresented. GABP was found to be involved in macromolecule metabolic processing, and some clear differences were seen between NRF-1 and NRF-2 peaks, e.g. only NRF-1 was found to be involved in apoptosis and cell death, a function that has previously been associated to NRF-1 overexpression in serum-depleted cells (32).
USF2 homodimers co-localize with FOXA2 and HNF4a
In a recent study, we investigated the genome-wide locations of USF1 and USF2 in HepG2 cells using high density oligonucleotide arrays (7). The binding sites for USF1 and USF2 were mostly shared but in 240 out of 1351 cases USF2 bound as a homodimer. In addition, these binding sites were located further away from TSSs than the USF1–USF2 heterodimers. Based on motif searches it was found that some HNFs could bind these regulatory elements, which was experimentally verified at selected locations. In the present study we find that more than 50% of the USF2 homodimers were close to HNF4a and FOXA2 (Figure 2A) and that peaks for FOXA2 and HNF4a were higher when USF2 was present (Supplementary Figure S8), indicating cooperative binding to these regions.
GABP peaks with the NRF-2 motif co-localize with HNF4a peaks at TSSs
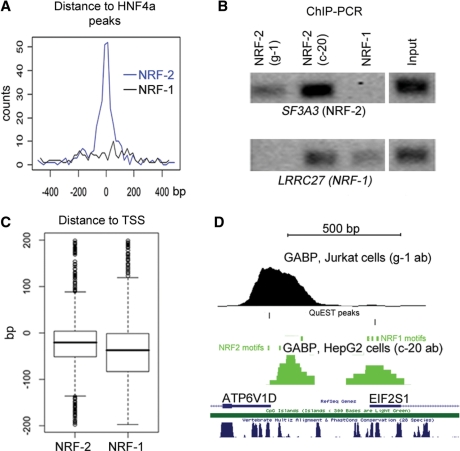
For the GABP dataset 85% of the peaks were within 500 bp of an UCSC gene and 95% were within 500 bp of either an UCSC gene or a CAGE-tag cluster. The identification of both NRF-1 and NRF-2 motifs in the GABP dataset was intriguing. Although pre-ChIP controls had been made by Western blot, the possibility remains that the polyclonal antibody recognizes both GABP (NRF-2) and NRF-1 in vivo. We therefore analyzed peaks containing NRF-1 or NRF-2 motifs separately and found the subsets to have distinct characteristics. Firstly, we found an enrichment of HNF4a peaks close to GABP only when the NRF-2 motif was present (Figure 5A). Secondly, peaks containing only the NRF-2 motif were closer to the TSS than those with only the NRF-1 motif (Figure 5C). We next compared our results to the GABP signals from Jurkat cells (20), where a monoclonal antibody directed towards the N-terminal part of the protein was used. Valouev et al. found only the NRF-2 motif in the GABP dataset and strikingly, 90% of our NRF-2 peaks were co-localizing with a peak in the Jurkat cells whereas the overlap was only 14% for peaks with the NRF-1 motif. To exclude the possibility that the differences are due to cell-type specific events or experimental variation we performed ChIP-PCR using the two GABP antibodies and a NRF-1 specific antibody. Figure 5B show that ChIPs using both NRF-2 antibodies but not the NRF-1 antibody was positive for an NRF-2 site in the SF3A3 promoter while both NRF-1 and the polyclonal NRF-2 antibody detected an NRF-1 site in the LRRC27 promoter. However, in some cases the NRF-1-only peaks did co-localize with weak binding sites in the Jurkat dataset (Figure 5D), indicating that this antibody also recognizes proteins binding to these sites but less efficiently compared to the polyclonal antibody.
Figure 5.
Analysis of NRF-1 and NRF-2 motifs in GABP peaks. (A) Distribution of peaks with NRF-2 (blue) and NRF-1 (black) motifs around HNF4a peaks. (B) ChIP-PCR for sites containing either NRF-2 or NRF-1 motifs using two different GABP antibodies and one NRF-1 specific antibody. (C) NRF-2 motifs are located closer to TSS than NRF-1. (D) GABP signal is found both in Jurkat and HepG2 in the ATP6V1D promoter (left) but with different peak centers. Signals are also found in both datasets at a known NRF-1 site in the EIF2S1 promoter (right).
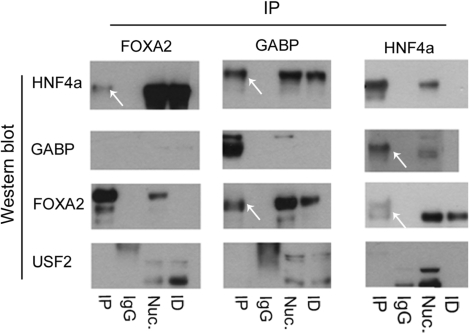
Interactions with GABP is verified by co-IP
To further study the interaction between these TFs, we performed co-immunoprecipitation experiments on nuclear extracts from HepG2 cells. Western blot of the HNF4a IP was positive for GABP and FOXA2 and similarly, HNF4a and FOXA2 were present in the GABP IP (Figure 6). Although the USF2 signal from the ChIP-chip study (7) was found to co-localize with FOXA2 and HNF4a in many cases, it was not as frequent and close as the GABP signal. We could not verify this interaction with the co-IP experiments, suggesting that these TFs do not physically interact.
Figure 6.
Co-immunoprecipitation results. Western blots are shown for HNF4a, NRF-2, USF2 and FOXA2 on IPs for the TFs and for IgG. Nuclear extract (Nuc) and Immuno depleted (ID) material are included as controls. White arrows indicate positive signals.
ChIP-seq facilitates detection of functional SNPs
Recently several genome wide association studies (GWAS) have identified many non-coding SNPs associated to liver mediated phenotypes such as plasma levels of liver enzymes, lipid and glucose levels and diabetes risk (33–39). In some cases a coding functional variant in LD with the associated SNPs have been identified but it is likely that some of the non-coding SNPs are causative and may exert the effect by affecting TF binding, either directly by changing the targeted DNA sequence or indirectly, e.g. by changing the methylation status of the DNA. We matched the SNPs present within our positive peaks to the lists of SNPs in these studies and to a collection of SNPs from GWAS studies (40) available from http://www.genome.gov/26525384 and found several matches to liver-mediated traits for HNF4a and FOXA2, with examples such as a SNP altering the plasma levels of ALT and two diabetes associated SNPs in MTNR1B and GCKR (Table 2). Although none of these SNPs were in the motif sequences they could be functional as they are located in regions with regulatory potential. Additionally, some SNPs associated to colorectal cancer and other traits were within the peaks (Supplementary Table S1), including the 8q24 cancer risk variant rs6983267 which is located 15 bp from an HNF4a site. This SNP was recently shown to be differentially bound by TCF7L2, a TF with similar sequence specificity as HNF4a, and to have long-range interaction with MYC in colorectal cancer (41).
Table 2.
SNPs from GWAS studies of liver-mediated traits present in peaks
| SNP | Height | Gene | Trait |
|---|---|---|---|
| rs11597390 | 116/49 | CPN1 | Plasma levels of ALT |
| rs2144300 | 75 | GALNT2 | HDL/triglycerides |
| rs3847303 | 75/14 | ABCA1 | HDL |
| rs561241 | 41/21 | F7 | Factor VII |
| rs1800588 | 28 | LIPC | HDL |
| rs10830963 | 20 | MTNR1B | T2D, fasting plasma glucose |
| rs11887534 | 15 | ABCG8 | Gallstones |
| rs780094 | 15 | GCKR | Triglycerides |
| rs12740374 | 15/13 | CELSR2 | LDL cholesterol |
Height denotes the maximum overlaps for HNF4a/FOXA2 in the peak
We also note that ChIP-seq datasets can be used to identify allele-specific enrichment of TF binding given sufficient base coverage within the peaks. We looked for this by comparing the sequences at SNPs for HNF4a and FOXA2 and identified some positions where the factors may bind different alleles; however in these datasets the number of reads covering the SNPs was generally too few to yield significant results (Supplementary Figure S8 and Supplementary Table S2).
DISCUSSION
The recent advances in microarray and sequencing technologies have made it possible to map the gene regulatory circuitry in living cells on a genome-wide basis and thus also to challenge some previous notions on gene regulation. Boyer et al. used the distribution of binding sites from the Transfac database to state that ‘Although some transcription factors are known to regulate genes from distances >8 kb, 98% of known binding sites for human transcription factors occur within 8 kb of target genes’ (42), and the preference for binding in promoter regions has also been claimed to be true for FOXA2 and other related factors (43). In this article, we present genome-wide maps of HNF4a and FOXA2 in human cells. In concordance with previous results from 1% of the genome (28) we found the vast majority of binding sites to be located far from TSSs, with more than 80% at distal positions even when a large set of CAGE-tags was included as putative TSSs. For sites close to TSS, enrichment was seen in the first 100–200 upstream bases of many liver-specific genes, however, the genome wide distribution of binding sites indicates that studies aiming to identify the effect of these TFs on specific genes should not be limited to promoter regions.
We and others have shown that it is possible to identify the base-pairs a TF interacts with by motif analysis in enriched peaks. However, identification of alternative motifs in subsets of peaks is often not easily done since most motif finders will only report variants of the best scoring motifs, and due to the prohibitively slow running time on large datasets they are often used only on the top scoring peaks (19). We report here a fast and accurate way of identifying independent motifs in ChIP-seq datasets. With this method we were able not only to identify the established motifs for the three different factors but also to find that the antibody used against GABP can identify the related factor NRF-1 and show that the two subsets of peaks have different properties, such as the interaction between NRF-2 and HNF4a and the differences in enriched GO-categories for bound genes.
As we and others found in the ENCODE project (1), and as further shown by Odom et al. (44) TFBS evolve faster than protein coding regions. We found here also a low overlap between FOXA2 sites in mouse and human. However, we find that many of the strongest binding events were evolutionary conserved. Thus sequence conservation can be used to highlight particularly important binding sites, but restricting functional studies to conserved elements will exclude a large proportion of the true binding sites.
We previously found that the HNF4a peaks at TSSs in the ENCODE regions had fewer matches to the motif, leading to the theory that this is due to indirect binding, e.g. by virtue of chromatin loops (28). In this study we identified a subset of HNF4a peaks at TSS that were also bound by GABP and found that these sites most often did not contain the HNF4a consensus. Apparently HNF4a is not using the traditional sequence to interact with these regions. This means that the interaction could be to a so far unrecognized motif, unspecifically to DNA or indirectly via another protein. The results from the co-IP experiments are compatible with the last hypothesis as HNF4a was found in complex with GABP. The interaction between GABP and HNF4a has previously been identified in silico from ChIP-chip data on promoter arrays (45) but to our knowledge this is the first study presenting in vivo data on their co-localization.
In conclusion, our study verify that ChIP-seq is a powerful method to map gene regulatory networks and has the potential to identify not only the bound bases but also the allelic preference of bindings to regulatory SNPs. We propose that this strategy can be used to find TFs binding to SNPs associated to several common diseases, thereby providing more knowledge of the mechanism leading to pathologic results. To do this systematically, cells or tissues from several individuals need to be studied so that the relevant alleles are sampled from the population. The rapid increase in sequencing throughput may soon make such projects feasible, but in the mean time studies like the one presented here will continue to contribute data on the regulatory wiring of important tissues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council for Natural Science and for Medicine, the Markus Bergström Foundation, The Cancer Foundation, Swedish Diabetes Foundation and The Family Ernfors Fund grants to C.W. Swedish Foundation for Strategic Research, The Knut and Alice Wallenberg Foundation, Uppsala University and the Swedish University for Agricultural Sciences grants to J.K. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the team lead by D. Bentley at Illumina, UK and Y. Zhao and R. Varhol at Genome Sciences Centre at the British Columbia Cancer Agency for performing large scale sequencing and alignments and L Rönnegård, Linneaus Center for Bioinformatics for statistical assistance.
REFERENCES
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genetics. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 6.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M, Bell GI. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 10.Holmkvist J, Almgren P, Lyssenko V, Lindgren CM, Eriksson KF, Isomaa B, Tuomi T, Nilsson P, Groop L. Common variants in maturity-onset diabetes of the young genes and future risk of type 2 diabetes. Diabetes. 2008;57:1738–1744. doi: 10.2337/db06-1464. [DOI] [PubMed] [Google Scholar]
- 11.Damcott CM, Hoppman N, Ott SH, Reinhart LJ, Wang J, Pollin TI, O'C;onnell JR, Mitchell BD, Shuldiner AR. Polymorphisms in both promoters of hepatocyte nuclear factor 4-alpha are associated with type 2 diabetes in the Amish. Diabetes. 2004;53:3337–3341. doi: 10.2337/diabetes.53.12.3337. [DOI] [PubMed] [Google Scholar]
- 12.Silander K, Mohlke KL, Scott LJ, Peck EC, Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G, et al. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 13.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 14.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaji H, Kasukawa T, Fukuda S, Katayama S, Kai C, Kawai J, Carninci P, Hayashizaki Y. CAGE Basic/Analysis Databases: the CAGE resource for comprehensive promoter analysis. Nucleic Acids Res. 2006;34:D632–D636. doi: 10.1093/nar/gkj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- 18.Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 2006;7:181. doi: 10.1186/1471-2164-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura H, Tomaru Y, Nakanishi M, Kondo S, Hayashizaki Y, Suzuki M. Identification of DNA regions and a set of transcriptional regulatory factors involved in transcriptional regulation of several human liver-enriched transcription factor genes. Nucleic Acids Res. 2009;37:778–792. doi: 10.1093/nar/gkn978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braidotti G, Borthwick IA, May BK. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J. Biol. Chem. 1993;268:1109–1117. [PubMed] [Google Scholar]
- 23.Tomaru Y, Nakanishi M, Miura H, Kimura Y, Ohkawa H, Ohta Y, Hayashizaki Y, Suzuki M. Identification of an inter-transcription factor regulatory network in human hepatoma cells by Matrix RNAi. Nucleic Acids Res. 2009;37:1049–1060. doi: 10.1093/nar/gkn1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antes TJ, Chen J, Cooper AD, Levy-Wilson B. The nuclear matrix protein CDP represses hepatic transcription of the human cholesterol-7alpha hydroxylase gene. J. Biol. Chem. 2000;275:26649–26660. doi: 10.1074/jbc.M002852200. [DOI] [PubMed] [Google Scholar]
- 25.Ellrott K, Yang C, Sladek FM, Jiang T. Identifying transcription factor binding sites through Markov chain optimization. Bioinformatics. 2002;18(Suppl. 2):S100–S109. doi: 10.1093/bioinformatics/18.suppl_2.s100. [DOI] [PubMed] [Google Scholar]
- 26.Wederell ED, Bilenky M, Cullum R, Thiessen N, Dagpinar M, Delaney A, Varhol R, Zhao Y, Zeng T, Bernier B, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp536. doi:10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rada-Iglesias A, Wallerman O, Koch C, Ameur A, Enroth S, Clelland G, Wester K, Wilcox S, Dovey OM, Ellis PD, et al. Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Hum. Mol. Genet. 2005;14:3435–3447. doi: 10.1093/hmg/ddi378. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol. Endocrinol. 2008;22:78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am. J. Human Genet. 2008;83:520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, Pramstaller PP, Penninx BW, Janssens AC, Wilson JF, Spector T, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, et al. A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat. Genet. 2009;41:89–94. doi: 10.1038/ng.277. [DOI] [PubMed] [Google Scholar]
- 36.Kathiresan S, Willer CJ, Peloso GM, Demissie S, Musunuru K, Schadt EE, Kaplan L, Bennett D, Li Y, Tanaka T, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, et al. Variants in MTNR1B influence fasting glucose levels. Nat. Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 39.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol. Syst. Biol. 2006;2:0017. doi: 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith AD, Sumazin P, Das D, Zhang MQ. Mining ChIP-chip data for transcription factor and cofactor binding sites. Bioinformatics. 2005;21(Suppl. 1):i403–i412. doi: 10.1093/bioinformatics/bti1043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.