Abstract
The LysR-family regulator MexT modulates the expression of the MexEF-OprN efflux system in the human pathogen Pseudomonas aeruginosa. Recently, we demonstrated that MexT regulates certain virulence phenotypes, including the type-three secretion system and early attachment independent of its role in regulating MexEF-OprN. In this study, transcriptome profiling was utilized to investigate the global nature of MexT regulation in P. aeruginosa PAO1 and an isogenic mexEF mutant. Twelve genes of unknown function were highly induced by overexpressing MexT independent of MexEF-OprN. A well-conserved DNA motif was identified in the upstream regulatory region of nine of these genes and upstream of mexE. Reporter fusion analysis demonstrated that the expression of the genes was significantly induced by MexT in P. aeruginosa and a heterogenous Escherichia coli strain and that the conserved sequence was required for this induction. The conserved DNA motif was further characterized as the MexT binding site by site-directed mutagenesis and electrophoretic mobility shift assays. Genes containing this conserved regulatory sequence were identified across other Pseudomonas species, and their expression was activated by MexT. Thus, a novel regulon directly modulated by MexT, that includes but is independent of mexEF-oprN, has been identified.
INTRODUCTION
Pseudomonas aeruginosa is a leading cause of hospital-acquired infections and remains the most important pathogen associated with chronic lung infections in cystic fibrosis patients. Pseudomonas aeruginosa is well known for its intrinsic resistance to a wide range of antimicrobial agents and its ability to develop multidrug resistance following antibiotic therapy (1). Resistance-Nodulation-Division (RND) efflux systems are responsible for much of the intrinsic and acquired multidrug resistance in P. aeruginosa and genes encoding 12 RND efflux pumps have been identified in its genome. To date, seven—MexAB-OprM, MexCD-OprJ, MexEF-OprN, MexGHI-OpmD, MexJK, MexXY and CzrAB-OpmN—have been functionally characterized and their expression is tightly regulated. Although these efflux pumps can exclude various antimicrobials, most of them are not induced by these substrates in wild-type strains (2). Nevertheless, genes encoding for RND efflux pumps are highly conserved in many living organisms (3) and, recently, increasing attention has been focused on understanding the physiological roles of efflux pumps, that probably precede antibiotics and are relevant to the behaviour of bacteria in their natural ecosystems (3–5). It is now evident that efflux pumps are involved in diverse cellular activities including functions such as the detoxification of intracellular metabolites, bacterial virulence in both animal and plant hosts, cell homeostasis and intercellular signal trafficking. However, little is known about how the expression of these efflux pumps is modulated within innate genetic networks to support these versatile cellular functions in response to changing environments.
MexEF-OprN is a unique RND pump that is modulated by a transcriptional activator, while other RND pumps are modulated by transcriptional repressors in P. aeruginosa (2). The expression of mexEF-oprN is activated by a LysR-type transcriptional regulator (LTTR) MexT, which is encoded by a gene located just upstream of mexEF-oprN in the same orientation in P. aeruginosa (6). The MexEF-OprN system is normally quiescent in wild-type cells under normal laboratory conditions, but is highly induced in nfxC-type phenotypic mutants. These mutants exhibit increased resistance to chloramphenicol, trimethoprim and fluoroquinolones as well as exhibiting susceptibility to certain β-lactam and aminoglycoside antibiotics (7,8). The underlying mechanisms leading to nfxC-type mutants are not fully understood but appear to be multifactorial (9–11). For example, the frequency of isolating nfxC-type mutants has been associated with mutations in the mexT gene of the parent strain that render the protein active or inactive (9). Indeed, several mutations in the nucleotide sequence of mexT were identified in different laboratory PAO1 strains rendering the protein inactive and leading to a low frequency in the isolation of nfxC-type mutants. In other PAO1 strains, the mexT sequence encoded an active protein leading to a high frequency of nfxC-type mutants. In this class of PAO1 strain, the authors suggest that the expression or activity of MexT is suppressed under normal laboratory growth conditions (9). More recently, a deletion in the adjacent mexS gene was shown to result in an nfxC-type mutation (11). mexS encodes a probable oxidoreductase but its role in modulating MexT function is as yet unknown. Nevertheless, in all cases, it appears a functional MexT is crucial for the generation of nfxC-type mutants and this function is suppressed under normal growth conditions. Accordingly, and in support of the demand theory of gene regulation whereby the design of molecular control mechanisms is generally dependent on the demand for expression of the regulated genes (12), it is predicted that the expression of mexEF-oprN and indeed other possible targets of MexT may be in high demand in natural environments of P. aeruginosa.
Besides the mexEF-oprN operon, MexT can activate the expression of mexS, previously referred to as qrh, in P. aeruginosa (6). mexS is located adjacent to mexT and transcribed in the opposite direction, which is a typical arrangement of genes controlled by LTTRs (13). A palindromic DNA sequence has been identified in the promoter region of target genes, to which LTTRs are known to bind. This LTTR box was first identified in Azorhizobium caulinodans as an interrupted palindrome with the sequence ATC-N9-GAT, referred to as the ‘nod-box’ (14). Indeed, a nod-box-like sequence in the promoter region of mexE has been proposed as the binding site of MexT (6). Interestingly, the promoter region of mexS does not contain a clear ‘nod-box’.
In addition to antibiotic resistance, overexpression of MexEF-OprN in nfxC-type mutants has been linked to reduced levels of homoserine lactone-dependent virulence traits, including pyocyanin, elastase, rhamnolipids and PQS (15) and to reduced expression of type-three secretion effector proteins (16). It was suggested that MexEF-OprN mediates these effects via efflux of cell-signalling intermediates, which ultimately commits the cell to a state of reduced virulence (15,17). Recently, we have observed that MexT downregulates several virulence determinants, such as TTSS gene expression, pyocyanin biosynthesis and early surface attachment, in a MexEF-OprN independent manner in P. aeruginosa PAO1 (18). We have suggested that MexT acts as a more global regulator than previously described and novel target genes may be directly activated by MexT in P. aeruginosa. This is supported by the fact that mexT is not tightly linked to the mexEF-oprN in the genomes of most Pseudomonas species, according to Pseudomonas Genome Database (www.pseudomonas.com). For instances, mexT (PP_2826) and mexE (PP_3425) are located in totally different loci in the genome of P. putida KT2440 strain.
Elucidating the transcriptional regulatory network linked to MexT will provide a new angle to understand the physiological roles of the MexEF-OprN pump in a broader context. In this study, we demonstrate the existence of a novel regulon incorporating mexEF-oprN in Pseudomonas species. MexT activates the expression of this novel regulon by binding to a conserved DNA motif in their promoter regions.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are shown in Table 1. All Escherichia coli and P. aeruginosa PAO1 strains were routinely grown at 37°C, while P. fluorescens PfO-1 and P. putida KT2440 strains were grown at 30°C in Luria–Bertani (LB) broth with aeration. When required, antibiotics were added at the following concentrations (µg/ml): E. coli, ampicillin (Ap, 50), kanamycin (Km, 50), chloramphenicol (Cm, 20), tetracycline (Tc, 10) or gentamicin (Gm, 10); P. aeruginosa PAO1, carbenicillin (Cb, 200), gentamicin (Gm, 50), tetracycline (Tc, 50) or streptomycin (Str, 200).
Table 1.
Strains and vectors used in this study
| Genotype or phenotype | Source | |
|---|---|---|
| Pseudomonas aeruginosa | ||
| PAO1 wild-type | Lab stock | |
| PAO1ΔmexEF | mexE-mexF deletion mutant of PAO1 | (18) |
| PAO1nfxC | mexEF-oprN overexpressing mutant of PAO1 | This work |
| PAO1nfxCΔmexT | mexT deletion mutant of PAO1nfxC | This work |
| Escherichia coli | ||
| DH5α | F-φ80 lacZΔM15 (ΔlacZYA-argF )U169 deoR recA1 endA1 hsdR17(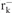 , , 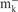 ) phoA supE44 thi-1 gyrA96 relA1Δ− ) phoA supE44 thi-1 gyrA96 relA1Δ−
|
Invitrogen |
| ST18 | hemA λpir lysogen of S17-1: pro, Res−, Mob+, RecA− derivative of E. coli 294 with Rp4-2 (Tc::mu) (Km::Tn7) in the chromosome | (22) |
| BL21-CodonPlus (DE3)-RIPL | protein expression host | Merck |
| Pseudomonas fluorescens | ||
| PfO-1 | wild-type | (41) |
| Pseudomonas putida | ||
| KT2440 | mt-2 hsdRl hsdM+ | (42) |
| Vectors | ||
| pCR2.1-TOPO TA | cloning vector, Apr, Kmr | Invitrogen |
| pME6032 | pVS1-p15A origin, lacIq-Ptac expression vector, Tcr | (43) |
| pME6032-mexT | pME6032-derived PAO1 mexT expression vector | (18) |
| pEX18Tc | oriT+ sacB+, gene replacement vector, Tcr | (20) |
| pPS856 | Gmr cassette flanked with FRT sequences, Apr, Gmr | (20) |
| pFLP2 | inducible Flp recombinase, Apr | (20) |
| pMP190 | IncQ origin, low-copy-number lacZ fusion vector; Cmr Strr | (44) |
| pMP-PAmexEp | pMP190-derived PAO1 mexE promoter-lacZ fusion plasmid | This work |
| pMP-PA1744p | pMP190-derived PAO1 PA1744 promoter-lacZ fusion plasmid | This work |
| pMP-PA2759p | pMP190-derived PAO1 PA2759 promoter-lacZ fusion plasmid | This work |
| pMP-PA3229p | pMP190-derived PAO1 PA3229 promoter-lacZ fusion plasmid | This work |
| pMP-PA4354p | pMP190-derived PAO1 PA4354 promoter-lacZ fusion plasmid | This work |
| pMP-PA4623p | pMP190-derived PAO1 PA4623 promoter-lacZ fusion plasmid | This work |
| pMP-PA4881p | pMP190-derived PAO1 PA4881 promoter-lacZ fusion plasmid | This work |
| pMP-PA4623m1p | conserved DNA motif mutated version of pMP-PA4623p | This work |
| pMP-PA4881m1p | conserved DNA motif mutated version of pMP-PA4881p | This work |
| pMP-Pfl2659p | pMP190-derived PfO-1 Pfl01_2659 promoter-lacZ fusion plasmid | This work |
| pMP-Pfl3748p | pMP190-derived PfO-1 Pfl01_3748 promoter-lacZ fusion plasmid | This work |
| pMP-PP3425p | pMP190-derived P. putida KT2440 PP_3425 promoter-lacZ fusion plasmid | This work |
| pMP-PP4858p | pMP190-derived P. putida KT2440 PP_4858 promoter-lacZ fusion plasmid | This work |
| pET28a | T7 promoter-driven His-tag protein expression vector, Kmr | Novagen |
| pETmexTH6C | pET28a-derived C-terminal His6-tag PAO1 mexT expression vector | This work |
| pETmexTm1H6C | pET28a-derived C-terminal His6-tag non-functional mexT expression vector | This work |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; Strr, streptomycin resistance; Gmr, gentamycin resistance.
Molecular biology procedures
All routine molecular biology procedures were carried out as described in the standard laboratory manual (19). All DNA primers used in this study are listed in Supplementary Table S1.
Expression profiling experiments
PAO1 (pME6032), PAO1 (pME6032-mexT) and PAO1ΔmexEF (pME6032-mexT) were grown at 37°C shaking (150 r.p.m.) in 20 ml of LB medium supplemented with tetracycline in 100-ml culture flasks. At an OD600 of 0.5, cell cultures were mixed with Qiagen RNAprotect Bacteria Reagent (Ratio of 1:2) to stabilize RNA. Total RNA was prepared with an RNeasy mini kit (Qiagen) and contaminating DNA was removed by RNase-free DNase I (Ambion). For each strain, RNA was prepared from three independent batch cultures. cDNA synthesis and hybridization to Affymetrix GeneChip P. aeruginosa genome arrays were carried out by a commercial Affymetrix Genechip service supplier (Conway Institute of Biomolecular & Biomedical Research, UCD, Ireland). Array data were normalized using the GC-RMA algorithm, and the data were then further analyzed using the microarray software package Genespring GX 10.0.1 (Agilent). To visualize the microarray data, MA plots (linear regression of log ratio, M, against average intensity, A) were generated by Genespring GX software, and 3D scatter plots on log2 based intensity data obtained from three samples were generated by SigmaPlot 11 software (Systat Software Inc.). Genes whose levels of expression were significantly influenced (Fold change ≥ 2, P ≤ 0.05) were identified with Genespring GX, using a Benjamini–Hochberg multiple-testing correction and a false detection rate of 5%. The full list of genes whose expression was significantly altered is shown in Supplementary Table S2.
To validate the microarray results, semi-quantitative reverse transcription PCR analysis was carried out using RNA samples prepared from biological replicates not used in the arrays. Two hundred nanograms of RNA samples were reverse transcribed using random primers and AMV reverse transcriptase (Promega). 1/100 aliquot of the reaction mixture was used as template in the subsequent PCR. PCR (20 µl reaction volume) was performed with the GoTaq® Green Master Mix (Promega) with the following conditions: 95°C for 3 min; 30 cycles of 30 s at 95°C; 30 s at 55°C and 30 s at 72°C; 10-min elongation at 72°C. For visualization, 10 µl of the resulting PCR was subjected to agarose gel electrophoresis and stained with ethidium bromide. The clpX gene was used as an internal control to ensure equal amounts of RNA were used in all samples.
Generation of PAO1nfxC mutant and PAO1nfxCΔmexT strains
PAO1nfxC was selected by plating the wild-type strain PAO1 on LB agar plates containing chloramphenicol at 600 µg/ml, a condition that exclusively selects nfxC-type mutants (7). The mexT ORF and 500-bp upstream region of this PAO1nfxC strain had no sequence alteration(s) compared to the parental PAO1 strain.
In order to construct PAO1nfxCΔmexT, a 1-kb SacI–KpnI upstream region including the start codon of mexT, and a 1-kb KpnI–BamHI downstream region containing the stop codon of mexT, were PCR-amplified and linked together. The resulting 2-kb fragment was cloned into the suicide plasmid pEX18Tc digested with SacI and BamHI. A 1.1-kb KpnI fragment containing the FRT gentamicin-resistance (Gm) cassette from plasmid pPS856 (20) was then inserted in-between the flanking regions on the plasmid. The mexT of PAO1nfxC was then replaced with the plasmid as described by Hoang et al. (20). The Gm-resistance sequence in the chromosome was removed by introducing plasmid pFLP2, which carries the Flp recombinase gene (20). Correct insertion in the constructed mutant was verified by PCR using primers bind to flanking chromosomal regions of the fragments cloned in pEX18Tc.
Construction of the promoter-lacZ reporter gene fusions and β-galactosidase assays
The promoter region of each putative MexT target gene was PCR-amplified and TA-cloned into pCR2.1-TOPO (Invitrogen). The site-directed mutagenesis of the conserved DNA motif was performed by the protocol described previously (21). Once confirmed by sequencing, the promoter regions were subcloned into the broad-host low-copy-number plasmid pMP190 (Table 1). The resulting plasmids were introduced into P. aeruginosa strains by conjugal transfer from E. coli donor strain ST18. ST18 is a hemA mutant of S17-1λpir strain (Table 1), and can be easily selected against as its growth is strictly dependent on the presence of 5-aminolevulinic acid (5-ALA), even in LB medium (22). Therefore, 50 µg/ml of 5-ALA was added to the medium during conjugation and omitted when selecting for trans-conjugants. Since PAO1 containing pME6032-mexT and PAO1nfxC strains have a high chloramphenicol resistance due to the nfxC phenotype, all trans-conjugant P. aeruginosa cells containing pMP190-derived plasmids were selected on LB agar supplemented with 200 µg/ml streptomycin. The plasmid transformation into E. coli strains in this study was done by routine chemical competent cell protocol (19).
For β-galactosidase assays, cells were grown overnight in LB broth supplemented with appropriate antibiotics, then 1:50 diluted into 10 ml fresh medium in 100-ml culture flasks and incubated at 37°C shaking at 150 r.p.m. Cells were recovered at logarithmic growth phase (OD600 0.5–1.2), and β-galactosidase assays were performed as described by Miller (23). Data are the mean of two independent experiments with triplicate samples.
Purification of His6-MexT
The plasmids used to overexpress the C-terminal His6-tagged MexT proteins were constructed by PCR-amplifying the MexT coding sequence and cloned into NcoI–XhoI sites of pET28a. The plasmid harbouring the functional mexT from PAO1 was designated as pETmexTH6C, while the plasmid harbouring the non-functional mexT due to a mutation of the 39th amino acid Ala to Val was designated as pETmexTm1H6C (Table 1). The plasmids were transformed into the E. coli expression host strain BL21-CodonPlus(DE3)-RIPL (Merck) and grown at 37°C with vigorous shaking in 200 ml of LB medium containing kanamycin (50 µg/ml) to an OD600 of 1. The cells were then induced with 1 mM IPTG and allowed to express for 1 h at 37°C. The cells were harvested by centrifugation and stored overnight at –20°C. The pellet was resuspended in CelLyticTM B II buffer (Sigma®) (10 ml per gram cell paste) with 5 µg/ml DNase and 200 µl per gram cell paste of Protease Inhibitor Cocktail (Sigma®). The soluble proteins were extracted according to manufacturer’s instructions (Sigma®, CelLyticTM B II, Bacterial Cell Lysis Extraction Reagent). The protein extract were applied to a Poly-Prep® Chromatography Column (Bio-Rad) containing 1 volume of HIS-SelectTM Nickel Affinity Gel (Sigma®). The resin was previously washed with 2 vol of deionized sterile water and equilibrated with 2 vol of Wash buffer (HEPES 100 mM, pH 7.5, 1 mM Imidazole). After all the protein extract was loaded, the column was washed twice with 2 vol of Wash buffer. The His-tag proteins were eluted with 1 vol of Elution buffer (HEPES 100 mM, pH 7.5, 500 mM Imidazole) and their homogeneity was verified by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE). The protein was then either immediately frozen with 20% of glycerol at –80°C for later use or promptly used for the electrophoretic mobility shift assay (EMSA). The protein concentrations were determined by the Bio-Rad method.
EMSAs
DNA probes of 207 bp containing the putative binding region were generated by PCR using pMP-PA4881p (intact motif) or pMP-4881m1p (mutated motif) as templates using the primer pair fullmexTboxF and fullmexTboxR. The two purified PCR products were 3′-end-labelled with digoxigenin following the manufacturer instruction (Roche Applied Science). The EMSA was carried out using the DIG Gel Shift Kit 2nd Generation (Roche Applied Science) as recommended, with some modifications. Ten fmoles of the DIG-labelled fragment and a range of 0–300 ng of MexT protein were added to the binding reaction. The mixture was allowed to proceed for 45 min at room temperature. The samples were separated by electrophoresis on 6% native polyacrylamide gels and transferred to Hybond-N blotting membrane (Amersham Life Science) by electroblotting. Protein–DNA complexes were visualized by NBT/BCIP according to manufacturer’s instructions (Roche Applied Science). Competition experiments were performed with 200 pmol of unlabelled double-stranded oligonucleotide. The labelled and non-labelled probes were mixed to the binding buffer before adding the protein for the binding reaction.
Identification of a conserved regulatory sequence
For the identification of a conserved regulatory sequence motif in the upstream region of co-regulated genes, the 500-bp sequence upstream of the translational start site of each gene induced by MexT was obtained from the ‘www.pseudomonas.com’ database. Sequence motifs common to the upstream regions were identified by the online MEME software (http://meme.sdsc.edu/meme4_1_1/cgi-bin/meme.cgi) (24) with a motif width range of 5–20 bp and the occurrences of a single motif distributed among the sequences between zero or one per sequence.
RESULTS
Transcriptome profiling of P. aeruginosa strains overexpressing MexT
In order to assess the global regulatory nature of the LysR-family transcriptional regulator MexT, independent of its induction of the MexEF-OprN efflux system, whole-genome transcriptome profiling was carried out on PAO1 wild-type and an isogenic mexEF mutant overexpressing MexT. In the PAO1 strain used in this study MexT is quiescent under normal laboratory growth conditions. However, when the entire ORF and 600-bp upstream of the mexT gene were cloned on plasmid pME6032 (designated as pME6032-mexT), MexT was fully functional in P. aeruginosa PAO1, increasing the minimum inhibitory concentration (MIC) of chloramphenicol from 32 to 2048 µg/ml, even without IPTG induction of the tac promoter on pME6032 (18). Affymetrix P. aeruginosa whole-genome microarrays were used to compare the transcription profiles of PAO1 wild-type containing either pME6032-mexT or the empty vector control pME6032 and an isogenic mexEF deletion mutant strain, PAO1ΔmexEF, containing pME6032-mexT, during Mid-logarithmic growth conditions. Thus, we identified a potential MexT regulon and differentiated those targets that were dependent or independent of the MexEF-OprN efflux pump.
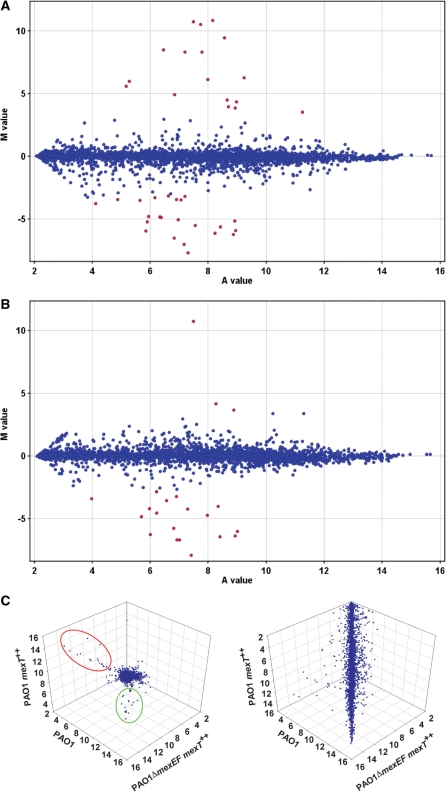
Genespring GX software was used to analyse the data (Supplementary Table S2) and MA and 3D scatter plots were used to visually compare the overall gene expression profiles of all three samples (Figure 1). Overexpressing mexT in PAO1 wild-type (PAO1 mexT++) increased the expression of 36 genes and decreased the expression of 76 genes compared to PAO1 harbouring the pME6032 empty vector (≥2-fold, P ≤ 0.05; see Supplementary Table S2). The most significantly altered genes (≥10-fold) are summarized in Table 2, and include 17 genes upregulated and 23 genes downregulated by overexpressing MexT in wild-type. These genes were also indicated as the most significantly altered genes in the MA plot in Figure 1A. To test if this modulation in gene expression was dependent on the MexEF-OprN pump, the transcriptome profile of PAO1ΔmexEF mexT++ was compared with PAO1 mexT++. Eighteen of the genes downregulated by MexT in wild-type were upregulated in PAO1ΔmexEF mexT++, indicating their regulation was dependent on mexEF expression. In contrast, none of the 17 genes (apart from the mexEF-oprN operon) induced by MexT in wild-type was altered in PAO1ΔmexEF mexT++ compared to PAO1 mexT++ (Table 2), indicating their induction by MexT was independent of mexEF expression. This was also evident from the changes in MA plot of PAO1 mexT++ compared to PAO1ΔmexEF mexT++ whereby only three genes (mexE, mexF and oprN) were indicated as being highly induced, while 18 genes were highly reduced (Figure 1B). The differentiation of regulatory effects of MexT with regard to their dependency on the MexEF-OprN efflux pump was also evident in the 3D scatter plot (Figure 1C). The expression of the majority of genes was similar among the three samples, forming the main cloud body. In contrast, genes whose expression was highly altered were positioned out of the main cloud body in the 3D scatter plot. These were further categorized into two groups: one incorporating the genes induced by overexpressing MexT independent of MexEF-OprN (in red oval) and the second incorporating the genes downregulated by overexpressing MexT dependent of MexEF-OprN (in green oval). This supports the MA plot analysis and demonstrates that the most significant induction in gene expression by MexT is independent of MexEF-OprN efflux pump, while the most significant reduction by MexT is dependent of MexEF-OprN, in the condition tested. These results support our hypothesis that the genes highly induced by MexT are direct targets of the regulator.
Figure 1.
(A and B) MA-plots showing the relationship between the logarithmic mean signal ratios, M value, and the logarithmic mean signal intensities, A value, are used to spot genes whose expression is significantly altered in (A) PAO1 mexT++ compared to PAO1 and (B) PAO1 mexT++ compared to PAO1ΔmexEF mexT++. The spots representing genes altered more than 10-fold are labelled in red. A positive M value indicates increased gene expression, while a negative value indicates reduced expression. (C) 3D scatter-plot projecting the logarithmic mean signal intensities obtained from PAO1, PAO1 mexT++ and PAO1ΔmexEF mexT++ is used to spot genes whose expression was altered by overexpressing MexT dependent or independent of the MexEF-OprN efflux pump. (Left) The orientation of the main cloud body is adjusted towards the reader, genes highly activated by overexpressing MexT independent of the MexEF-OprN efflux pump are grouped in the red oval and genes highly reduced by overexpressing MexT dependent of the MexEF-OprN efflux pump are grouped in the green oval. (Right) The orientation of the main cloud body is adjusted to show the full scale.
Table 2.
Genes of expression altered more than 10-fold in response to MexT overexpression
| Gene No.a | Gene name | PAO1 mexT++ versus PAO1 controlb | PAO1ΔmexEF mexT++ versus PAO1 mexT++c | Protein description |
|---|---|---|---|---|
| PA0852 | cbpD | −9.4 | 20 | Chitin‐binding protein CbpD precursor |
| PA0996 | pqsA | −64 | 95 | Probable coenzyme A ligase |
| PA0997 | pqsB | −51 | 72 | Homologous to β-keto-acyl-acyl-carrier protein synthase |
| PA0998 | pqsC | −219 | 241 | Homologous to β-keto-acyl-acyl-carrier protein synthase |
| PA0999 | pqsD | −72 | 86 | 3-Oxoacyl-[acyl-carrier-protein] synthase III |
| PA1000 | pqsE | −87 | 76 | Quinolone signal response protein |
| PA1001 | phnA | −126 | 95 | Anthranilate synthase component I |
| PA1002 | phnB | −34 | 23 | Anthranilate synthase component II |
| PA1657 | −36 | <2 | Conserved hypothetical protein | |
| PA1658 | −12 | <2 | Conserved hypothetical protein | |
| PA1718 | pscE | −12 | <2 | Type III export protein PscE |
| PA1744 | 48 | <2 | Hypothetical protein | |
| PA1869 | −54 | 11 | Probable acyl carrier protein | |
| PA1970 | 237 | <2 | Hypothetical protein | |
| PA2193 | hcnA | −52 | 11 | Hydrogen cyanide synthase HcnA |
| PA2194 | hcnB | −32 | <2 | Hydrogen cyanide synthase HcnB |
| PA2195 | hcnC | −11 | <2 | Hydrogen cyanide synthase HcnC |
| PA2386 | pvdA | <2 | 10 | l-ornithine N5-oxygenase |
| PA2486 | 338 | <2 | Hypothetical protein | |
| PA2491 | mexS | 75 | <2 | Probable oxidoreductase |
| PA2492 | mexT | 23.3 | <2 | Transcriptional regulator MexT |
| PA2493 | mexE | 2033 | −1714 | RND multidrug efflux membrane fusion protein MexE precursor |
| PA2494 | mexF | 1382 | −9.5 | RND multidrug efflux transporter MexF |
| PA2495 | oprN | 340 | −10 | Multidrug efflux outer membrane protein OprN precursor |
| PA2759 | 72 | <2 | Hypothetical protein | |
| PA2811 | 34 | <2 | Probable permease of ABC‐2 transporter | |
| PA2812 | 12 | <2 | Probable ATP‐binding component of ABC transporter | |
| PA2813 | 20 | <2 | Probable glutathione S‐transferase | |
| PA3205 | 11 | <2 | Hypothetical protein | |
| PA3229 | 1789 | <2 | Hypothetical protein | |
| PA3326 | −75 | 26 | Probable Clp-family ATP-dependent protease | |
| PA3331 | −26 | 5.0 | Cytochrome P450 | |
| PA3332 | −18 | <2 | Conserved hypothetical protein | |
| PA3333 | fabH2 | −13 | 3.7 | 3-Oxoacyl-[acyl-carrier-protein] synthase III |
| PA4141 | −32 | 19 | Hypothetical protein | |
| PA4354 | 11 | <2 | Conserved hypothetical protein | |
| PA4623 | 74 | <2 | Hypothetical protein | |
| PA4770 | lldP | −43 | 46 | l-lactate permease |
| PA4771 | lldD | −28 | 70 | l-lactate dehydrogenase |
| PA4772 | −7.0 | 19 | Probable ferredoxin | |
| PA4881 | 751 | <2 | Hypothetical protein |
aGene number from the Pseudomonas Genome Project (http://www.pseudomonas.com).
bFold change in gene expression of PAO1 (pME6032-mexT) compared to PAO1 (pME6032); positive value means expression increased and negative value means expression decreased.
cFold change in gene expression of PAO1ΔmexEF (pME6032-mexT) compared to PAO1 (pME6032-mexT ).
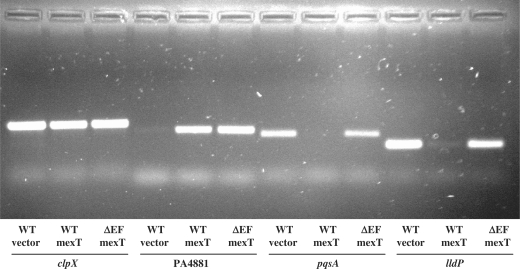
To test the validity of the microarray data, genes whose expression was significantly altered (PA4881, pqsA and lldP) were selected for semi-quantitative reverse transcription PCR analysis (Figure 2). RNA was prepared from biological replicates not used in the arrays. The housekeeping gene clpX was used as the internal control. The transcript level of PA4881 was induced by overexpressing MexT in both PAO1 and PAO1ΔmexEF strains but was not detectable in the PAO1 strain containing pME6032. The transcript levels of pqsA and lldP were reduced by overexpressing MexT in PAO1 but not in PAO1ΔmexEF strains. Furthermore, the expression from promoter fusions of a selected number of genes were analysed in the wild-type PAO1 and a heterogeneous E. coli background, with and without pME6032-mexT, and all genes were highly induced by overexpressing MexT in both backgrounds (Table 3). In all cases, the results confirmed the microarray data.
Figure 2.
Semi-quantitative reverse transcription PCR validation of selected genes identified by microarray analysis as significantly altered by MexT. Total RNA was isolated from PAO1 containing pME6032 (WT vector), PAO1 containing pME6032-mexT (WT mexT) or PAO1ΔmexEF containing pME6032-mexT (ΔEF mexT). The identity of the genes targeted in each set of PCRs is indicated at the bottom of the figure.
Table 3.
The regulatory effect of MexT on the expression of the promoter-lacZ fusions of putative target genes
| Strain (plasmid)→ | PAO1 | PAO1 | PAO1 nfxC | E. coli DH5a | E. coli DH5a | |
|---|---|---|---|---|---|---|
| Promoter-lacZ↓ | (pME6032) | (pME6032-mexT) | PAO1 nfxCΔmexT | (pME6032)a | (pME6032-mexT)a | |
| pMP-PAmexEp | 45 ± 1 | 3991 ± 912 | 954 ± 21 | 33 ± 2 | 9 ± 1 | 230 ± 1 |
| pMP-PA1744p | 120 ± 32 | 2801 ± 143 | NAb | NA | 7 ± 3 | 7520 ± 849 |
| pMP-PA2759p | 52 ± 5 | 896 ± 104 | NA | NA | 2 ± 1 | 4147 ± 666 |
| pMP-PA3229p | 76 ± 19 | 4852 ± 120 | NA | NA | 4 ± 1 | 1148 ± 96 |
| pMP-PA4354p | 824 ± 83 | 3385 ± 462 | NA | NA | 3 ± 1 | 143 ± 1 |
| pMP-PA4623p | 23 ± 3 | 4302 ± 30 | 1714 ± 86 | 29 ± 4 | 41 ± 4 | 6479 ± 724 |
| pMP-PA4881p | 34 ± 11 | 1217 ± 91 | 716 ± 43 | 20 ± 1 | 8 ± 2 | 7783 ± 465 |
| Mutated DNA motif: | ||||||
| pMP-PA4623m1p | 30 ± 10 | 39 ± 6 | 25 ± 2 | 26 ± 1 | 21 ± 19 | 18 ± 12 |
| pMP-PA4881m1p | 38 ± 9 | 12 ± 5 | 22 ± 3 | 20 ± 2 | 13 ± 3 | 12 ± 1 |
| Homologous genes across Pseudomonas species: | ||||||
| pMP-Pfl2659p | NA | NA | NA | NA | 9 ± 1 | 801 ± 15 |
| pMP-Pfl3748p | NA | NA | NA | NA | 7 ± 1 | 10 474 ± 202 |
| pMP-PP3425p | NA | NA | NA | NA | 4 ± 1 | 2231 ± 168 |
| pMP-PP4858p | NA | NA | NA | NA | 2 ± 1 | 3288 ± 43 |
aIPTG (1 mM final conc.) was added in the medium.
bNA, not assayed.
Due to the limited number and lack of good annotation of the significantly altered genes in the expression profiles (Supplementary Table S2), the pathway mapping through main pathway databases such as KEGG (www.genome.jp/kegg/pathway.html) provided limited information. However, the expression of several operons with known cellular functions was significantly altered by MexT in a mexEF-oprN-dependent way. The transcript levels of the pqs and phn operon genes, encoding enzymes involved in the synthesis of PQS signal molecules, showed a 30- to 220-fold reduction in PAO1 wild-type, when mexT was overexpressed and a 23- to 240-fold induction in PAO1ΔmexEF mexT++ relative to PAO1 mexT++. Similarly, the hcn operon genes, encoding hydrogen cyanide synthase, showed a 11- to 52-fold reduction in PAO1 wild-type overexpressing mexT, while hcnA was upregulated 11-fold in PAO1ΔmexEF mexT++ relative to PAO1 mexT++. Previously we demonstrated a considerable reduction in hcnB expression by overexpressing mexT in PAO1 strain but not in PAO1ΔmexEF strain using semi-quantitative reverse transcription PCR (18). Our results indicated that pqs and hcn operons were indirect targets of MexT, as the modulation in expression was dependent on the overexpression of the MexEF-OprN efflux pump. A reduction in expression of pqs and hcn operons was also observed previously in the transcriptome profiling of an nfxC-type mutant strain overexpressing mexEF-oprN (10). The microarray data presented here also suggest that the lldPD operon (PA4770-PA4772), PA3326, and the PA3331-PA3334 operon are novel MexEF-OprN-dependent indirect targets of MexT.
The set of genes upregulated by MexT independent of MexEF included mexS, previously shown to be activated by MexT in PAO1 (6). The mexS gene is located adjacent to mexT and divergently transcribed, which is a typical arrangement of genes controlled by LysR-type activators (13). In our transcriptome data, the mexS transcript level increased 75-fold in PAO1 overexpressing MexT and was not changed in PAO1ΔmexEF mexT++ relative to PAO1 mexT++ (Table 2). These data supported the hypothesis that mexS is an MexE-independent target of MexT in P. aeruginosa PAO1.
Twelve additional genes were upregulated by overexpressing mexT independent of MexEF-OprN (Table 2). Most of these genes were annotated as of unknown function but included genes with probable transporter functions; PA2813-PA2811 encode components of a probable ABC transporter and the PA4354-PA4356 operon encodes a putative repressor, a probable major facilitator superfamily (MFS) transporter and a xenobiotic reductase, respectively. We hypothesized that this set of genes includes direct positively regulated targets of MexT and suggests that MexT may be a key regulator of cellular homeostasis.
Identification of a conserved DNA motif required for MexT regulated expression.
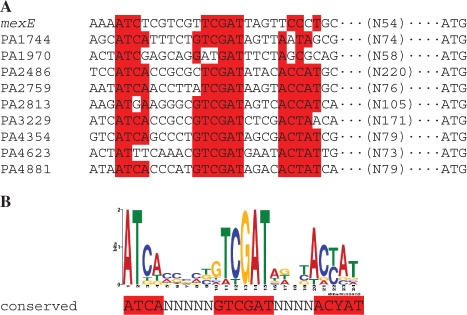
In order to identify possible direct targets of MexT and a potential consensus sequence for MexT binding, the genes that were most highly induced (≥10-fold) by MexT in PAO1 independent of mexEF were selected for further analysis (Table 2). In addition to mexS and mexEF-oprN, the 12 genes showing induction ≥10-fold were PA1744, PA1970, PA2486, PA2759, PA2813-PA2811, PA3205, PA3229, PA4354, PA4623 and PA4881. The DNA sequence of the upstream regulatory region (−500 bp from the ATG translational start site) of each gene (including PA2813 only of the PA2813-PA2811 operon) was aligned using MEME Suite online software (24). A well-conserved DNA motif ATCA-N5-GTCGAT-N4-ACYAT was identified in the upstream regions of 9 of the 12 genes and mexE (Figure 3). Interestingly, a clear consensus sequence was not identified in the mexS promoter region. The DNA motif overlapped the ATC-N9-GAT sequence, referred to as a ‘nod-box’ and proposed as the general motif of the binding sites for LysR-family proteins (13). Previously, this ‘nod-box’ was proposed to be part of the binding site of MexT in the upstream region of mexE (6).
Figure 3.
Alignment of the predicted conserved DNA motif in the upstream regulatory regions of P. aeruginosa genes highly induced by MexT (A) and sequence logo for the conserved DNA motif reflecting position-specific probability matrixes (B) deduced from MEME software analysis. Nucleotides with high probability (≥70%) are highlighted by red. The number of nucleotides between the conserved DNA motif and the start codon ATG is shown in brackets.
To confirm the transcriptional activation of these putative target genes by MexT, promoter-lacZ fusions of selected genes were constructed, transformed into strains of different genetic backgrounds and the level of expression monitored when MexT was overexpressed (Table 3). In the wild-type PAO1 strains, the expression level of all the promoters containing the newly deduced conserved DNA motif was highly induced by co-transforming pME6032-mexT, compared to the empty vector control (Table 3). As MexEF-OprN is normally quiescent in wild-type cells, but highly induced in nfxC-type mutants (7), we investigated if the genes were also induced in an nfxC-type mutant background. Therefore, an nfxC-type mutant was generated by plating the wild-type strain PAO1 on LB agar plates containing chloramphenicol at 600 µg/ml, a condition previously shown to select nfxC-type mutants (7). A selection of promoter-lacZ fusion constructs were transformed into the nfxC mutant and high levels of expression were observed from all constructs tested, including mexE (Table 3). When the mexT gene was deleted in this nfxC-type mutant the expression of these genes was attenuated (Table 3). We also tested other nfxC-type mutant strains from different sources, and all of them showed high induction of PA4881 (data not shown). These results indicated that all the genes with the conserved DNA motif were highly induced in the nfxC-type mutant strains and thus, induced by MexT in physiologically relevant conditions.
To further investigate if these genes are direct targets of MexT, we assessed if MexT could activate their expression in a heterogenous E. coli background. The promoter-lacZ fusion plasmids and pME6032-mexT or pME6032 were co-transformed into the E. coli DH5α strains. The mexT gene cloned in pME6032-mexT was highly expressed in P. aeruginosa PAO1 without IPTG induction (Table 2). However, in E. coli, the native mexT promoter was not induced and IPTG induction was essential for full activation (data not shown). Therefore, a saturated concentration of IPTG (1 mM) was used to induce mexT expression from pME6032-mexT in E. coli. As shown in Table 3, low expression levels of the putative MexT target promoters, including mexE was observed in the E. coli strains containing empty vector pME6032. In contrast, when mexT was overexpressed, the expression of the promoters were significantly induced (Table 3). These results indicate that MexT may directly activate the expression of the newly identified target genes in P. aeruginosa. This was further supported by the observation that an overexpressing mexT construct, cloned from a different PAO1 wild-type available in the laboratory collection, and which contained a single base-pair mutation that resulted in an alanine to valine (A to V) change in the 39th residue of the MexT protein, failed to induce the expression of these promoter fusions in either PAO1 or E. coli DH5α backgrounds (data not shown). According to the Pfam database alignment (25), the 39th residue is located in the helix–turn–helix motif of MexT, the DNA-binding motif of the protein. Indeed our results suggested that a mutation in this domain rendered the protein non-functional. While other inactivating mutations in the mexT gene have been reported (9) this single base pair mutation has not been previously identified in wild-type strains and appears to be a novel mechanism of inactivating mexT. Subsequently, to confirm that the conserved DNA motif in the promoter region was required for MexT activation of the putative target genes, site-directed mutagenesis was carried out to mutate GTCGAT to GTCTGT in the DNA motif of PA4623 and PA4881 promoters. This alteration in the DNA motif completely attenuated activation of the promoters by MexT in the wild-type PAO1, PAO1nfxC and the heterogenous E. coli DH5α (Table 3). Thus, the conserved DNA motif is required for the induced expression of these genes by MexT.
MexT protein binds to the conserved DNA motif in target promoters
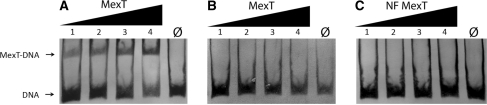
In order to verify if the MexT protein directly binds the conserved DNA motif, a plasmid expressing C-terminal His-tagged MexT protein was constructed. This tagged MexT protein activated the expression of PA4881 promoter-lacZ fusion in E. coli, indicating the protein was functional (data now shown). EMSAs were performed to detect binding of MexT to two 207-bp DNA fragments containing the unaltered or mutated PA4881 promoter region. The MexT protein clearly bound to the DNA fragments containing the intact conserved DNA motif (Figure 4A), but not to the DNA fragments containing the mutated conserved DNA motif (Figure 4B). The specificity of MexT binding to its DNA target was confirmed by a competition assay with an excess of unlabeled target DNA (Supplementary Figure S1). These results demonstrated, for the first time, that MexT protein bound to the conserved DNA motif and that a mutation in two of the most conserved nucleotides in the motif abolished the binding of the MexT protein.
Figure 4.
Purified MexT protein binds to the conserved DNA motif in the promoter region of PA4881. EMSAs were carried out applying 0 ng, 37.5 ng, 75 ng, 150 ng and 300 ng (lanes Ø, 1, 2, 3 and 4) of purified His6-tagged functional (A and B) or non functional (C) MexT proteins to 10 fmol of DIG-labelled DNA target containing the intact conserved DNA motif (A and C), or the mutated DNA target with the conserved DNA motif disrupted (B). Arrow indicates the free DNA probes and the band shifts of the MexT–DNA complex.
To further investigate MexT binding to this conserved motif, the residue 39 A to V mutated non-functional MexT was purified using the His-tag expression plasmid previously used for the wild-type MexT protein. EMSAs were performed to detect binding of the mutated MexT protein to the conserved DNA motif and no binding was detected (Figure 4C). These results demonstrated that this particular mutation in the helix–turn–helix motif abolished the binding of MexT protein to the target DNA.
MexT activates the expression of target genes across other Pseudomonas species
Orthologues of the mexEF-oprN operon can be found in all Pseudomonas genomes sequenced to date. The mexT gene is located immediately upstream and transcribed in the same direction as mexEF-oprN in all P. aeruginosa strains and P. stutzeri A1501 strain but not in other species. For instance, in the genome of P. fluorescens PfO-1 mexT (Pfl01_2666) and mexE (Pfl01_2659) are separated by six open reading frames (ORFs) and in the genome of P. putida KT2440 mexT (PP_2826) and mexE (PP_3425) are located in totally different loci. To date, it has not been shown if the expression of these mexE genes is activated by MexT. In order to investigate this, promoter-lacZ fusions of Pfl01_2659 and PP_2826 were constructed and transformed into E. coli strains containing pME6032-mexT. The expression of both mexE fusions was highly induced by MexT in E. coli (Table 3).
Two newly identified MexT target genes of particular note were PA4881 and PA4623, which encode hypothetical proteins of 113 amino acids and 129 amino acids, respectively (Pseudomonas Genome Database). PA4881 and PA4623 actually encode homologous proteins, each containing four tandem repeat motifs of average 23 amino acids (Figure 5A and B). Genes encoding tandem repeat proteins are widely distributed in the genomes of different organisms (26). Indeed, the genomes of various Pseudomonas species harbour a number of genes encoding tandem repeat proteins, which share some conserved residues with PA4881 and PA4623 encoded proteins (Figure 5 and Supplementary Figure S2). Among them, several genes contain the conserved MexT-binding motif in their upstream regulatory regions (Figure 5C–F). To address the question if these genes can be induced by MexT, promoter-lacZ fusions of two tandem repeat proteins, P. fluorescens PfO-1 Pfl01_3748 and P. putida KT2440 PP_4858 were constructed and transformed into E. coli strains containing pME6032-mexT. The expression of both promoter fusions was highly induced by MexT in E. coli (Table 3). These data suggest that MexT activates the expression of a novel regulon across Pseudomonas species by binding to a conserved DNA motif in the promoter regions of target genes.
Figure 5.
Upstream regulatory DNA sequences and amino acid sequences of PA4623 (A) and PA4881 (B) and tandem repeat proteins in other Pseudomonas (C–F). Locus IDs are from the Pseudomonas Genome Project (http://www.pseudomonas.com). The conserved DNA motif is underlined and the start codon ATG is boxed. The amino acid sequence of each protein is organized showing alignment of tandem repeats with the position number of the first amino acid residue at the beginning of each line.
DISCUSSION
Identification of a novel MexT regulon
Previously, MexT was known to be a transcriptional activator of mexEF-oprN, an operon encoding a multidrug efflux pump in P. aeruginosa (6). Based on our observation that MexT modulated certain virulence factors in a mexEF-oprN-independent manner (18), we hypothesized that MexT has a wider function than previously thought and plays a more global regulatory role in P. aeruginosa. To test this hypothesis, we aimed to identify and characterize other regulatory targets in P. aeruginosa. Here, we present a genomics-based strategy for uncovering novel genes regulated by MexT in P. aeruginosa PAO1. The transcriptome profiling allowed the genes regulated by MexT to be divided into those that were indirectly modulated via MexEF-OprN efflux and those that were regulated by MexT independent of this efflux pump. The MexEF-dependent operons identified were all previously shown to be controlled by quorum sensing signalling (27–29). This correlates with the previous report that overexpression of MexEF-OprN per se in nfxC-type mutant strains caused a reduction of virulence determinants controlled by cell-to-cell signalling, possibly due to decreased intracellular PQS levels resulting from the efflux of either PQS or a PQS precursor (3,15).
Our main interest, however, was in the observation that in addition to mexE and mexS, 12 genes were highly induced by MexT independent of MexEF-OprN (Table 2). When the upstream regulatory region of these genes was compared, a well-conserved DNA motif was identified (Figure 3). The conserved motif contained the previously described LysR regulator ‘nod-box’, ATC-N9-GAT, but also conserved nucleotides immediately 5′ of the GAT and an additional conserved sequence 4 nucleotides 3′ of the GAT, consisting of ACT/CAT. This sequence was highly conserved in most of the promoters but with some changes in mexE, PA1744 and PA1970. The motif was confirmed experimentally as a MexT-binding site (Figure 4) and that the GA of the ‘nod-box’ was essential for MexT binding under the conditions tested. Interestingly, when a P. aeruginosa genome-wide search (http://bioinfo.hku.hk/GenoList/index.pl?database=aerulist) was performed to identify genes that carried a similar sequence motif in their upstream regions, a number of genes were identified that were not altered by MexT in the array analysis. This information warrants further investigation and may be valuable in follow on studies of the MexT-binding site itself and of physiological conditions that may influence binding.
Two of the MexT regulated genes identified in the array analysis do not contain the consensus sequence; PA3205, which was induced just 11-fold, but more interestingly, mexS, the divergently transcribed gene previously shown to be induced by MexT (6) does not even contain the core LysR ATC-N9-GAT consensus motif. Furthermore, no binding of the MexT protein to the mexS promoter region was detected by EMSA under the conditions used for PA4881 promoter binding (data not shown). This was surprising as, although it is possible that mexS is indirectly regulated by MexT, available data suggest that mexS should be a direct target of MexT. Firstly, the genomic arrangement of mexS-mexT is a typical arrangement of a LysR-type regulator and its target gene, and it is well conserved across Pseudomonas species. Secondly, in addition to its induction by MexT in P. aeruginosa, mexS can be activated by MexT in a heterogeneous E. coli background (6). This anomaly warrants further study, not least to investigate the kinetics of MexT binding under different experimental conditions and to investigate the possibility that MexT may also bind to a modified binding site under different conditions. It has been shown that different co-factors can modify the conformation of the same LysR-type regulator protein and therefore the protein could have different targets according to one or either conformation (13). To date the co-factor(s) of MexT has not been identified but may indeed influence its binding to target genes.
It has also been shown that a LysR-type regulator can be an activator or a repressor for different target genes (13). The transcriptome data revealed that a number of genes were downregulated >2-fold by MexT independent of mexEF expression (Supplementary Table S2). Interestingly, one of these was a gene (pscE) encoding a type-three secretion protein. Previously, we demonstrated that MexT exerted a negative effect on virulence traits, including type-three secretion, independent of MexEF-OprN, when grown under TTSS inducing conditions (18). Furthermore, a LysR-type regulator YtxR has been shown to have a global regulatory role in Yersinia enterocolitica, including direct suppression of the transcriptional expression of TTSS (30). However, none of the downregulated genes contained an ATC-N9-GAT DNA motif and it remains to be clarified whether they are direct or indirect targets of MexT regulation.
Nevertheless, a novel regulon including mexEF-oprN directly activated by MexT has been identified and this is an important step towards understanding the global regulatory role of MexT and the physiological role of MexEF-OprN multidrug efflux pump as a component of this regulon in P. aeruginosa.
Possible cellular function of the novel MexT regulon
In addition to mexEF-oprN, the novel MexT regulon identified in this study contains two additional putative operons (PA2813-PA2812-PA2811 and PA4354-PA4355-xenB) (Table 2 and Supplementary Table S2). PA2813-PA2812-PA2811 encodes a probable glutathione S-transferase, a probable ATP-binding component of ABC transporter, and a probable permease of ABC-2 transporter, respectively. Glutathione S-transferases constitute a large family of enzymes, which catalyze the addition of glutathione to endogenous or xenobiotic, often toxic electrophilic chemicals. In Pseudomonas species, multiple glutathione S-transferases are specifically involved in mineralization of recalcitrant compounds, while glutathione S-transferase was known to be involved in detoxification in eukaryotes (31). The putative operon PA4354-PA4355-xenB encodes an ArsR-type transcriptional regulator, a probable major facilitator superfamily (MFS) transporter and a xenobiotic reductase, respectively. The ArsR-type transcriptional regulators are prokaryotic metalloregulatory transcriptional repressors. They repress or derepress the expression of operons involved in the detoxification of di- and multivalent heavy metal ions in the absence or presence of these toxics (32). The MFS is the largest characterized family of transporters, capable of transporting various substrates, such as sugars, polyols, drugs, neurotransmitters, amino acids, peptides, and inorganic anions (33). The xenobiotic reductase is a member of the Old Yellow Enzyme family (flavoprotein oxidoreductases) and catalyze the degradation of explosive compounds such as nitroglycerin, 2,4,6-trinitrotoluene and hexahydro-1,3,5-trinitro-1,3,5-triazine in Pseudomonas species (34–36). From the compilation of these possible functions, the MexT regulon is likely involved in a broad-spectrum detoxification or nutrient-scavenging in P. aeruginosa.
The MexT regulon also contains six genes (PA1744, PA1970, PA2759, PA3229, PA4623 and PA4881) encoding small proteins (70–130 amino acids) of unknown function but with either type I or type II export signal peptides (37). This suggested that MexT may induce a family of cell envelope or secreted proteins. Of particular note are PA4623 and PA4881, two homologue genes encoding tandem repeat proteins (Figure 5A and B). While the function of these proteins is unknown, recently, it was reported that Ehrlichia chaffeensis, an obligately intracellular bacterium, exploits the host cell by a secreted, differentially expressed, tandem repeat protein interacting with multiple host proteins associated with cell signalling, transcriptional regulation and vesicle trafficking (38). Although there is no primary sequence similarity between P. aeruginosa tandem repeat proteins encoded by PA4881 and PA4623 and this E. chaffeensis tandem repeat protein, it is tempting to speculate that these P. aeruginosa tandem repeat proteins may be involved in the interaction with host proteins and warrants further investigation. Furthermore, PA4623 and PA4881 as well as PA3229 and mexEF-oprN were shown to be induced in P. aeruginosa, after a 12-h interaction with primary normal human airway epithelial cells (39). The role of these proteins in host–cell interactions remains to be elucidated but the report that nfxC-type mutants were readily recovered from an experimental model of rat pneumonia in the absence of antibiotic selection (40) indicates that there is some advantage to MexT regulon expression in vivo.
This is supported by the fact that many Pseudomonas species have a number of genes encoding tandem repeat proteins, which contain the identical conserved MexT binding motif in their upstream regulatory regions (Figure 5) and can be activated by MexT (Table 3). Moreover, the component genes of the MexT regulon are widely distributed on the genome of P. aeruginosa PAO1. These facts strongly suggest that the MexT regulon may serve primary functions in Pseudomonas species and understanding their function warrants further investigation. This may also provide new insights into the physiological role of MexEF-OprN pump in Pseudomonas species as the pump may function in an integrated way with other components in the MexT regulon.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Commission (MTKD-CT-2006-042062; O36314); the Science Foundation of Ireland (SFI 07/IN.1/B948; 08/RFP/GEN1295); the Department of Agriculture and Food (DAF RSF 06 321; DAF RSF 06 377); Irish Research Council for Science, Engineering and Technology (IRCSET) (05/EDIV/FP107); the Health Research Board (RP/2006/271; RP/2007/290); the Environmental Protection Agency (EPA 2006-PhD-S-21; 2008-PhD-S-2); and the Natural Science Foundation of China (to Y.-P.W.) (30830005). Funding for open access charge: Science Foundation Ireland Principal Investigator grant: SFI 07IN.1/B948.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Max Schobert for providing the E. coli strain ST18. We acknowledge Hazel O’Connor and Marlies Mooij for their scientific contributions, Pat Higgins for his technical assistance and Jerry Reen for his comments on the manuscript.
REFERENCES
- 1.Poole K, Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 2.Schweizer HP. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet. Mol. Res. 2003;2:48–62. [PubMed] [Google Scholar]
- 3.Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, Alvarez-Ortega C. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS. Microbiol. Rev. 2009;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- 4.Piddock LJ. Multidrug-resistance efflux pumps – not just for resistance. Nat. Rev. Microbiol. 2006;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- 5.Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köhler T, Epp SF, Curty LK, Pechere JC. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 1999;181:6300–6305. doi: 10.1128/jb.181.20.6300-6305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Curty LK, Pechère JC. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 8.Köhler T, Michéa-Hamzehpour M, Plesiat P, Kahr AL, Pechère JC. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maseda H, Saito K, Nakajima A, Nakae T. Variation of the mexT gene, a regulator of the MexEF-OprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2000;192:107–112. doi: 10.1111/j.1574-6968.2000.tb09367.x. [DOI] [PubMed] [Google Scholar]
- 10.Ramsey MM, Whiteley M. Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 2004;53:1075–1087. doi: 10.1111/j.1365-2958.2004.04181.x. [DOI] [PubMed] [Google Scholar]
- 11.Sobel ML, Neshat S, Poole K. Mutations in PA2491 (mexS) promote MexT-dependent mexEF-oprN expression and multidrug resistance in a clinical strain of Pseudomonas aeruginosa. J. Bacteriol. 2005;187:1246–1253. doi: 10.1128/JB.187.4.1246-1253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savageau MA. Design of molecular control mechanisms and the demand for gene expression. Proc. Natl Acad. Sci. USA. 1977;74:5647–5651. doi: 10.1073/pnas.74.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 14.Goethals K, Van Montagu M, Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc. Natl Acad. Sci. USA. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köhler T, van Delden C, Curty LK, Hamzehpour MM, Pechère JC. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J. Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linares JF, Lopez JA, Camafeita E, Albar JP, Rojo F, Martinez JL. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 2005;187:1384–1391. doi: 10.1128/JB.187.4.1384-1391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, Van Delden C, Curty LK, Köhler T. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 2002;184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Z-X, Mac Aogáin M, O'C;onnor HF, Fargier E, Mooij MJ, Adams C, Wang Y.-P, O'G;ara F. MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb. Pathog. 2009;21:440–446. doi: 10.1016/j.micpath.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 20.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 21.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–574. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 22.Thoma S, Schobert M. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol Lett. 2009;294:127–132. doi: 10.1111/j.1574-6968.2009.01556.x. [DOI] [PubMed] [Google Scholar]
- 23.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.Bailey TL, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- 25.Finn RD, Tate J, Mistry J, Coggill PC, Sammut JS, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katti MV, Sami-Subbu R, Ranjekar PK, Gupta VS. Amino acid repeat patterns in protein sequences: their diversity and structural-functional implications. Protein Sci. 2000;9:1203–1209. doi: 10.1110/ps.9.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolfgang MC, Jyot J, Goodman AL, Ramphal R, Lory S. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl Acad. Sci. USA. 2004;101:6664–6668. doi: 10.1073/pnas.0307553101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alxer-DiPerte GL, Hinchliffe SJ, Wren BW, Darwin AJ. YtxR acts as an overriding transcriptional off switch for the Yersinia enterocolitica Ysc-Yop type 3 secretion system. J. Bacteriol. 2009;191:514–524. doi: 10.1128/JB.01305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vuilleumier S, Pagni M. The elusive roles of bacterial glutathione S-transferases: new lessons from genomes. Appl. Microbiol. Biotechnol. 2002;58:138–146. doi: 10.1007/s00253-001-0836-0. [DOI] [PubMed] [Google Scholar]
- 32.Busenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 33.Flumana N, Bibi E. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim. Biophys. Acta. 2009;1794:738–747. doi: 10.1016/j.bbapap.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 34.Blehert DS, Fox BG, Chambliss GH. Cloning and sequence analysis of two Pseudomonas flavoprotein xenobiotic reductases. J. Bacteriol. 1999;181:6254–6263. doi: 10.1128/jb.181.20.6254-6263.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pak JW, Knoke KL, Noguera DR, Fox BG, Chambliss GH. Transformation of 2, 4, 6-trinitrotoluene by purified xenobiotic reductase B from Pseudomonas fluorescens I-C. Appl. Environ. Microbiol. 2000;66:4742–4750. doi: 10.1128/aem.66.11.4742-4750.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuller ME, McClay K, Hawari J, Paquet L, Malone TE, Fox BG, Steffan RJ. Transformation of RDX and other energetic compounds by xenobiotic reductases XenA and XenB. Appl. Microbiol. Biotechnol. 2009;84:535–544. doi: 10.1007/s00253-009-2024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewenza S, Gardy JL, Brinkman FS, Hancock RE. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 2005;15:321–329. doi: 10.1101/gr.3257305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wakeel A, Kuriakose JA, McBride JW. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signalling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 2009;77:1734–1745. doi: 10.1128/IAI.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frisk A, Schurr JR, Wang G, Bertucci DC, Marrero L, Hwang SH, Hassett DJ, Schurr MJ. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect. Immun. 2004;72:5433–5438. doi: 10.1128/IAI.72.9.5433-5438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Join-Lambert OF, Michéa-Hamzehpour M, Köhler T, Chau F, Faurisson F, Dautrey S, Vissuzaine C, Carbon C, Pechère J. Differential selection of multidrug efflux mutants by trovafloxacin and ciprofloxacin in an experimental model of Pseudomonas aeruginosa acute pneumonia in rats. Antimicrob. Agents Chemother. 2001;45:571–576. doi: 10.1128/AAC.45.2.571-576.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeFlaun MF, Tanzer AS, McAteer AL, Marshall B, Levy SB. Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. Appl. Environ. Microbiol. 1990;56:112–119. doi: 10.1128/aem.56.1.112-119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franklin FC, Bagdasarian M, Bagdasarian MM, Timmis KN. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl Acad. Sci. USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'G;ara F, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. J. Mol. Plant Microb. Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- 44.Spaink HP, Okker RJH, Wijffelman CA, Pees E, Lugtenberg BJJ. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1J1. Plant Mol. Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.