Abstract
A subtelomeric region, 4q35.2, is implicated in facioscapulohumeral muscular dystrophy (FSHD), a dominant disease thought to involve local pathogenic changes in chromatin. FSHD patients have too few copies of a tandem 3.3-kb repeat (D4Z4) at 4q35.2. No phenotype is associated with having few copies of an almost identical repeat at 10q26.3. Standard expression analyses have not given definitive answers as to the genes involved. To investigate the pathogenic effects of short D4Z4 arrays on gene expression in the very gene-poor 4q35.2 and to find chromatin landmarks there for transcription control, unannotated genes and chromatin structure, we mapped DNaseI-hypersensitive (DH) sites in FSHD and control myoblasts. Using custom tiling arrays (DNase-chip), we found unexpectedly many DH sites in the two large gene deserts in this 4-Mb region. One site was seen preferentially in FSHD myoblasts. Several others were mapped >0.7 Mb from genes known to be active in the muscle lineage and were also observed in cultured fibroblasts, but not in lymphoid, myeloid or hepatic cells. Their selective occurrence in cells derived from mesoderm suggests functionality. Our findings indicate that the gene desert regions of 4q35.2 may have functional significance, possibly also to FSHD, despite their paucity of known genes.
INTRODUCTION
One of the best cell culture models for mammalian differentiation is the induction of myotube formation from cultured myoblasts. However, only one high-resolution study of chromatin has been reported for human myoblasts, namely, analysis of histone H4 hyperacetylation on a single fetal myoblast cell strain using a commercial SNP tiling array (1). High-resolution analyses of other human cell types for chromatin epigenetics and annotation-neutral searches for transcripts are revealing evidence for many new differentiation-associated genes, alternative transcription start sites and transcription control elements within genes or located distant from them (2–6).
We have begun analysis of chromatin structure in myoblasts by focusing on 4q35.2, the subtelomeric chromosomal region that contains a muscular dystrophy-linked repeat array, D4Z4. The relationship of D4Z4 to pathogenic gene dysregulation in facioscapulohumeral muscular dystrophy (FSHD) is still enigmatic. FSHD is the only known disease caused by having additional few copies of a long, tandemly repeated sequence (7). More than 95% of FSHD patients have only 1–10 copies of this 3.3-kb repeat unit on one allelic 4q35.2 (Figure 1A), while unaffected individuals have 11–100 copies on both 4q35.2 alleles. This progressive and painful disease is usually diagnosed in the teens. It is initially confined to certain groups of skeletal muscles and has no efficacious treatment. An extremely low copy number of 4q35 D4Z4 repeats (1–3) often correlates with an earlier onset and more severe disease but no FSHD-linked array has been found to have zero copies of the repeat unit (7).
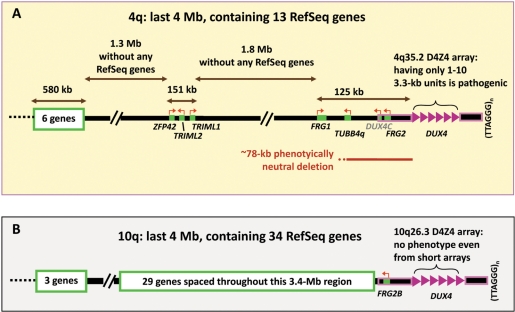
Figure 1.
Schematic comparison of the subtelomeric region of 4q containing FSHD-linked D4Z4 arrays (A) and that of 10q, whose D4Z4 arrays are always non-pathogenic (B). Near their distal end, the 4q35.2 and 10q26.3 regions contain ∼98% identical 4q and 10q D4Z4 arrays (8) and other highly homologous sequences outlined in pink. DUX4, within each D4Z4 repeat unit (pink triangle), has no polyA signal except for the most distal copy, which borrows a poly(A) signal from sequences distal to the array (34,35). DUX4 is weakly expressed, has mostly truncated transcripts, is highly polymorphic in number of copies of the repeat (1–100) and shows no more expression from longer than shorter arrays (34,35). Therefore, it was counted as a single gene for the tally of RefSeq genes. DUX4C (grey font) is a predicted gene, not a RefSeq gene.
Several findings implicate the involvement in FSHD of sequences in cis to a short D4Z4 array on 4q35.2. A short D4Z4 array by itself does not cause FSHD because almost identical arrays on 10q26.3 have no phenotypic effect even when they are equally short (7). This 4q-dependence of pathogenicity is found despite 98% homology between 4q and 10q within D4Z4 and >95% homology centromere proximally for 42 kb and distally for ∼15–25 kb up to the telomere (Figure 1) (7,8). A very small number of single-base sequence variations distinguish canonical 4q35.2 D4Z4 repeat units from those of 10q26.3 (7). A short canonical 4q35-type D4Z4 array on 10q26.3 does not lead to FSHD while a short 10q26.3-type array on 4q35.2 does (9). Therefore, the chromosomal position of the 4q35.2 D4Z4 array links it to FSHD. Nonetheless, there is only controversial or incomplete evidence for functional linkage of FSHD to a 4q35-specific gene despite many expression microarray and real-time PCR (RT-PCR) studies (10–14).
We and others have focused attention on the proximal side of the D4Z4 array in subtelomeric 4q because no genes have been found in the short partially sequenced subregion distal to the array (7). Moreover, the short D4Z4-distal region shares high homology between 4q and 10q (15). Sequences at 4q35.2, which may be implicated in the disease, should be >78 kb proximal to D4Z4 because naturally occurring deletions of sequences within this ∼78-kb proximal region (Figure 1) do not impact the phenotype of FSHD patients carrying a short D4Z4 array in cis (16). The furthest proximal sequence that is considered a disease candidate gene is SLC25A4 (ANT1), which is located on 4q35.1 ∼5 Mb proximal to D4Z4. Disease-linked SLC25A4 overexpression was seen in some studies (17,18) but not others (11–13).
We proposed that there is a long-distance interaction between a short D4Z4 array and some unidentified proximal gene present on subtelomeric 4q, but not on 10q, that results in FSHD-specific transcription dysregulation (19,20). Long-range control of disease-related gene expression in humans can occur by looping interactions of ∼1 Mb and even longer interactions may occur (21,22). The hypothesized long-range pathogenic interaction of short D4Z4 arrays and other 4q sequences in cis should involve a ‘molecular ruler’ that recognizes differences in sizes of D4Z4 arrays slightly above or below a near threshold of 33 kb (10 3.3-kb repeat units). Because it is the most likely region for the postulated cis-interactions, the 4 Mb of 4q35.2 with the addition of the SLC25A4 gene at 4q35.1 were the focus of the present study of chromatin in FSHD and control myoblasts. About 80% of 4q35.2 is devoid of known genes, including no reported micro RNA (miRNA) genes. Approximately 25% of the genome consists of gene-poor regions >500 kb termed gene deserts (23). Studying 4q35.2, which is mostly gene desert, should enhance our understanding of the underlying chromatin structure of the human genome as well as the molecular genetics of FSHD.
Using FSHD and control myoblast cell strains, we mapped DNaseI-hypersensitive (DH) sites at high resolution. DH sites are associated with nucleosome-free chromatin and various gene regulatory elements (24). Approximately 30% of them are in promoters of genes that are active or poised for activity (6,24). Crawford and colleagues (2,6,25) developed two specific high-throughput methods to identify large numbers of DH sites at once, using tiled arrays (DNase-chip) or high-throughput sequencing (DNase-seq). We analyzed myoblasts with DNase-chip, which employs custom tiling arrays and is ideally suited for analysis of targeted regions of the genome. Because DNase-chip and DNase-seq are highly correlated and display similarly high levels of sensitivity (92%) and specificity (94%) (6), we could also compare data from DNase-chip on FSHD and control myoblasts and with DNase-seq mapping of DH sites from other cells types.
MATERIALS AND METHODS
Cell culture
Myoblast cell strains from FSHD patients (F1, 2 and 3; 28-year-old male, 18-year-old female and 14-year female, respectively) were derived from moderately affected, deltoid or quadriceps biopsies of FSHD patients. Their disease-linked D4Z4 arrays had three, three and two 3.3-kb 3 U, respectively. The control myoblasts (C1, 31-year female, 31 years; C2 and C3, two different batches of myoblasts from a 27-year-old male) were from similar biopsies of unaffected individuals. These individuals were unrelated except for the 27-year-old unaffected male, who was the brother of patient F1. Duly signed patient consent forms were obtained that had been approved by the Institutional Review Boards of Tulane Health Science Center and the University of Mississippi Medical Center in Jackson. Myoblasts were propagated and checked by immunocytochemistry for desmin (20), a marker for muscle cells; >90% of the cells in the batches used for these experiments were desmin-positive.
DNA and RNA isolation
Total RNA was extracted with TRIzol reagent (Invitrogen) and treated with DNaseI (Turbo DNA-free, Ambion). cDNA was synthesized (SuperscriptIII, Invitrogen) using random hexamer primers. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed (SYBR Green Detection; iQ5, BioRad) with the following parameters: 95°C, 30 s; 63°C, 30 s; 72°C, 30 s for 45 cycles. For each sample analyzed, RT-minus controls were included. For each primer-pair (Supplementary Table S2), a standard curve with serial 10-fold dilutions of genomic DNA and the melting curve of the product were generated. The slopes of the standard curves were −3.3 ± 0.4 and the correlation coefficients were >0.98. The RNA level is represented as 1000 times the quantity (in arbitrary units) relative to that for human hypoxanthine phosphoribosyltransferase (HPRT).
DNase-chip
DNase-chip was performed as previously described (25). Briefly, myoblasts from normal control and FSHD patients were lysed with NP40 and nuclei were lightly digested with optimal concentrations of DNaseI (Roche). High-molecular weight DNase-treated DNA was prepared, and DNase-digested ends were repaired by T4 DNA polymerase (New England Biolabs). Biotinylated linkers were ligated to the DNase ends and the ligation product was sonicated to an average size of 300–700 bp. DNase ends were captured on streptavidin beads (Invitrogen), sheared ends were blunted with T4 DNA polymerase and repaired ends were ligated to a second set of linkers. DNase-enriched material was amplified by PCR, labeled and hybridized to custom tiling arrays (NimbleGen). These tiling arrays contained probes from across 4q35.2 [chromosome 4 (chr4): 187 300 001–191 273 063; all positions are relative to hg18, UCSC Genome Browser] and the ANT1 locus (chr4:186 291 392–186 315 418). Because there are many repetitive sequences within these regions, we included probe sets that overlapped moderately repeated sequences, including probe sets from the 3.3-kb D4Z4 repeat, but not highly repeated sequences (http://www.repeatmasker.org/). All probes (average ∼30-nt overlap) were designed using an isothermal probe selection strategy, where probes were 45–75 nt in length and were size-adjusted to give a Tm of 76°C. Due to the extremely high GC content (73%) and repetitive nature of the D4Z4 region, two additional sets of isothermal probes were designed to have a Tm of 79 or 82°C. DNase-chip data were analyzed as previously described (25,26). Data were visualized in custom tracks of the UCSC Genome Browser (http://genome.ucsc.edu/) or the Integrated Genome Browser (http://www.affymetrix.com/).
RESULTS
Gene deserts in 4q35.2
The terminal 3 Mb of the q arm of chr4 in 4q35.2 has the lowest gene density of all the autosomal q arms (Supplementary Figure S1). Within 4q35.2, there are two central gene deserts occupying 3.1 Mb and separated by three genes, ZFP42, TRIML1 and TRIML2 (Figure 2). ZFP42 is a marker for pluripotent stem cells (27). TRIML1/Triml1, which encodes a RING finger protein, is expressed in preimplantation mouse embryos (28). Similarly, the related provisional geneTRIML2 seems to be very restricted in its expression (http://biogps.gnf.org). Consistent with previous findings about the regions of low gene density (29), the 4q35.2 gene desert regions consist predominantly of low (G + C) isochores [IsoFinder (30)] and have low concentrations of short interspersed repeats (SINEs; Supplementary Figure S2).
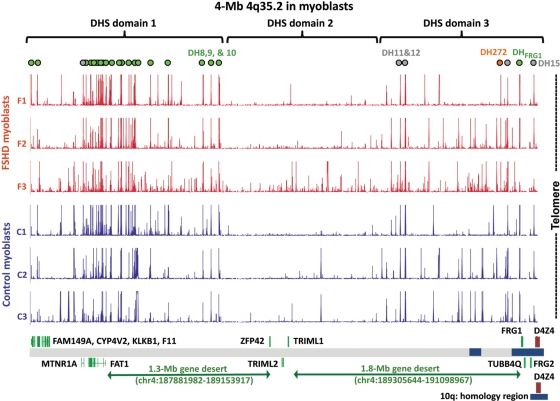
Figure 2.
DH sites at 4q35.2 in myoblast cell strains are present in gene deserts as well as in the vicinity of known genes. Green dots, unique DH sites, seen in all six myoblast cultures; orange dot, DH272, seen preferentially in the three FSHD myoblast cell strains and grey dots, DH sites overlapping STRs (70) and seen in all six myoblast cultures. DH site domains are indicated as described in the text. Blue boxes near the bottom of the figure are regions of segmental duplications shared with various chromosomes; the 4q/10q segmental duplication is shown at the bottom of the figure. SLC25A4 (ANT1), which is 5 Mb proximal to D4Z4 and included in the probe set, is not indicated in this figure because it is located on 4q35.1.
DH sites in gene deserts in myoblasts
We mapped DH sites in vivo in three FSHD and three normal-control myoblast cell cultures at 4q35.2 by DNase-chip. The DH fraction (labeled with Cy5) and the randomly sheared DNA (labeled with Cy3) were cohybridized to custom tiling arrays. Our DH mapping results indicated that 4q35.2 in myoblasts has three chromatin domains differing in the density of DH sites. The most proximal (DHS domain 1, Figure 2) had the highest density of DH sites seen at 4q35.2, in accord with its higher gene content. However, DH sites in this domain extended far into the adjacent gene desert. The distal domain (DHS domain 3) contains several genes near its telomeric end. Only one of these, FRG1, is expressed at a substantial level in myoblasts or other tested cell types (Table 1). Surprisingly, the DH sites observed in all myoblast strains in this domain included sites that extended 0.9 Mb from FRG1 into the neighboring gene desert. DHS domain 2, which is located in the middle of 4q35.2, had a much lower density of DH sites, and none was detected in more than two of the six myoblast cultures. The only genes in this domain are ZFP42, TRIML1 and TRIML2, which are associated with stem cells or early embryogenesis, as mentioned earlier.
Table 1.
DH sites and Refseq genes in the 4q35.2 region
| Gene | DH sites within gene region | Reported RNA in skeletal musclea | Status of geneb | Distance to D4Z4 (kb) | Gene coordinates (hg18) |
|||
|---|---|---|---|---|---|---|---|---|
| Start | End | Strand | ||||||
| FAM149A | Promoter | Moderate | Curated | 3893 | 187 307 320 | 187 330 811 | + | |
| CYP4V2 | First intron | Moderate | Curated | 3852 | 187 349 667 | 187 371 611 | + | |
| KLKB1 | None | Little or no | Curated | 3807 | 187 385 665 | 187 416 619 | + | |
| F11 | None | Little or no | Curated | 3776 | 187 424 111 | 187 447 829 | + | |
| MTNR1A | First intron | Little or no | Curated | 3510 | 187 713 531 | 187 691 802 | − | |
| FAT1 | One in the promoter and six in introns | Moderate | Curated | 3342 | 187 881 981 | 187 745 930 | − | |
| ZFP42 | None | Little or no | Curated | 2060 | 189 153 918 | 189 163 193 | + | |
| TRIML2 | None | Little or no | Provisional | 1960 | 189 263 402 | 189 249 420 | − | |
| TRIML1 | None | Little or no | Provisional | 1918 | 189 297 591 | 189 305 643 | + | |
| FRG1 | Promoter | Moderate | Curated | 102 | 191 098 967 | 191 121 353 | + | |
| TUBB4Qc | None | Little or no | Pseudogene | 81 | 191 143 018 | 191 140 671 | − | |
| DUX4Cd | None | None | Predicted | 43 | 191 180 814 | 191 177 249 | − | |
| FRG2 | None | Little or no | Provisional | 38 | 191 185 406 | 191 182 516 | − | |
| DUX4e | None | Low | Curated | Inside D4Z4 | 191 226 073 | 191 227 684 | + | |
aRelative RNA levels are based on gene expression array data from nine reconstructive surgery skeletal muscle samples [http://bioinfo.amc.uva.nl/HTMseq/controller, (31)] and four skeletal muscle samples (http://biogps.gnf.org). All genes listed as having moderate expression in skeletal muscle are expressed in a variety of tissues. ZFP42 and TRIML1 have very tissue specific and development-restricted expression, as described in the text. TRIML2 has low expression in skeletal muscle.
bThe status of these gene is based on RefSeq data from National Center for Biotechnology Information (NCBI).
cTUBB4Q is considered to be a pseudogene based upon the sequence of its open reading frame and lack of transcripts (32).
dAlthough related to DUX4 and considered as an FSHD candidate gene (33), DUX4C is a predicted gene (http://www.genecards.org) not included in RefSeq genes.
eNo data are available from microarray expression analyses for this gene due to the lack of specific probes. From RT-PCR, its expression level is low, most transcripts are not full-length transcripts and most are not polyadenylated; moreover, transcripts were also found from other parts of the 3.3-kb D4Z4 repeat unit outside of the 1.6-kb DUX4 region and include antisense transcripts (34).
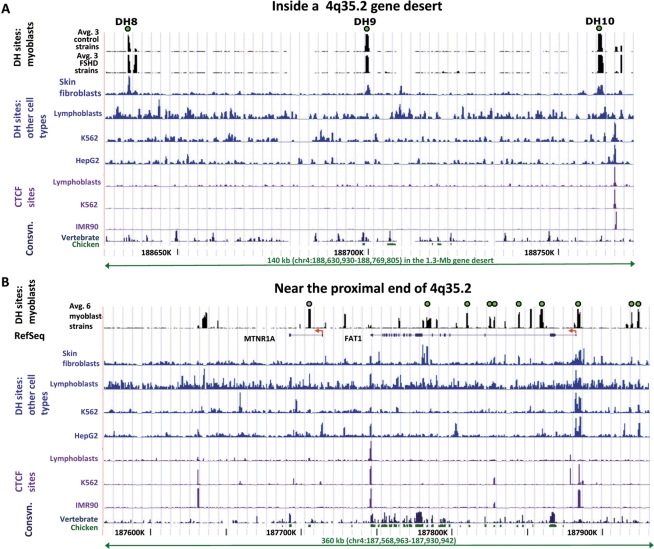
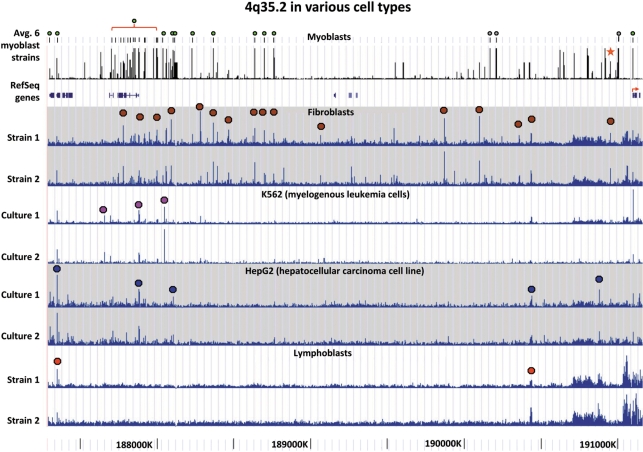
Three DH sites, DH8, -9 and -10, were located near the boundary of DHS domains 1 and 2 and were situated ∼0.4–0.5 Mb proximal to ZFP42 and ∼0.75–0.9 Mb distal to FAT1, the nearest gene known to be expressed in the muscle (Figure 2). These DH sites contain unique DNA sequences and were of similar intensity in all FSHD and control myoblast cell strains (Figures 2 and 3A). Recently, DH sites were identified across the whole genome from a number of human cell types by DNase-seq. This is a similar strategy to DNase-chip but uses next-generation sequencing (6). Even though DNase-seq and DNase-chip rely on different readout platforms, they have been shown to be highly correlated. The DNase-seq data have been generated as part of the ENCODE project and made available on the UCSC Genome Browser (http://genome.ucsc.edu/, Open Chromatin track). DNase-seq data from K562 (myeloid leukemia cell line), HepG2 (hepatocellular carcinoma cell line), GM12878 (lymphoblastoid cells), HeLa S3 cells and primary CD4+ T-cells, revealed no appreciable DH peak at the genomic positions of DH8, 9 or 10; however, two skin fibroblast cell strains did exhibit these peaks [Figure 3A and data not shown; all but the CD4+ data (6) are previously unpublished]. In some cell types, ∼4 kb distal to DH10, a DH site was observed that overlapped a CCCTC-binding factor (CTCF) binding site identified by chromatin immunoprecipitation (ChIP) followed by next-generation sequencing (ChIP-seq) or tiling array analysis (Figure 3A).
Figure 3.
DH sites in the proximal half of 4q35.2. (A) DH sites 8, 9 and 10 in the 1.3-Mb gene desert of 4q35.2 were seen in myoblasts and fibroblasts but not other cell types. DH sites in the middle of a gene desert are shown for FSHD and control myoblast cell strains and other cell types. For myoblasts, the average data for DH peak height for three FSHD and three control myoblast cultures from DNase-chip are displayed. Individual representative samples from DNase-seq are shown for a skin fibroblast cell strain (GM05879), lymphoblastoid cell line (GM12878), K562 cells (myelogenous leukemia cell line) and HepG2 cells (hepatocellular carcinoma cell line). CTCF binding data for the non-myoblast cells were mapped in IMR90 (lung fibroblasts) by CTCF ChIP following by tiling array analysis (4) and in the other cell types by CTCF ChIP-seq. The single CTCF site in this region is 4 kb distal to DH10. High-resolution mapping of CTCF sites has not yet been done in myoblasts. Consvn, vertebrate (17-way vertebrate alignment) or chicken/human conservation (http://genome.ucsc.edu/). (B) DH sites in the FAT1 and MTNR1A regions are cell type-specific. Tracks and DH sites are labeled as in Panel A.
DH sites at FSHD candidate genes in myoblasts
Five genes in 4q35.2 (DUX4, DUX4C, FRG1, FRG2 and TUBB4Q; Figure 4) and one at 4q35.1 (SLC25A4/ANT1) have been considered as candidates for the 4q-specific pathogenicity of short D4Z4 arrays (10,17,32,33,35,36). Of these FSHD candidate genes, only FRG1 has easily detectable expression at the RNA level (Table 1). Overexpression of FRG2, FRG1 and SLC25A4 RNA in FSHD versus control muscle was reported to be >60-, >25- and ∼10-fold, respectively (10,17). In addition, FSHD-associated elevation of protein levels was detected for SLC25A4 (18). Among the FSHD candidate genes on 4q35, DH sites were detected only at the promoters of FRG1 and SLC25A and no significant differences in DH peak intensity were observed between FSHD and control myoblasts (Figure 4, inset and data not shown). However, the biological significance of DH signal intensity is unknown and different DH sites display a wide range of openness (6). Moreover, by qRT-PCR, there was not a significant association of the relative concentration of FRG1 or SLC25A4 RNA with disease status in myoblasts. The relative steady-state RNA levels for FSHD versus control myoblasts were 1.4 for FRG1 and 2.0 for ANT1 (P > 0.5; assays of three FSHD versus three control myoblast cell strains in duplicate).
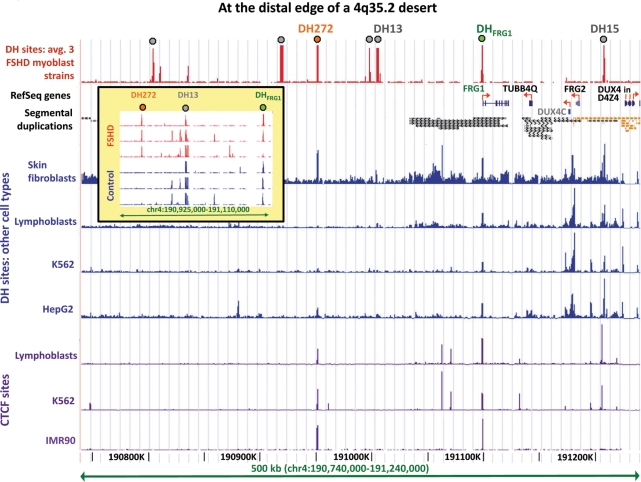
Figure 4.
DH272 and DHFRG1 in the terminal 0.4 Mb of 4q of myoblasts overlap DH sites and CTCF sites observed in other cell types. The average data from three FSHD myoblast cell strains are shown; control myoblasts gave the same results except that the DH272 peak was missing or much smaller (inset). Segmental duplications from the UCSC Genome Browser: orange, >99% similarity (mostly to 10q26.3) and grey, 90–98% similarity. Grey dots, DH peaks that overlap STRs (see legend to Figure 2) the only peaks overlapping STRs and seen in all six myoblast cultures in this subregion of 4q35.2 were DH13 and DH15. CTCF data are shown as in Figure 3. DUX4C (grey font) is a predicted gene. Only a few copies of DUX4 within each D4Z4 3.3-kb unit are shown (see Figure 1). The inset displays the mapped DH sites in each of the six studied myoblast cultures in the indicated subregion.
The ability to detect DH sites at promoters of TUBB4Q, DUX4C, FRG2 and DUX4 in the terminal 0.25 Mb of4q35.2 is complicated by this region containing many segmental duplications (Figure 4). With the exception of the 5′-end of FRG1 (37), analysis of all known 4q-terminal genes is difficult by almost any experimental method because of the near-absence of unique sequences. A further complication is that some of the sequences cross-hybridizing to this region of 4q35.2, especially within D4Z4 itself, are contained within incompletely sequenced regions of the genome, notably the short arms of the acrocentric chromosomes (38,39). Consequently, while there was sufficient probe coverage for TUBB4Q, DUX4C and FRG2 regions on the array, there was no coverage with unique probes.
Repeat-masked coverage for D4Z4, including its internal DUX4 gene, consisted of only scattered probes. DUX4 is the 1.6-kb homeobox-containing gene within each 3.3-kb D4Z4 repeat unit. Given the poor coverage with repeat-masking, we attempted to analyze the D4Z4 region by including probe sets that corresponded to these repetitive segments. DH sites in D4Z4 might be detected if all paralogs behaved similarly. However, the signal from D4Z4 probes did not identify any DH sites in this subregion (data not shown).
Other myoblast DH sites near genes in 4q35.2
Among the six genes in 4q35.2 at the proximal end (Figure 2 and Table 1), DH sites in or near the promoter region were observed for FAT1, a cadherin gene involved in cell migration; CYP4V2, a cytochrome P450 family member and FAM149A, a RefSeq gene of unknown function encoding a hypothetical protein (Figure 3B, Supplementary Figures S3 and S4). Multiple DH sites in FAT1 introns were consistently seen in the myoblasts cultures (Supplementary Figure S3). No FSHD-associated differences were observed at any of these DH sites. The adjacent gene MTNRIA, a melatonin receptor gene, had a highly reproducible DH site within the first intron, but not at the 5′-end (Supplementary Figure S3). Accordingly, MTNRIA RNA was not detectable in FSHD or control skeletal muscle, as determined on cDNA expression microarrays (11). No DH sites were observed at two other proximal 4q35.2 genes, KLKB1 and F11, which encode proteins involved in blood coagulation. One of the few described mRNAs from the terminal 1 Mb of 4q is AY956760 (590-kb proximal to the D4Z4 array), the reported product of the heat shock protein gene HSP90AA4P. No DH site was observed in its vicinity in myoblasts or the other investigated cell types (data not shown).
Tissue-specific patterning of DH sites along 4q35.2
We next compared the positions of DH sites along the length of 4q35.2 between myoblasts and diverse cell types. Many reproducible cell type-specific differences were observed, including differences in DH sites in gene deserts (Figure 5). Importantly, myoblast-specific differences in DH sites at documented genes were seen most prominently within the large FAT1 gene (Figure 5 and data not shown), which is subject to complicated alternative splicing and implicated in modulating cell contacts (40) and repair of vascular smooth muscle injury (41). For replicates of a given cell type, the positions and relative signal intensities of DH sites were remarkably consistent, even when the replicates were from different individuals (as for myoblasts and fibroblasts). This indicates that genetic polymorphisms are not responsible for the observed cell type-specific differences. Myoblasts were most similar to skin fibroblasts in their distribution of DH sites along subtelomeric 4q, although these two cell types did display some differences (Figure 5). The muscle specificity of DH sites in 4q35.2 was confirmed in preliminary whole-genome DNase-seq analysis of control myoblasts (Crawford, G.E. and Ehrlich, M. et al., unpublished data).
Figure 5.
Cell type-specific global patterns of DH sites along 4q35.2 even in gene deserts. The average data from three FSHD and three normal control myoblast cultures are shown; green dots, unique-sequence DH sites observed in all six cultures; grey dots, DH sites overlapping STRs and star, DH272. For the other cell types, colored dots indicate the DH peak combination that is most characteristic for a given cell type. The fibroblast tracks are from two skin fibroblast cell strains (GM02185 and GM05879) and the lymphoblast tracks from two lymphoblastoid cell lines (GM19238 and GM19239). The most distal portion of 4q35.2 (∼0.1 Mb in hg18) is not shown because its DH peaks were not reliable, as seen in large variations from sample to sample that we attribute to the many sequences throughout the genome displaying >93% identity to this subregion.
Primary and secondary structure of DH sites
In 4q35.2, there were 28 DH sites found in all six myoblasts cell cultures (Supplementary Table S1). Five were located in the 5′-gene regions (within 2-kb upstream through the first intron; Supplementary Table S1, boldface). These 5′-region DH sites were more GC-rich than the overall human genome, as is often found for immediate 5′-regions. Most of the other DH sites did not share this property, but rather had GC percentages similar to that of bulk human DNA (42% G + C).
We looked for primary and secondary structure motifs that might be associated with the DH sites (including DH272) that mapped within the gene deserts of 4q35.2 in myoblasts (Supplementary Table S1). This set of sites was compared to four analogous sets of randomly chosen sequences of similar G + C content and the length as for the DH sites (35– 48% G + C; 338–1642 bp). No significant differences were seen between DH sites and random control sequences in the frequencies of transcription factor consensus motifs, the possible 4–6 k-mers (DSGene, http://accelrys.com), the free energy of predicted secondary structures [Mfold web server, (42)] or the frequency of potential intramolecular G quadruplexes [GQRS Mapper, (43)].
One striking characteristic of DH sites in 4q35.2 was that ∼20% of those observed in all six myoblast cultures (Supplementary Table S1) overlapped a simple tandem repeat [STR, (44)]. BLAST searches indicated, with one exception, that the sequences of these DH-STR sites were located at a single chromosomal position, as tandem repeats in the reference genome. The exception, DH15, was located 15-kb proximal to D4Z4 (Figure 2) and had 84–97% identity to sequences on chromosomes 3, 7 and 17. Four out of five of the most distal DH sites (80%) observed in all six myoblast cultures overlapped an STR while only one out of the remaining 23 DH sites (5%) showed such overlap (Figure 2, grey circles). The STRs overlapping DH sites in 4q35.2 had consensus lengths of 27–142 nt and were tandemly repeated 3–59 times (Supplementary Table S1). They are unlikely to be artifacts because all DH sites in DNase-chip were identified by determining enrichment of DNase treated versus randomly sheared material from the same individual (25). DH15, located 15-kb proximal to D4Z4, was the only DH site overlapping an STR that was also detected in whole genome DNase-seq data from other cell types (data not shown). However, such STR-containing DH sites may be missed because DNase-seq experiments filter sequence tags that map to more than four places in the reference genome (hg18). Interestingly, the repeat overlapping DH15 has only three tandem copies in the reference genome.
At the 1-Mb terminus of 4q, there were seven DH sites observed in three to five of the six myoblast cultures, rather than in all six of them (Figure 4 and data not shown). Only one of these did not overlap an STR, namely, DH272. It is located 272-kb proximal to D4Z4, outside the region of high homology between subtelomeric 4q and 10q. It was also beyond the region shown to be deleted in rare FSHD families with no effect on the phenotype [Figures 1 and 4, (16)].
A DH site 272 kb from D4Z4 and qRT-PCR analyses in its vicinity
DH272 was observed preferentially in FSHD versus control myoblast cell strains (Figure 4). In other cell types examined by DNase-seq, this DH site was usually found at least as a small peak (Figure 4 and data not shown). Data on CTCF binding to this region were available from ChIP followed by next-generation sequencing for K562, HepG2 and GM12878 lymphoblasts (V. Iyer and B.-K. Lee, recently released data) or by tiling array analysis for lung fibroblasts (4). CTCF binding overlapped DH272 (as well as DHFRG1) in all of these cell types (Figure 4 and data not shown). Myoblasts have not yet been analyzed for CTCF binding in this region but most CTCF binding sites seem to be invariant among different human cell types (4). Sequence conservation among vertebrates was observed in the DH272 peak, including at the consensus sequence CTCF site, and was greater than that at DH8, 9 or 10 (Supplementary Figures S2 and S5; Figure 3A; and data not shown). In a preliminary comparative genome hybridization, using high-density tiling arrays to compare two FSHD samples with one control sample, we found no copy number variations in the region of DH272.
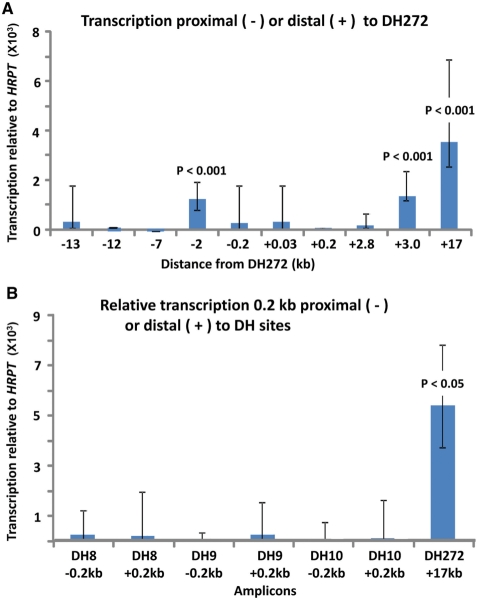
Because DH sites sometimes mark promoters of unannotated transcripts, we searched for evidence of transcription surrounding DH272, DH8, DH9 and DH10. Amplicons for qRT-PCR were chosen on the basis of sequence conservation, proximity to the DH site and locations of predicted genes (Supplementary Figure S5). We compared different unique amplicons with similar PCR efficiency (as determined on genomic DNA) by qRT-PCR using cDNA synthesized by random-hexamer priming. Several subregions in the vicinity of DH272 had significantly higher levels of RNA than others (Figure 6A). The more highly expressed amplicons were located 3.0- or 17-kb distal or 2.0-kb proximal to the center of DH272. This interval spans 19 kb. It is interrupted by amplicons that gave significantly less RT-PCR product (Figure 6A), possibly due to posttranscriptional processing or the use of several transcription units. The RNA levels for these three amplicons were ∼200- to 500-fold lower than that for the HPRT standard, a moderately transcribed gene. Therefore, these RNA levels were low but within the range of weakly expressed, but well-documented, genes (45).
Figure 6.
Some sequences in 4q35.2 gene deserts near DH sites are transcribed much more than others. (A) Amplicons around DH272 (Supplementary Figure S5) were compared for their average steady-state RNA levels among six myoblast cell strains (pooled data for FSHD and control cell strains, three each) as determined by qRT-PCR. +, distal; −, proximal. P-values (t-test) are given for the comparison of the indicated amplicon to the amplicon 12-kb proximal to DH272. (B) qRT-PCR determinations were done on amplicons located 0.2-kb proximal or distal to the midpoint of the indicated DH site and on the amplicon located 17-kb distal to DH272. P-values are for comparison of the amplicon 17-kb proximal to DH272 to the others.
In control fibroblast cell strains and lymphoblastoid cell lines (two each), transcripts from the three most highly expressed amplicons in the vicinity of DH272 were also significantly (P < 0.05) more abundant than those of neighboring amplicons (data not shown). No significant tissue-specific differences were seen among myoblasts, fibroblasts and lymphoblasts. No FSHD-specific differences were observed when comparing RNA from four FSHD patients and three normal controls (data not shown).
DISCUSSION
FSHD is associated with short arrays of the macrosatellite D4Z4 at subtelomeric 4q but not at subtelomeric 10q. It is still uncertain why, despite the near identity of 4q and 10q D4Z4 and much homology proximally and distally, FSHD is a 4q-specific disease. This dominant disease is caused by the reduction in size of a 4q D4Z4 array past a near-threshold of ∼36 kb (Figure 1). For example, contraction of a 40-kb array (with 12 3.3-kb repeat units) to one of 30 kb (with 9 3.3-kb repeat units) can result in the disease. We proposed that FSHD involves pathogenic long-range looping in cis of the centromere-proximal end of D4Z4 chromatin with 4q-specific sequences at 4q35.2 that is enabled by changes in intra-array chromatin looping dependent on the array size (20,46). The importance of pathogenic chromatin structure changes to this disease is indicated by recent evidence for FSHD-specific chromatin alterations in the D4Z4 array itself in FSHD patient's cells (47,48). In addition, the most proximal D4Z4 repeat unit apparently has a more open structure than the bulk of the array (20,48,49). Many experimental studies of the molecular genetics of FSHD do not duplicate the unusual chromatin environment of 4q35.2, which is likely to be critical for this disease in view of its 4q specificity. We used DNase-chip to examine 4q35.2 for chromatin features suggestive of a distinctive higher order structure. Given the lack of definitive findings about cis effects of short D4Z4 arrays at 4q35.2 on gene expression (10–14,34,35), DNase-chip also served as an annotation-neutral method of finding evidence for undocumented genes that may be important to FSHD in this gene-sparse 4-Mb region.
At 4q35.2, we found 28 DH sites detectable in all six examined myoblast cultures from FSHD patients or normal controls. As expected, most were located in the proximal 1 Mb of 4q35.2, the most gene-rich subregion. Surprisingly, within the bifurcated 3.1-Mb gene desert at 4q35.2 (Figure 2), 12 DH sites were observed in all tested myoblast cultures >100 kb from the nearest gene. For some of these DH sites, notably DH8, DH9 and DH10, the distances to the closest genes active in myoblasts were very large, >0.7 Mb. Nonetheless, these sites may identify long-distance enhancers, silencers or locus control regions (50–53). Alternatively, they might be associated with unannotated genes or structural elements, such as looping hubs (54). That DH8, 9 and 10 were observed in myoblasts and fibroblasts, both of mesodermal origin, but not in cells of the lymphoid, myeloid and hepatic lineages, suggests functionality.
In the D4Z4-proximal 1-Mb region, which is mostly gene desert, we found nine DH sites present in at least three of the six myoblast cell cultures. Only two of these, namely, DHFRG1 (in the promoter of FRG1) and DH272 (in the distal gene desert) did not overlap a DNA repeat. The other seven overlapped tandem repeats of short units (STRs). We also observed that DH sites frequently overlap STRs also in the terminal 1 Mb of 10q by DNase-chip analysis (unpublished data). While the biological significance of these DH-STRs remains to be determined, there are precedents for shorter tandem repeats influencing nucleosome positioning and excluding nucleosomes (55,56). With respect to DHFRG1, the DH site at the FRG1 promoter, one group reported overexpression of FRG1 RNA in FSHD muscle (17) but several others were unable to confirm this (11–13). In this study, we found no difference between control and FSHD myoblasts in this DH site and no significant difference in the amount of RNA product. DH272, the unique DH site located 150 kb proximal to FRG1, was observed preferentially in FSHD versus control myoblast cultures. Preliminary results from DNase-seq on three other control myoblast cell strains also revealed little or no DH peak at the position of DH272. We found nearby unannotated transcripts (probably non-coding RNAs, Supplementary Figure S5) that were not FSHD-specific in myoblasts. However, further study is needed of both myotubes and myoblasts to test the possibility of disease-linked expression of amplicons in the vicinity of DH272 and other DH sites in the 4q35.2 gene desert.
Even DH sites in 4q35.2 that did not display FSHD-related differences might be involved in pathogenic chromatin looping interactions. DH sites could be identical in both normal and disease cells, but the 3D structure (looping) and protein complexes that bind to them could differ between them. Given that DH sites can be associated with loci at which chromatin looping occurs (54), our results suggest subregions of 4q35.2 with the potential for these chromatin interactions that should be investigated. Our study points to the DH272 region as particularly attractive for searching for FSHD-related sequences because of the overlap of DH272 with a CTCF sequence found in many cell types [Figure 4 and (4)]. Moreover, the potential CTCF binding site identified in this ChIP-positive region of lung fibroblasts by Kim et al. (4) matched the CTCF consensus sequence at 19 out of 20 nt. CTCF is a sequence-specific DNA-binding protein with diverse functions, including as an insulator and organizer of chromatin looping (54,57). CTCF might play a role in our proposed pathogenic looping of 4q35.2-specific sequences to a short pathogenic D4Z4 array because D4Z4 was recently shown to have a CTCF-binding sequence (47). Some evidence was presented for increased binding of CTCF to D4Z4 in FSHD versus control myoblasts (47).
In addition to the revealing candidate DNA sequences for FSHD involvement in gene deserts, our data indicate that FAT1 transcription warrants further study of possible differences in FSHD and control muscle cells beyond the few studies involving expression microarrays (11,13). FAT1 is the only annotated 4q35.2 gene with evidence for complex tissue-specific expression and, in this study, a muscle-specific pattern of DH sites. Many myoblast-specific DH sites were found in and around this large gene in both FSHD and control cell strains, suggesting that this subregion contains active regulatory elements associated with the muscle lineage. The cell type-specific differences in chromatin that we observed are consistent with tissue-specific production of multiple FAT1 RNA and protein isoforms from predicted gene-internal promoters and by alternative splicing (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/). FAT1, which contains >25 exons, encodes a cadherin-type integral membrane protein which is implicated in diverse developmental and signaling pathways, including in vascular smooth muscle remodeling (58).
It has been proposed that a short disease-linked D4Z4 array at 4q35.2, but not at 10q26.3, triggers abnormal transcription of DNA sequences within the array itself in affected FSHD muscle cells (34,35,39,59). Overexpression of DUX4 RNA, derived from the 1.6-kb gene inside each 3.3-kb D4Z4 repeat unit, was reported in FSHD myotubes relative to control myotubes (35) but truncated transcripts or transcripts from other portions of the D4Z4 repeat unit are more prevalent than full-length DUX4 transcripts (34). Currently, definitive conclusions as to the relationship of D4Z4 transcription and pathogenicity are precluded by low expression levels, small numbers of samples, many cross-hybridizing sequences and the variety of small transcripts (34). If dysregulated expression of some D4Z4 sequence from short arrays initiates abnormal gene expression in FSHD, it remains to be explained why it is only short 4q arrays that cause the disease despite the ∼98% identity between 4q and 10q D4Z4 (8) and homology outside the arrays (Figure 1). In addition, exchanges between the almost (but not completely) identical 4q and 10q D4Z4 arrays are rather frequent and can result in an array with 4q-type repeat units replacing all the 10q units (60). Nonetheless, short D4Z4 arrays cause disease only when they reside on 4q (61). Therefore, polymorphisms that were found to be associated with canonical 4q-type D4Z4 units, but not canonical 10q-type D4Z4 units (8), are unlikely to explain the 4q linkage of FSHD.
We propose that the chromosomal environment of 4q35.2 plays a key role in the 4q-specific nature of FSHD, whether abnormal expression from 4q containing a short D4Z4 array initiates from within or outside D4Z4. Both at the DNA and the chromosome levels, 4q35.2 is unusual. It has the lowest gene density in its terminal 3 Mb of any of the q arms. It is distal to a large bifurcated gene desert punctuated centrally by a few genes that appear to be critical in early embryogenesis. Like some other genes (62), especially those important in the control of development (63), these inter-desert genes may be flanked by gene deserts to help keep their expression tightly restricted to certain stages in development. They might be part of large blocks chromatin with distinguishing epigenetic features. In CD4+ cells, this gene desert region has histone modifications [(5) and http://genome.ucsc.edu] indicative of inactive euchromatin rather than constitutive heterochromatin. This is consistent with our previous immunocytochemical and DNA replication analyses of FSHD and control myoblasts (64). However, given the complexity of epigenetic modification of chromatin, there can be a variety of types of large distinctive chromatin blocks within euchromatin (65).
One of the properties that distinguishes subtelomeric 4q (which can have pathogenic D4Z4 arrays) and 10q (whose D4Z4 arrays are always phenotypically neutral) is that only the 4q subtelomere (and not 10q or 4q) has a strong association with the nuclear rim in FSHD and control myoblasts and myotubes (66). A marker that was 0.22 Mb from D4Z4 on 4q35.2 (close to DH272) showed a significantly closer association with the nuclear periphery than did D4Z4. The unusual localization of subtelomeric 4q to the nuclear periphery might be necessary for pathogenicity. This localization may result partly from its uncommonly large region of inactive euchromatin (67,68) in a distinctive conformation, as reflected in its low concentration of DH sites. Our results emphasize the underappreciated importance of considering the regional chromatin context of D4Z4 in analysis of the mechanism by which contraction of D4Z4 to a size of <36 kb can lead to disease (69).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NS04885 to M.E., HG003169 to G.E.C.); the FSH Society (to M.E.); Fields Center for FSHD and Neuromuscular Research (R.T. and J.S.). Funding for open access charge: The National Institutes of Health [NS04885 to M.E.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr V. Vedanarayanan for several of the muscle samples from which myoblast cell strains were generated and to Dr Vishy Iyer and Bum-Kyu Lee, who generated the ENCODE CTCF ChIP-seq data.
REFERENCES
- 1.McCann JA, Muro EM, Palmer C, Palidwor G, Porter CJ, Andrade-Navarro MA, Rudnicki MA. ChIP on SNP-chip for genome-wide analysis of human histone H4 hyperacetylation. BMC Genomics. 2007;8:322. doi: 10.1186/1471-2164-8-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Maarel SM, Frants RR, Padberg GW. Facioscapulohumeral muscular dystrophy. Biochim. Biophys. Acta. 2007;1772:186–194. doi: 10.1016/j.bbadis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Lemmers RJ, Wohlgemuth M, van der Gaag KJ, van der Vliet PJ, van Teijlingen CM, de Knijff P, Padberg GW, Frants RR, van der Maarel SM. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, Padberg GW, Frants RR. Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum. Mol. Genet. 1996;5:1997–2003. doi: 10.1093/hmg/5.12.1997. [DOI] [PubMed] [Google Scholar]
- 10.Gabellini D, D’Antona G, Moggio M, Prelle A, Zecca C, Adami R, Angeletti B, Ciscato P, Pellegrino MA, Bottinelli R, et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- 11.Winokur ST, Chen YW, Masny PS, Martin JH, Ehmsen JT, Tapscott SJ, van der Maarel SM, Hayashi Y, Flanigan KM. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 12.Jiang G, Yang F, van Overveld PG, Vedanarayanan V, van der Maarel S, Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- 13.Osborne RJ, Welle S, Venance SL, Thornton CA, Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 14.Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, Ehrlich M. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochem. Biophys. Acta. 2007;1769:29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- 16.Deak KL, Lemmers RJ, Stajich JM, Klooster R, Tawil R, Frants RR, Speer MC, van der Maarel SM, Gilbert JR. Genotype-phenotype study in an FSHD family with a proximal deletion encompassing p13E-11 and D4Z4. Neurology. 2007;68:578–582. doi: 10.1212/01.wnl.0000254991.21818.f3. [DOI] [PubMed] [Google Scholar]
- 17.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 18.Laoudj-Chenivesse D, Carnac G, Bisbal C, Hugon G, Bouillot S, Desnuelle C, Vassetzky Y, Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J. Mol. Med. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 19.Jiang G, Sanchez C, Yang F, Ehrlich M. Histone modification in constitutive heterochromatin vs. unexpressed euchromatin in human cells. J. Cell Biochem. 2004;93:286–300. doi: 10.1002/jcb.20146. [DOI] [PubMed] [Google Scholar]
- 20.Tsumagari K, Qi L, Jackson K, Shao C, Lacey M, Sowden J, Tawil R, Vedanarayanan V, Ehrlich M. Epigenetics of a tandem DNA repeat: chromatin DNaseI sensitivity and opposite methylation changes in cancers. Nucleic Acids Res. 2008;36:2196–2207. doi: 10.1093/nar/gkn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun M, Ma F, Zeng X, Liu Q, Zhao XL, Wu FX, Wu GP, Zhang ZF, Gu B, Zhao YF, et al. Triphalangeal thumb-polysyndactyly syndrome and syndactyly type IV are caused by genomic duplications involving the long range, limb-specific SHH enhancer. J. Med. Genet. 2008;45:589–595. doi: 10.1136/jmg.2008.057646. [DOI] [PubMed] [Google Scholar]
- 22.Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochem. Biophys. Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 24.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 25.Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, Erdos MR, Green R, Meltzer PS, Wolfsberg TG, Collins FS. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 27.Masui S, Ohtsuka S, Yagi R, Takahashi K, Ko MS, Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev. Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian L, Wu X, Lin Y, Liu Z, Xiong F, Han Z, Zhou Y, Zeng Q, Wang Y, Deng J, et al. Characterization and potential function of a novel pre-implantation embryo-specific RING finger protein: TRIML1. Mol. Reprod. Dev. 2009;76:656–664. doi: 10.1002/mrd.20997. [DOI] [PubMed] [Google Scholar]
- 29.Costantini M, Clay O, Auletta F, Bernardi G. An isochore map of human chromosomes. Genome Res. 2006;16:536–541. doi: 10.1101/gr.4910606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver JL, Carpena P, Hackenberg M, Bernaola-Galvan P. IsoFinder: computational prediction of isochores in genome sequences. Nucleic Acids Res. 2004;32:W287–292. doi: 10.1093/nar/gkh399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, et al. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 32.van Geel M, van Deutekom JC, van Staalduinen A, Lemmers RJ, Dickson MC, Hofker MH, Padberg GW, Hewitt JE, de Jong PJ, Frants RR. Identification of a novel beta-tubulin subfamily with one member (TUBB4Q) located near the telomere of chromosome region 4q35. Cytogenet. Cell Genet. 2000;88:316–321. doi: 10.1159/000015518. [DOI] [PubMed] [Google Scholar]
- 33.Bosnakovski D, Lamb S, Simsek T, Xu Z, Belayew A, Perlingeiro R, Kyba M. DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp. Neurol. 2008;214:87–96. doi: 10.1016/j.expneurol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum. Mol. Genet. 2009;18:2414–2430. doi: 10.1093/hmg/ddp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixit M, Ansseau E, Tassin A, Winokur S, Shi R, Qian H, Sauvage S, Matteotti C, van Acker AM, Leo O, et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl Acad. Sci. USA. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clapp J, Mitchell LM, Bolland DJ, Fantes J, Corcoran AE, Scotting PJ, Armour JA, Hewitt JE. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Deutekom JC, Lemmers RJ, Grewal PK, van Geel M, Romberg S, Dauwerse HG, Wright TJ, Padberg GW, Hofker MH, Hewitt JE, et al. Identification of the first gene (FRG1) from the FSHD region on human chromosome 4q35. Hum. Mol. Genet. 1996;5:581–590. doi: 10.1093/hmg/5.5.581. [DOI] [PubMed] [Google Scholar]
- 38.Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR, et al. The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res. 1994;2:225–234. doi: 10.1007/BF01553323. [DOI] [PubMed] [Google Scholar]
- 39.Lyle R, Wright TJ, Clark LN, Hewitt JE. The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics. 1995;28:389–397. doi: 10.1006/geno.1995.1166. [DOI] [PubMed] [Google Scholar]
- 40.Tanoue T, Takeichi M. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J. Cell Biol. 2004;165:517–528. doi: 10.1083/jcb.200403006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou R, Sibinga NE. Atrophin proteins interact with the fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells. J. Biol. Chem. 2009;284:6955–6965. doi: 10.1074/jbc.M809333200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nowbakht P, Ionescu MC, Rohner A, Kalberer CP, Rossy E, Mori L, Cosman D, de Libero G, Wodnar-Filipowicz A. Ligands for natural killer cell-activating receptors are expressed upon the maturation of normal myelomonocytic cells but at low levels in acute myeloid leukemias. Blood. 2005;105:3615–3622. doi: 10.1182/blood-2004-07-2585. [DOI] [PubMed] [Google Scholar]
- 46.Ehrlich M. In: FSHD Facioscapulohumeral Muscular Dystrophy: Molecular Cell Biology & Clinical Medicine. Cooper DN, Upadhyaya M, editors. New York, NY: BIOS Scientific Pub.; 2004. pp. 253–276. [Google Scholar]
- 47.Ottaviani A, Rival-Gervier S, Boussouar A, Foerster AM, Rondier D, Sacconi S, Desnuelle C, Gilson E, Magdinier F. The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet. 2009;5:e1000394. doi: 10.1371/journal.pgen.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng W, de Greef JC, Chen YY, Chien R, Kong X, Gregson HC, Winokur ST, Pyle A, Robertson KD, Schmiesing JA, et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Greef JC, Lemmers RJ, van Engelen BG, Sacconi S, Venance SL, Frants RR, Tawil R, van der Maarel SM. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009;30:1–11. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 50.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 51.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 52.Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, et al. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat. Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- 53.Bagheri-Fam S, Barrionuevo F, Dohrmann U, Gunther T, Schule R, Kemler R, Mallo M, Kanzler B, Scherer G. Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 2006;291:382–397. doi: 10.1016/j.ydbio.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–1094. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stein A, Bina M. A signal encoded in vertebrate DNA that influences nucleosome positioning and alignment. Nucleic Acids Res. 1999;27:848–853. doi: 10.1093/nar/27.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang YH, Gellibolian R, Shimizu M, Wells RD, Griffith J. Long CCG triplet repeat blocks exclude nucleosomes: a possible mechanism for the nature of fragile sites in chromosomes. J. Mol. Biol. 1996;263:511–516. doi: 10.1006/jmbi.1996.0593. [DOI] [PubMed] [Google Scholar]
- 57.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou R, Liu L, Anees S, Hiroyasu S, Sibinga NE. The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J. Cell Biol. 2006;173:417–429. doi: 10.1083/jcb.200508121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gabriels J, Beckers MC, Ding H, de Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 60.van Overveld PG, Lemmers RJ, Deidda G, Sandkuijl L, Padberg GW, Frants RR, van der Maarel SM. Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum. Mol. Genet. 2000;9:2879–2884. doi: 10.1093/hmg/9.19.2879. [DOI] [PubMed] [Google Scholar]
- 61.Buzhov BT, Lemmers RJ, Tournev I, Dikova C, Kremensky I, Petrova J, Frants RR, van der Maarel SM. Genetic confirmation of facioscapulohumeral muscular dystrophy in a case with complex D4Z4 rearrangments. Hum. Genet. 2005;116:262–266. doi: 10.1007/s00439-004-1237-0. [DOI] [PubMed] [Google Scholar]
- 62.Hillier LW, Graves TA, Fulton RS, Fulton LA, Pepin KH, Minx P, Wagner-McPherson C, Layman D, Wylie K, Sekhon M, et al. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434:724–731. doi: 10.1038/nature03466. [DOI] [PubMed] [Google Scholar]
- 63.Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang F, Shao C, Vedanarayanan V, Ehrlich M. Cytogenetic and immuno-FISH analysis of the 4q subtelomeric region, which is associated with facioscapulohumeral muscular dystrophy. Chromosoma. 2004;112:350–359. doi: 10.1007/s00412-004-0280-x. [DOI] [PubMed] [Google Scholar]
- 65.Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Masny PS, Bengtsson U, Chung SA, Martin JH, van Engelen B, van der Maarel SM, Winokur ST. Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum. Mol. Genet. 2004;13:1857–1871. doi: 10.1093/hmg/ddh205. [DOI] [PubMed] [Google Scholar]
- 67.Murmann AE, Gao J, Encinosa M, Gautier M, Peter ME, Eils R, Lichter P, Rowley JD. Local gene density predicts the spatial position of genetic loci in the interphase nucleus. Exp. Cell Res. 2005;311:14–26. doi: 10.1016/j.yexcr.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 68.Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 69.Ottaviani A, Schluth-Bolard C, Rival-Gervier S, Boussouar A, Rondier D, Foerster AM, Morere J, Bauwens S, Gazzo S, Callet-Bauchu E, et al. Identification of a perinuclear positioning element in human subtelomeres that requires A-type lamins and CTCF. EMBO J. 2009;28:2428–2436. doi: 10.1038/emboj.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gelfand Y, Rodriguez A, Benson G. TRDB–the tandem repeats database. Nucleic Acids Res. 2007;35:D80–87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.