Abstract
Core promoters and chromatin insulators are key regulatory elements that may direct a transcriptional enhancer to prefer a specific promoter in complex genetic loci. Enhancer and insulator flank the sea urchin (Paracentrotus lividus) α-histone H2A transcription unit in a tandem repeated cluster containing the five histone genes. This article deals with the specificity of interaction between the H2A enhancer-bound MBF-1 activator and histone gene promoters, and with the mechanism that leads the H1 transcripts to peak at about one-third of the value for nucleosomal H3 and H2A mRNAs. To this end, in vivo competition assays of enhancer and insulator functions were performed. Our evidence suggests that the MBF-1 transcription factor participates also in the expression of the H3 gene and that the sns5 insulator buffers the downstream H1 promoter from the H2A enhancer. Altogether, these results provide a clear demonstration of the enhancer-blocking function of a chromatin insulator in a natural gene context. In addition, they suggest that both the H2A enhancer and the sns5 insulator may account for the diverse accumulation of the linker H1 versus the core nucleosomal histones during early development of the sea urchin embryo.
INTRODUCTION
The gene regulatory information encoded in the primary DNA sequence is interpreted and transmitted to the transcription machinery, that it is assembled at the promoter, by the binding of transcription factors to the appropriate sequence elements (1,2). Among these, enhancers are described as DNA elements that increase the level of transcription of the associated gene in a position- and distance-independent manner relative to the transcription start site (3). As a consequence, in complex genetic loci with multiple promoters, mechanisms ought to be put in place to make an enhancer prefer one specific promoter. Indeed, some transcriptional enhancers can discriminate between core promoters that contain either a TATA box or a DPE element (4). For instance, in Drosophila, the AE1 enhancer preferentially interacts with the ftz promoter rather than with the Scr promoter, in spite of its intergenic position and comparable distance from both promoters; in transgenic embryo, promoter competition dictates the IAB5 enhancer, placed between two divergently transcribed transgenes, to preferentially activate transcription from the TATA-containing promoter (5,6). In addition, in the sea urchin Hemicentrotus pulcherrimus, a combination of the upstream and either TATA-containing or TATA-less core promoter sequences seems to contribute to the establishment of the specific spatial and temporal expression profiles of developmentally regulated genes (7,8).
Besides promoter competition, chromatin insulators may also be involved in promoter selectivity by a given enhancer. Most of these regulatory elements may have two activities: (i) a boundary function blocks the spread of the heterochromatin into the euchromatic region and protects the transgenes from the negative influence of chromatin at the site of insertion; and (ii) an enhancer-blocking activity that restricts enhancer function in one direction and only when interposed between the enhancer and promoter (8–13). The DNA replication-dependent sea urchin early or α-histone gene cluster represents an interesting model system to investigate the specificity of function of an enhancer element in close proximity of different gene promoters. These genes are development-regulated and organized in a single large cluster made up of ∼2000 tandem repeat units, each containing one copy of each of the five histone genes, in the order 5′-H2B-H3-H2A-H1-H4-3′ (14,15). As embryogenesis occurs, transcription of these genes is limited to the rapid early cleavage and reaches its maximum at the morula/early blastula stage. After hatching, they become repressed and are maintained in such a transcriptional state for the whole life cycle of the animal (16).
We have previously described the cis-regulatory sequences and the necessary transcription factor for the timing of transcription of the α-H2A gene during the embryogenesis of the sea urchin P. lividus. In the 5′ region, a 30-nucleotide-long regulatory sequence termed modulator or simply M30 (17,18) specifically binds the MBF-1 activator (19). The H2A modulator has a bidirectional enhancer activity, both in the homologous and heterologous (Xenopus laevis oocytes) system (20,21). Worth mentioning is the capability of tandem copies of the MBF-1-binding sites to activate transcription from a viral promoter independent of distance and orientation (21,22). As shown in this article (Figure 1), this enhancer function is maintained also by a single MBF-1 recognition sequence. Remarkably, the MBF-1 regulator, although essential for H2A transcriptional activation, is constitutively bound to the H2A enhancer, which in a transgene construct can elicit transcription from a viral promoter also after silencing of the endogenous α-histone genes at the gastrula stage (23,24).
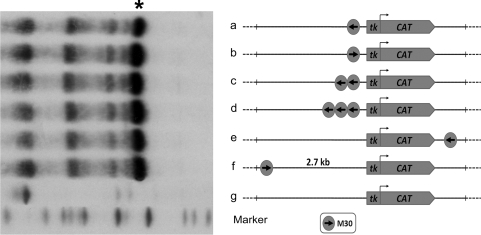
Figure 1.
A single copy of the MBF-1 activator binding site enhances transgene expression. The M30-tk-CAT transgenes, bearing one, two or three copies of the 30 bp H2A modulator sequence in different location and orientation, were microinjected into sea urchin zygotes. Total RNA from 30 to 50 gastrula stage embryos, microinjected with the indicated transgenes, were hybridized with a 32P-labelled CAT antisense probe and processed for the RNase protection assay described in ‘Materials and Methods’ section. Asterisk indicates the protected RNA band for the CAT transcript.
Down-regulation at the gastrula stage relies on the functional interaction between the 5′ dispersed GA repeats, located upstream of the enhancer, and the sns5 insulator placed at the 3′-end of the H2A transcription unit (24). The repressed H2A gene is characterized by the specific positioning of two nucleosomes in the promoter/enhancer region, histone de-acetylase recruitment, and histone H3K9 dimethylation in the insulator and 5′ regulatory sequences (25). Interestingly, the sns5 element contains the enhancer blocker sns that, in an enhancer-blocking assay, restricts enhancer function in a directional and polar manner in both sea urchin and mammalian cells (22,26,27). In addition, sns5 exhibits the other property of insulators, the ability to block repressive chromatin effects on the flanking regions of transgenes (28).
The five histone genes are coordinately expressed during early development. Despite this need for nucleosome assembly in the newly replicated chromatin, the number of H1 linker mRNA molecules is less than the value for histone nucleosomal mRNAs, being about half, as determined by kinetic and hybridization studies (29,30) or even less (this article). In this article, we have investigated the specificity of the H2A enhancer and the molecular mechanism that allows differential transcription of linker versus core histones genes during sea urchin development. In principle, the H2A enhancer-bound MBF-1 activator could interact, at least, with the promoters of the neighboring H3 and H1 genes, unless promoter competition and the 3′ located sns5 insulator restrain the activity of the H2A enhancer specifically to the associated H2A gene. Our results exclude competition between core promoter elements and indicate that the MBF-1 transcription factor participates also in the expression of the H3 gene in the resident chromatin. In addition, we show that the enhancer-blocking activity of the sns5 insulator buffers the downstream H1 promoter from the H2A enhancer. These results provide evidence for insulator action in a normal genomic context and suggest that both the H2A enhancer and the sns5 insulator are involved in the different regulation of nucleosomal core and linker histone transcription during early development of the sea urchin embryo.
MATERIALS AND METHODS
DNA constructs
The ΔpH3-H2A-ΔpH1 DNA plasmid, containing the core promoters of H3 and H1, and a wild-type H2A transcription unit, was constructed by polymerase chain reaction (PCR) amplification of histone DNA cluster Ph70 and cloning in the pBS vector. The M30-CAT plasmids were constructed by shotgun cloning of ligated double-stranded oligonucleotides bearing the H2A modulator sequence either upstream or downstream the chloramphenicol acetyltransferase (CAT) coding sequences of the tk-70 pBL2 vector under the control of the thymidine kinase gene (tk) promoter (31).
A dominant negative construct was obtained by fusing the MBF-1 DNA-binding domain encoding sequences (19) to those of the Engrailed repressor domain cloned in the CS2+nls expression vector. All DNA clones were checked by sequencing.
Microinjection of DNA constructs, double-strand oligonucleotides and synthetic RNA
Microinjection in P. lividus and Sphaerechinus granularis was conducted as previously described (32). Approximately 5000 molecules of the desired plasmid DNA were injected into the zygote, together with Texas Red-conjugated dextran added at a concentration of 5% in a 2 pl volume of 30% glycerol.
In the in vivo competition experiments, double-stranded M30, M30mut, or BoxA oligonucleotides were ligated with T4 DNA ligase and fractionated onto polyacrylamide gel. DNA fragments containing four to six tandem copies were eluted from the gel and mixed with the plasmid solution to be microinjected at a molar ratio of the specific genetic element to construct of 50:1. In the competition experiments for the endogenous histone genes, the purified double-strand oligonucletides were mixed with glycerol and Texas Red-conjugated dextran, and injected at a final concentration ranging from 1 to 15 ng/µl.
The oligonucleotide sequences used in the competition assays are listed in Supplementary Table S1A.
For mRNA injection, dn-MBF-1 and control CS2+nlsEn (33) constructs were linearized and transcribed in vitro using the Sp6 mMessage mMachine kit (Ambion). Capped mRNAs were resuspended in ultrapure RNase-free water (Gibco) and 2 pl, corresponding to the amounts of 0.1, 0.5 and 1.0 pg, respectively, were injected. Injected embryos at the desired stage were harvested and total RNA extracted.
RNase protection assay
Total RNA samples, extracted from microinjected embryos, were hybridized with antisense 32P-labelled RNA probes. The H3, H2A and H1 antisense RNA probes did not protect the endogenous S. granularis RNA bands. Hybridization conditions, RNase digestion and gel fractionation of the RNase resistant hybrids were as described (22).
Real-time quantitative PCR
The amounts of histone gene transcription in control and injected embryos at morula stage were evaluated as described (34). Briefly, total RNA from batches of 150 microinjected embryos was extracted by using the High Pure RNA Isolation kit (Roche). RNA samples were treated with reagents provided by the Turbo DNA-free kit (Ambion) and resuspended in a final volume of 30 μl. Reverse transcription into cDNA was performed in an 80 μl reaction using random hexamers and the TaqMan Reverse Transcription Reagents kit (Applied Biosystems). The resulting cDNA sample was further diluted and the equivalent amount corresponding to one embryo was used as template for Q-PCR analysis. Q-PCR experiments were performed from two different batches and all reactions were run in triplicate on the 7300 Real-Time PCR system (Applied Biosystems) using SYBR Green detection chemistry (Applied Biosystems). ROX was used as a measure of background fluorescence and MBF-1 mRNA (19) was used to normalize all data, in order to account for fluctuations among different preparations. At the end of the amplification reactions, a ‘melting-curve analysis’ was run to confirm the homogeneity of all Q-PCR products. Calculations from QPCR raw data were performed by the RQ Study software version 1.2.3 (Applied Biosystems), using the comparative Ct method (ΔΔCt). The oligonucleotide sequences, length and predicted amplicon size are described in Supplementary Table S1B.
RESULTS
A single copy of the MBF-1 activator-binding site enhances transgene expression
As reported previously, the MBF-1 activator has been identified as being capable of specifically binding a 30-bp sequence of the modulator, and tandem copies of this sequence, denoted as M30, enhanced transcription from a viral promoter in a position- and orientation-independent manner in transgenic sea urchin embryos (22). To gain more details on the promoter specificity of the MBF-1 activator, we assessed, in the first place, whether a single copy of the M30 sequence was capable of activating the basal tk promoter. The constructs schematically drawn in Figure 1 were microinjected into sea urchin zygotes, embryos were raised, and expression of the reporter CAT gene was determined at the gastrula stage by RNase protection assays with RNA samples from the same number of injected embryos. The results shown in Figure 1 demonstrate that the MBF-1-binding sequence enhanced transcription from the viral promoter to a similar extent as did multiple copies. Furthermore, this enhancer activity was displayed independently of location and orientation and occurred also at 2.7-kb distance from the tk promoter.
Promoter specificity of the MBF-1 activator
To elucidate the role of the MBF-1 activator in the expression of the early histone genes, we inhibited its binding to the H2A enhancer by performing an in vivo competition assay. This assay involves the titration of a given DNA-binding factor by molar excess of tandem copies of a cis-regulatory sequence. We have routinely used this approach to knockdown the function of regulatory sequences of the sns5 insulator and of histone and Hbox12 gene promoters. As demonstrated in several instances, the effect on transgene expression is identical to that obtained by the mutation of the same sites (24,27,34).
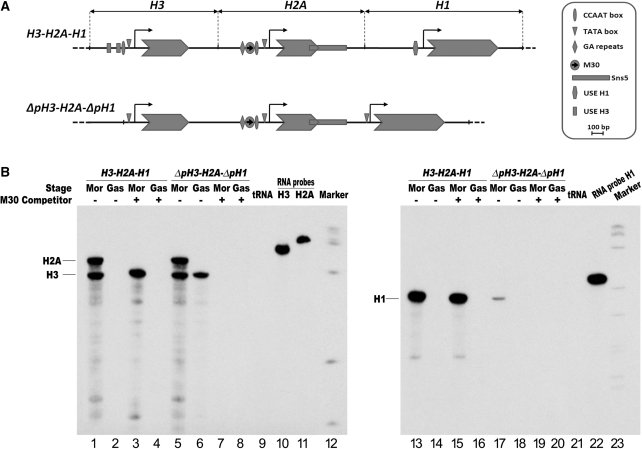
Concatameric ligation products containing on average six tandem copies of the M30 oligonucleotide were co-injected with the P. lividus H3-H2A-H1 histone gene constructs, depicted in Figure 2A, into the sea urchin S. granularis zygotes, to distinguish between endogenous and transgene histone transcripts. Embryos were raised and the expression of the injected genes was detected by RNAse protection assays. All three injected genes followed the embryonic temporal expression profile of the endogenous histone genes, i.e. they are highly transcribed at morula stage and silenced at gastrula stage (lanes 1, 2, 13 and 14). As a control, we used the mutant M30 sequence (M30 mut), which as reported cannot bind the MBF-1 protein translated in vitro (19,21). Injection of M30 mut at the same doses as M30 had no effect on histone gene transcription (Supplementary Figure S1). Conversely, the injected M30 sequence selectively inhibited transcription of the H2A gene, while it did not affect the expression of both H3 and H1 genes at the morula stage (lanes 3 and 15). These results would suggest that the H2A enhancer prefers to interact with the cognate promoter.
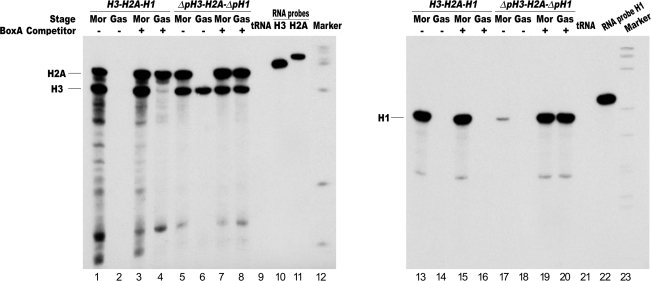
Figure 2.
In vivo competition assay to knock-down the H2A enhancer function in transgenic embryos. (A) Annotated map of the P. lividus wild type and deletion mutants early H3, H2A and H1 histone genes, highlighting the cis-regulatory sequence elements. The horizontal black line and arrow-shaped boxes represent, respectively, the genomic DNA and coding sequences, while the bent arrows denote the putative transcription start site. (B) The P. lividus histone gene constructs, orientated as in the endogenous histone gene repeat, were co-injected with excess of the modulator binding site (M30) into S. granularis zygotes. RNase protection was carried out by hybridizing antisense 32P-labelled RNA, transcribed in vitro from H3, H2A and H1 subclones, with total RNA exctracted from 25 injected embryos at morula (Mor) and gastrula (Gas) stages. The two H2A and H3 probes were hybridized together. The protected 409, 357 and 209 nt RNA bands, respectively, for the H2A, H3 and H1 transcripts are indicated.
To assess whether competition between core promoter elements is one of the molecular mechanisms that lead the MBF-1 activator to selectively interact with the H2A promoter, we performed the in vivo competition assay for the H2A enhancer function on the deletion mutant ΔpH3-H2A-ΔpH1 (Figure 2A). In such a construct, the H2A transcription unit is a wild type. All the regulatory sequence elements upstream the TATA box were deleted from the H3 promoter (31,35), while the 5′ deletion of the linker H1 promoter occurred up to half of the essential regulatory sequence USE (36).
We have already described that the expression of H3 driven by only the TATA box and other core promoter elements is up-regulated in a H3-H2A transgene (24). The result displayed in Figure 2B extends this observation, by showing that the H2A enhancer-bound MBF-1 transcription factor is most probably responsible for the up-regulation. In fact, while the H2A gene followed the time of expression of the endogenous gene, the core promoter of the H3 gene, which by itself displays a barely detectable transcriptional activity (not shown), gave rise to comparable levels of transcripts. Significantly, co-injection of an excess of the M30 enhancer abolishes transcription of both genes (Figure 2B, lanes 5–8), suggesting that the TATA boxes and other core promoter elements of H3 and H2A genes do not compete for the interaction with the MBF-1 activator.
A different result was obtained with the H1 gene driven by the mutated H1 promoter. The core promoter elements displayed a very low transcriptional activity and did not respond to the trans-activating signal emanated by the MBF-1 transcription factor (Figure 2B, lanes 17–20), suggesting that H1 gene expression is autonomously regulated. This independence from the H2A enhancer can be explained either with a core promoter competition mechanism or by the action of the sns5 insulator located at 3′ of the H2A gene, between the H2A enhancer and H1 promoter.
The MBF-1 activator is involved also in the transcription of the early H3 gene
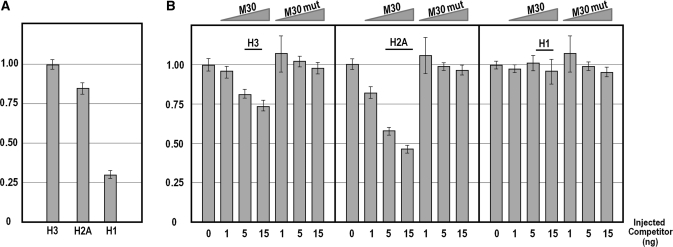
The results displayed in Figure 2 seem to be in apparent contrast. In one case, we have observed a lack of influence of the M30 competitor on the function of the wild type H3 promoter. In the other case, we showed the interaction of the MBF-1 activator with the H3 core promoter. To gain insights on this issue, we have determined by quantitative PCR (Q-PCR) whether competition of the H2A enhancer activity affected, and to what magnitude, the expression of the endogenous H2A, H3 and H1 genes. The histone mRNAs prevalence in the un-competed embryos, shown in Figure 3A, indicates that, in the sea urchin P. lividus at morula stage, the H1 linker histone peaks at about one-third of the value for the nucleosomal H3 and H2A. As expected, the mRNA levels for H2A decreased with the microinjection of increasing amounts of the wild type MBF-1-binding site. Once again, the mutant M30 sequence had no effect. Conversely, the number of mRNA molecules for the linker histone did not change with the rise of either M30 or M30 mut competitors, confirming the independence of H1 transcription from the H2A enhancer. Very interestingly, we found that inhibition of MBF-1 binding reduced also the H3 gene transcripts, although to a lesser extent than the H2A mRNA.
Figure 3.
In vivo competition assay for endogenous H3, H2A and H1 histone gene expression. (A) Relative abundance of histone mRNAs in the P. lividus embryo at morula stage. A similar prevalence is detected for the two nucleosomal H3 and H2A mRNAs, while the H1 linker histone mRNA peaks at about one-third of the value for the formers. (B) Endogenous histone gene expression analysis carried out in embryos at morula stage microinjected with excess of the M30 sequence element or the mutated M30 mut oligonucleotide as a control. Graphs show n-fold changes in mRNA expression level of histone genes based on the threshold cycle number (Ct) of injected embryos compared to that of the uncompeted control embryos. Ct numbers were normalized for the endogenous MBF-1 in the same sample. Data were derived from two independent microinjection experiments and each bar represents the average of triplicate samples from the two batches of embryos.
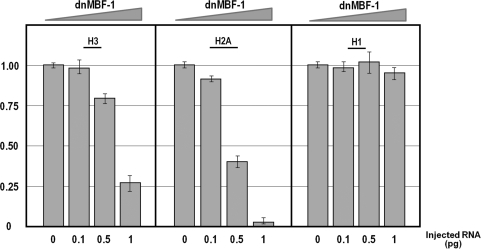
In order to obtain clear-cut evidence on the involvement of the MBF-1 activator in the expression of the H3 gene, we performed a more direct experiment to knock down its function. To this end, we made a dominant negative construct, termed dn-MBF-1, in which the DNA-binding domain of MBF-1 (19) was joined to the repression domain of Drosophila engrailed (33). An in vitro transcribed mRNA was injected into the sea urchin zygote and the expression of H3, H2A and H1 genes analyzed by Q-PCR at morula stage. The results obtained (Figure 4) are in accordance with the in vivo competition analysis described above. In fact, we found a dose-dependent negative effect of the MBF-1 forced repressor on the expression of the nucleosomal H2A and H3, but not H1 genes. Once again, transcription of H3 was less affected compared with H2A.
Figure 4.
Knock-down MBF-1 function by microinjection of a synthetic mRNA encoding for as dominant repressor (dnMBF-1). Increasing amounts (0.1–1 pg) of the chimeric RNA were injected in P. lividus zygotes and RNA extracted from embryos at morula stage. Graphs show n-fold changes in mRNA expression level of histone genes based on the threshold cycle number (Ct) of dnMBF-1 injected embryos compared to that of the uninjected control embryos. Ct numbers were normalized for the endogenous MBF-1 in the same sample, by amplifying a fragment of the coding region external to the DNA binding domain. Data were derived from two independent microinjection experiments and each bar represents the average of triplicate samples from the two batches of embryos.
In summary, these results confirmed that the MBF-1 transcription factor is absolutely necessary for the activation of the H2A gene. In addition, they suggest that the MBF-1 activator is involved also in the transcription of the upstream H3 gene and has no role on the expression of the linker H1 gene.
The sns5 insulator confers transcriptional independence to the linker H1 histone gene promoter
The location of the sns5 insulator between the H2A enhancer and H1 promoter (Figure 1) prompted us to examine its possible involvement in the mechanism of enhancer specificity. Wild type and mutated H3-H2A-H1 three gene constructs were microinjected, respectively, in the absence and in the presence of excess BoxA-ligated oligonucleotides. As reported, BoxA is one of the cis-acting sequences absolutely required for the enhancer blocking and silencing function of the sns5 insulator at gastrula stage (24,27). This is further shown in Figure 5. The competitor BoxA up-regulated the H2A, while the wild type H3 gene followed the temporal regulatory program of the endogenous gene (lanes 1–4). In addition, the inhibition of the insulator activity did not influence the constitutive trans-activation of the H3 core promoter (lanes 5–8). What is most important, however, is the effect of the competition of the insulator activity on the mutant H1 promoter. In this case, although it appeared that the expression of the wild type H1 was not affected, we did find a strong trans-activation of the mutated H1 transgene, at both early and late developmental stages, caused most probably by the interaction of MBF-1 with the H1 core promoter (lanes 13–20).
Figure 5.
In vivo competition assay to inhibit the sns5 insulator function in transgenic embryos. Wild-type histone gene construct H3-H2A-H1 and the deletion mutant ΔpH3-H2A-ΔpH1 (showed in Figure 2A), were microinjected with excess of the BoxA sequence element into S. granularis zygotes. RNase protection was carried out as for Figure 2. The protected 409, 357 and 209 nt RNA bands, respectively, for the H2A, H3 and H1 transcripts are indicated.
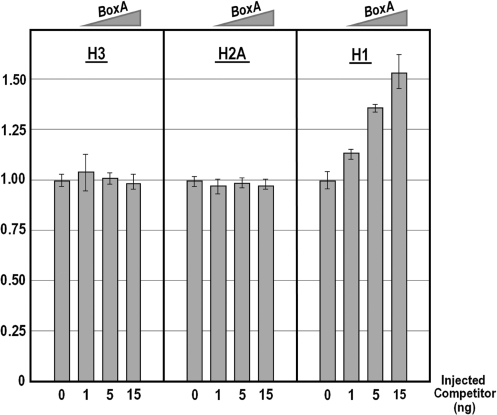
It should be noted that, as for the wild-type H3 transgene, the RNase protection assay did not reveal possible subtle differences in the transcription of the wild type H1 transgene between the un-competed and competed samples. For this reason, to eventually validate the regulatory role of the sns5 insulator, we looked at the expression by Q-PCR of the endogenous histone genes upon microinjection of the competitor BoxA sequence. As can be seen in Figure 6, both nucleosomal histone H2A and H3 mRNAs did not vary their prevalence at the different doses of competitor. By contrast, the number of molecules of the linker H1 mRNA increased monotonically with the augmentation of the BoxA oligonucleotide. Altogether, these results represent a strong indication that the sns5 insulator blocks the H2A enhancer in the interaction with the downstream H1 promoter.
Figure 6.
Endogenous gene expression analysis upon impairment of the sns5 insulator function by in vivo competition assay with excess of the BoxA sequence element. Graphs show n-fold changes in mRNA expression level of histone genes based on the threshold cycle number (Ct) of injected embryos compared to that of the uncompeted control embryos. Ct numbers were normalized for the endogenous MBF-1 in the same sample. Data were derived from two independent microinjection experiments and each bar represents the average of triplicate samples from the two batches of embryos.
DISCUSSION
The experiments described in this article highlight the regulatory function of the modulator/enhancer and insulator flanking the H2A transcription unit in the expression of S-phase-dependent histone genes during sea urchin development. These genes are organized in a large cluster and are coordinately regulated during embryogenesis. Notwithstanding this need, the transcripts of linker and core histones accumulate at the morula stage at different levels, with the H1 mRNA being, in P. lividus, ∼30% of the abundance of the two nucleosomal H3 and H2A mRNAs. This differential regulation occurs despite the presence of a strong enhancer in the 5′-flanking region of the H2A gene. Although some evidence suggests that the enhancer has a bipartite organization (16), our results indicate that a single binding site for the MBF-1 activator suffices the trans-activation of a distant promoter and in both orientations (Figure 1). Because of this property, we raised the issue of whether competition between promoter cis-regulatory sequences specifically directs the MBF-1 transcription factor toward the cognate H2A promoter.
Although the activity of the wild type promoter of the H3 transgene seemed refractory to the inhibition of MBF-1 binding, several lines of evidence substantiate the involvement of the MBF-1 transcription factor in the expression also of the α-H3 gene, as previously suggested (31). First, we found a robust activation of the H3 minimal core promoter that is inhibited by the titration of the MBF-1 factor. Second, we found a reduction of the number of endogenous H3 and H2A mRNA molecules with microinjection of increasing amounts of M30, but not the M30 mutant, competitor (Figure 3). The third evidence is even stronger, in that, the expression of a forced MBF-1 repressor had a severe impact on the transcription level of the two nucleosomal core histone genes (Figure 4). Clearly, the accumulation of the H2A transcripts is more affected than that of H3, indicating a different responsiveness of their gene promoters to MBF-1 knockdown. A straightforward interpretation of this observation is that the MBF-1 activator is the essential transcription factor for H2A gene, as suggested by previous experiments (19), but it only participates, together with other factor(s), in the transcription of the nucleosomal H3 histone gene. We speculate that one of the factors with which the enhancer-bound MBF-1 activator interacts is the protein complex containing the homeodomain CDP/cut that, as described (37), binds to the CCAAT sequence element for the maximum expression of the H3 gene.
The results described in this article demonstrate that the linker H1 histone gene is differentially regulated relative to the patterns of core histone gene transcription. In Drosophila, the cell cycle-dependent histone genes are tandemly arrayed and coordinately regulated, and the ratio of linker and core histones varies during embryonic development (38). However, the regulatory mechanism involved in the lower accumulation of H1 gene transcripts, with respect to the nucleosomal histones, are profoundly different between fly and sea urchin. In Drosophila, two distinct sets of core promoter recognition factors, the TBP and TBP-related factors TRF2 (39), are responsible for directing transcription, respectively, of the TATA-containing nucleosome core histone genes and the TATA-less linker histone H1 (40). Although the molecular mechanism is not clear, upstream promoter-bound regulatory factors are probably involved in the interaction with the proper core promoter transcription complex. In the sea urchin P. lividus, the H1 core promoter lacks a canonical TATA box and yet, as shown here, there is no competition between core promoter elements for the H2A enhancer. Instead, we obtained compelling evidence for the involvement of a chromatin insulator in making the linker H1 promoter independent from the action of the H2A enhancer.
Chromatin insulators are genetic regulatory elements that may possess both a boundary function and a directional enhancer-blocking activity (41). Thus, insulators, by restricting enhancer function, may impart promoter selectivity to a given enhancer in the eukaryotic genome. This role has been demonstrated in several cases. The Drosophila SF1 insulator, for instance, of the Antennapedia complex, placed in the scr-ftz intergenic region, restricts promoter selection by the ftz-distal enhancer in transgenic embryos (42) and separates the fushi tarazu from the neighboring Hox gene (43). As an additional example, the human and mouse imprinting controlling region (ICR) contain an insulator activity that depends on the binding of the CTCF regulator. In the maternal allele, the CTCF factor binds to the unmethylated ICR and prevents the downstream enhancer from interacting with the upstream IGF2 promoter. The blocked enhancer can activate transcription of the H19 gene (44).
Intriguingly, the P. lividus sns5 DNA fragment, located at the 3′ of the H2A transcription unit, has been identified as an essential element for the silencing of the α-H2A gene at the gastrula stage (27). In addition, as for the most known vertebrate insulator, the HS4 insulator of the chicken β-globin locus (13), the sns5 sequence element displays, on transgene constructs, both enhancer-blocking and boundary activities (22,28). The directional enhancer blocking activity is achieved by the cooperative interactions between all three different protein factors bound to their specific cis-regulatory sequences, in that, titration of any of them abolishes the function (24,27). Here, we have shown that microinjection of molar excess of the cis-acting BoxA element allowed trans-activation of the H1 transgene, driven by the core promoter elements. Most importantly, the BoxA competitor injected at the maximum dose (higher concentration is toxic for the embryo) specifically increased the expression of only the endogenous H1 gene. The most obvious interpretation of this result is that the chromatin insulator sns5 buffers the downstream H1 promoter from the activity of H2A enhancer-bound MBF-1 transcription factor. The sns5 insulator should leave also the expression of H4 and H2B genes of the repeating unit not linked to the action of MBF-1 input from the H2A promoter. In fact, the high level of expression of these two genes depends on strong promoter upstream elements (45,46).
Altogether, the results presented in this article suggest that the sns5 insulator is most probably responsible for the different level of accumulation of nucleosomal and linker transcripts during sea urchin embryogenesis. Finally, our findings provide a clear demonstration of the enhancer-blocking function of a chromatin insulator in a natural gene context.
An additional important issue to be clarified concerns the mechanism that prevents the activation of the H1 gene by the H2A enhancer in the adjoining histone gene repeat. As a possibility, the enhancer might not elicit a perceptible effect at the resulting genomic distance of more than 5 kb. Alternatively, it could be speculated that the H1 gene is flanked, at the 3′, by a supplementary insulator element. Experiments have been planned to distinguish between these two possibilities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The University of Palermo (ex 60%). Funding for open access charge: University of Palermo (ex 60%).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The participation of F. Palla and D. Di Caro in the experiment described in Figure 1 is acknowledged.
REFERENCES
- 1.Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 2.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 3.Khoury G, Gruss P. Enhancer elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- 4.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai HN, Zhang Z, Adams JR, Shen P. Genomic context modulates insulator activity through promoter competition. Development. 2001;128:4339–4347. doi: 10.1242/dev.128.21.4339. [DOI] [PubMed] [Google Scholar]
- 6.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi A, Akasaka K, Kawaichi M, Kokubo T. Functional interaction between TATA and upstream CACGTG elements regulates the temporally specific expression of Otx mRNAs during early embryogenesis of the sea urchin, Hemicentrotus pulcherrimus. Nucleic Acids Res. 2002;30:3034–3044. doi: 10.1093/nar/gkf439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi A, Watanabe Y, Akasaka K, Kokubo T. Real-time monitoring of functional interactions between upstream and core promoter sequences in living cells of sea urchin embryos. Nucleic Acids Res. 2007;35:4882–4894. doi: 10.1093/nar/gkm519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 10.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 11.Gerasimova TI, Corces VG. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 12.Geyer PK, Clark I. Protecting against promiscuity: the regulatory role of insulators. Cell Mol. Life Sci. 2002;59:2112–2127. doi: 10.1007/s000180200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentschel CC, Birnstiel ML. The organization and expression of histone gene families. Cell. 1981;25:301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- 15.Marzluff WF, Sakallah S, Kelkar H. The sea urchin histone gene complement. Dev. Biol. 2006;300:308–320. doi: 10.1016/j.ydbio.2006.08.067. [DOI] [PubMed] [Google Scholar]
- 16.Spinelli G, Birnstiel ML. The modulator is a constitutive enhancer of a developmentally regulated sea urchin histone H2A gene. Bioessays. 2002;24:850–857. doi: 10.1002/bies.10143. [DOI] [PubMed] [Google Scholar]
- 17.Grosschedl R, Birnstiel ML. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc. Natl Acad. Sci. USA. 1980;77:7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosschedl R, Machler M, Rohrer U, Birnstiel ML. A functional component of the sea urchin H2A gene modulator contains an extended sequence homology to a viral enhancer. Nucleic Acids Res. 1983;11:8123–8136. doi: 10.1093/nar/11.23.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alessandro C, Di Simone P, Buscaino A, Anello L, Palla F, Spinelli G. Identification of the enhancer binding protein MBF-1 of the sea urchin modulator alpha-H2A histone gene. Biochem. Biophys. Res. Commun. 2002;295:519–525. doi: 10.1016/s0006-291x(02)00708-8. [DOI] [PubMed] [Google Scholar]
- 20.Palla F, Casano C, Albanese I, Anello L, Gianguzza F, Di Bernardo MG, Bonura C, Spinelli G. Cis-acting elements of the sea urchin histone H2A modulator bind transcriptional factors. Proc. Natl Acad. Sci. USA. 1989;86:6033–6037. doi: 10.1073/pnas.86.16.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palla F, Bonura C, Anello L, Di Gaetano L, Spinelli G. Modulator factor-binding sequence of the sea urchin early histone H2A promoter acts as an enhancer element. Proc. Natl Acad. Sci. USA. 1994;91:12322–12326. doi: 10.1073/pnas.91.25.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palla F, Melfi R, Anello L, Di Bernardo M, Spinelli G. Enhancer blocking activity located near the 3' end of the sea urchin early H2A histone gene. Proc. Natl Acad. Sci. USA. 1997;94:2272–2277. doi: 10.1073/pnas.94.6.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palla F, Melfi R, Di Gaetano L, Bonura C, Anello L, Alessandro C, Spinelli G. Regulation of the sea urchin early H2A histone gene expression depends on the modulator element and on sequences located near the 3' end. Biol. Chem. 1999;380:159–165. doi: 10.1515/BC.1999.024. [DOI] [PubMed] [Google Scholar]
- 24.Di Caro D, Melfi R, Alessandro C, Serio G, Di Caro V, Cavalieri V, Palla F, Spinelli G. Down-regulation of early sea urchin histone H2A gene relies on cis regulative sequences located in the 5' and 3' regions and including the enhancer blocker sns. J. Mol. Biol. 2004;342:1367–1377. doi: 10.1016/j.jmb.2004.07.101. [DOI] [PubMed] [Google Scholar]
- 25.Di Caro V, Cavalieri V, Melfi R, Spinelli G. Constitutive promoter occupancy by the MBF-1 activator and chromatin modification of the developmental regulated sea urchin alpha-H2A histone gene. J. Mol. Biol. 2007;365:1285–1297. doi: 10.1016/j.jmb.2006.10.098. [DOI] [PubMed] [Google Scholar]
- 26.Di Simone P, Di Leonardo A, Costanzo G, Melfi R, Spinelli G. The sea urchin sns insulator blocks CMV enhancer following integration in human cells. Biochem. Biophys. Res. Commun. 2001;284:987–992. doi: 10.1006/bbrc.2001.5082. [DOI] [PubMed] [Google Scholar]
- 27.Melfi R, Palla F, Di Simone P, Alessandro C, Cali L, Anello L, Spinelli G. Functional characterization of the enhancer blocking element of the sea urchin early histone gene cluster reveals insulator properties and three essential cis-acting sequences. J. Mol. Biol. 2000;304:753–763. doi: 10.1006/jmbi.2000.4273. [DOI] [PubMed] [Google Scholar]
- 28.D'A;polito D, Baiamonte E, Bagliesi M, Di Marzo R, Calzolari R, Ferro L, Franco V, Spinelli G, Maggio A, Acuto S. The sea urchin sns5 insulator protects retroviral vectors from chromosomal position effects by maintaining active chromatin structure. Mol. Ther. 2009;17:1434–1441. doi: 10.1038/mt.2009.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxson RE, Jr, Wilt FH. The rate of synthesis of histone mRNA during the development of sea urchin embryos (Strongylocentrotus purpuratus) Dev. Biol. 1981;83:380–386. doi: 10.1016/0012-1606(81)90485-1. [DOI] [PubMed] [Google Scholar]
- 30.Mauron A, Kedes L, Hough-Evans BR, Davidson EH. Accumulation of individual histone mRNAs during embryogenesis of the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 1982;94:425–434. doi: 10.1016/0012-1606(82)90359-1. [DOI] [PubMed] [Google Scholar]
- 31.Palla F, Bonura C, Anello L, Casano C, Ciaccio M, Spinelli G. Sea urchin early histone H2A modulator binding factor 1 is a positive transcription factor also for the early histone H3 gene. Proc. Natl Acad. Sci. USA. 1993;90:6854–6858. doi: 10.1073/pnas.90.14.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavalieri V, Di Bernardo M, Spinelli G. Functional studies of regulatory genes in the sea urchin embryo. Methods Mol. Biol. 2009;518:175–188. doi: 10.1007/978-1-59745-202-1_13. [DOI] [PubMed] [Google Scholar]
- 33.Cavalieri V, Spinelli G, Di Bernardo M. Impairing Otp homeodomain function in oral ectoderm cells affects skeletogenesis in sea urchin embryos. Dev. Biol. 2003;262:107–118. doi: 10.1016/s0012-1606(03)00317-8. [DOI] [PubMed] [Google Scholar]
- 34.Cavalieri V, Di Bernardo M, Anello L, Spinelli G. cis-Regulatory sequences driving the expression of the Hbox12 homeobox-containing gene in the presumptive aboral ectoderm territory of the Paracentrotus lividus sea urchin embryo. Dev. Biol. 2008;321:455–469. doi: 10.1016/j.ydbio.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 35.DiLiberto M, Lai ZC, Fei H, Childs G. Developmental control of promoter-specific factors responsible for the embryonic activation and inactivation of the sea urchin early histone H3 gene. Genes Dev. 1989;3:973–985. doi: 10.1101/gad.3.7.973. [DOI] [PubMed] [Google Scholar]
- 36.Fei H, Childs G. Temporal embryonic expression of the sea urchin early H1 gene is controlled by sequences immediately upstream and downstream of the TATA element. Dev. Biol. 1993;155:383–395. doi: 10.1006/dbio.1993.1037. [DOI] [PubMed] [Google Scholar]
- 37.Medina R, Paredes R, Puchi M, Imschenetzky M, Montecino M. Developmentally-regulated interaction of a transcription factor complex containing CDP/cut with the early histone H3 gene promoter of the sea urchin Tetrapygus niger is associated with changes in chromatin structure and gene expression. Gene. 2001;272:237–248. doi: 10.1016/s0378-1119(01)00534-0. [DOI] [PubMed] [Google Scholar]
- 38.Ruddell A, Jacobs-Lorena M. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc. Natl Acad. Sci. USA. 1985;82:3316–3319. doi: 10.1073/pnas.82.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature. 2002;420:439–445. doi: 10.1038/nature01167. [DOI] [PubMed] [Google Scholar]
- 40.Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 42.Belozerov VE, Majumder P, Shen P, Cai HN. A novel boundary element may facilitate independent gene regulation in the Antennapedia complex of Drosophila. EMBO J. 2003;22:3113–3121. doi: 10.1093/emboj/cdg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majumder P, Roy S, Belozerov VE, Bosu D, Puppali M, Cai HN. Diverse transcription influences can be insulated by the Drosophila SF1 chromatin boundary. Nucleic Acids Res. 2009;37:4227–4233. doi: 10.1093/nar/gkp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 45.Lee IJ, Tung L, Bumcrot DA, Weinberg ES. UHF-1, a factor required for maximal transcription of early and late sea urchin histone H4 genes: analysis of promoter-binding sites. Mol. Cell Biol. 1991;11:1048–1061. doi: 10.1128/mcb.11.2.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell J, Char BR, Maxson R. An octamer element is required for the expression of the alpha H2B histone gene during the early development of the sea urchin. Dev. Biol. 1992;150:363–371. doi: 10.1016/0012-1606(92)90248-f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.