Abstract
In mammals, the dosage compensation of sex chromosomes between males and females is achieved by transcriptional inactivation of one of the two X chromosomes in females. However, a number of genes escape X-inactivation in humans. It remains poorly understood how the transcriptional activity of these ‘escape genes’ is maintained despite the chromosome-wide heterochromatin formation. To address this question, we analyzed a putative chromatin boundary between the inactivated RBM10 and an escape gene, UBA1/UBE1. Chromatin immunoprecipitation revealed that trimethylated histone H3 lysine 9 and H4 lysine 20 were enriched in the last exon through the proximal downstream region of RBM10, but were remarkably diminished at ∼2 kb upstream of the UBA1 transcription start site. Whereas RNA polymerase II was not loaded onto the intergenic region, CTCF (CCCTC binding factor) was enriched around the boundary, where some CpG sites were hypomethylated specifically on inactive X. These findings suggest that local DNA hypomethylation and CTCF binding are involved in the formation of a chromatin boundary, which protects the UBA1 escape gene against the chromosome-wide transcriptional silencing.
INTRODUCTION
The unbalanced gene dosage of sex chromosomes between males (XY) and females (XX) represents an impediment to normal development. In mammals, X-chromosome inactivation (XCI) is achieved by transcriptional silencing of all but one of the X chromosomes in a diploid female cell, to equalize the gene dosage of X chromosomes between males and females (1). The XIST/Xist gene, which maps to the X-inactivation center, is expressed from the inactive X-chromosome (Xi) in female somatic cells (2). XIST/Xist RNA is essential for the initiation of XCI (3,4), playing a key role as a cis-acting factor in recruiting the silencing machineries. Once XCI is initiated, genes on the X chromosome undergo inactivation by sequential epigenetic modifications, and an inactive chromatin status characteristic of heterochromatin is stably maintained through cell generations. A number of genes, however, escape XCI in human cells and are transcriptionally active (5,6); more than 15% of human X-linked genes are reported to escape the inactivation (7). These escape genes can be categorized into two groups based on the presence or absence of Y-homologs.
The distal ends of the short and long arms of the human sex chromosomes are known as pseudoautosomal regions 1 and 2 (PAR1 and PAR2), which have homologous sequences with Y chromosome and are required for X–Y pairing in male meiosis. Most of the genes mapped on PARs escape XCI (7,8).
A number of genes that do not have Y-homologs, on the other hand, also escape inactivation in human cells. However, these genes are mostly inactivated in mice (9). Such different sets of escape genes between human and mice may reflect evolutionary differences and could explain the distinct phenotypes when one X is missing. The absence of a single X-chromosome in humans (45, X) results in Turner syndrome, which shows a more severe phenotype compared to X0 mice (10). This suggests that double dose of transcription found in escape genes without Y-homolog is crucial for normal female development in human but not in mice, but the significance of the dosage of escape genes remains poorly understood.
Post-translational histone modifications like acetylation and methylation play important roles in the epigenetic control of gene expression in eukaryotes, in addition to DNA methylation (11–13). Hypoacetylation of histone H4 and methylation at lysine 9 and 27 of H3 (H3K9 and H3K27), for instance, are associated with chromatin compaction and gene repression. In contrast, euchromatic domains display higher levels of histone acetylation (14–17). Recent studies established that the methylation of H3K9 and H3K27 is an early event in the XCI process (18–22) proceeding to DNA methylation on the promoters of inactivated genes (23). In contrast to the inactivated genes, the escape genes lack DNA methylation and H3K9 methylation and are associated with euchromatic features like histone acetylation, H3K4 methylation, and early replication (24,25).
Here we analyse the transcriptional status of escaped genes and their neighbouring genes to analyze chromatin boundaries in detail. For this, we used human–mouse hybrid cells carrying either active or inactive human X chromosome in mouse background. We then focused on the RBM10-UBA1/UBE1 locus on Xp11.23, where the inactivated RBM10 and escape UBA1 are separated by only four kilobases of intergenic sequences. By profiling histone modifications using chromatin immunoprecipitation (ChIP), we detect a chromatin boundary in the RBM10-UBA1 intergenic region. Trimethylated H3K9 and H4K20 (H3K9me3 and H4K20me3) were enriched in the last exon through the proximal downstream region of RBM10 but were strongly diminished at ∼2 kb upstream of UBA1 on Xi. As previously found in other boundaries on Xi (26), ChIP also revealed association of CTCF to the intergenic region, suggesting the involvement of this zinc finger protein in maintaining the transcriptional activity of UBA1 and its downstream escape genes.
MATERIALS AND METHODS
Cells and cytogenetics
A9 (7149)-5 (27) and CF150 (28) cells harboring human active and Xi chromosomes, respectively, were generous gifts of Dr M. Oshimura and Dr T.K. Mohandas. All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum in a 5% CO2 incubator.
For cytogenetic analysis, cells were incubated in 100 µg/ml 5-bromo-2′-deoxyuridine (BrdU) for 6 h, 1 µg/ml colcemid was added to the culture medium, and cells were further incubated for 1 h. Chromosome spreads and staining were prepared according to the method described previously (29). Briefly, cells were treated with 75 mM KCl for 10 min, fixed with 3:1 methanol: glacial acetic acid on ice, and air-dried on clean glass slides. Chromosome spreads were stained with freshly prepared acridine orange (Sigma) and examined under a fluorescence microscope (BX-81; Olympus) using an oil-immersion 100× UPlanApo objective lens (NA: 1.35) equipped with a cooled CCD (ORCA-ER; Hamamatsu Photonics).
RNA extraction and RT–PCR
Total RNA was prepared from each cell line using TRIzol (Invitrogen) and treated with RNase-free DNase I (Roche). To check the transcriptional status of X-linked genes, cDNA was synthesized from 1 µg RNA using SuperScript VILO cDNA synthesis kit (Invitrogen) as described by the manufacturer. To detect the sense/antisense transcripts in RBM10-UBA1 intergenic region, cDNA was synthesized from 2 µg RNA using One-step RT–PCR kit (Qiagen) using a strand-specific primer as described by the manufacturer. To avoid primer-independent reverse transcription due to the secondary structure, the reaction mixture was incubated at 60°C.
Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) using a 7500 FAST (Applied Biosystems). Each PCR was run in triplicate to control PCR variation. All primers used here (summarized in Table 1) have proven to be species-specific.
Table 1.
PCR primers
| Application | Gene name | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|---|
| RT–PCR | POLA | aaaggggcagatgaggaac | ttcccaaacagaaataccacac |
| PGK1 | aaagggaagcgggtcgttatc | tgcccagcagagatttgagt | |
| XIST | gatgggttgccagctatc | ggatcctcatgccccatctc | |
| RBM10 | ggacctatgttcccgccc | ctcgtcatctcgcaggga | |
| UBA1 TSS1 | ttccgagctccgcgcaaact | gcacttcggacaacacggactg | |
| UBA1 TSS1′ | ttgtgtcggcggctcggctgtaag | gcacttcggacaacacggactg | |
| USP11 | catgaaacatccattcttcctcagtc | ctgagtcaggcatcgacgaa | |
| ZNF157 | aggaccctctacacacagacagc | ctgtcgggtgaaatccacagccac | |
| KLHL15 | atcattcagaatatccggttttgct | ttcaatgcttggtcaacttcgtaa | |
| EIF2S3 | ctgtcctggccacgatattt | aggtgttccgatgtctgagg | |
| ZFX | ctgatcctctgactaccgacgtagtt | aatcataaggtagtcctcacagttgcc | |
| Mouse Tbp | accacccccttgtacccttc | tttacagccaagattcacggtag | |
| ChIP and strand-specific RT-PCR | RBM10 Ex20 | gaggactggtggcagccta | ccagtcggtgagcttctcct |
| RBM10 Ex22 | cagctgaacgcagagaaaag | atatgccgccgtacttcctc | |
| RBM10 Ex24 | gggcagggaaggacagagtg | gatttggtcaagccctctctg | |
| RU1 | ctagcaccaccaggcatgaa | tccagatctagggatggcaaa | |
| RU2 | ccagcaatttaggaggctga | tgcgcccagcctaattttat | |
| RU3 | ggaacaacccaaatgtccaa | caccatgtgaacaatctcacca | |
| RU4 | tcaaatgccaaaactgttcca | cacatcatctgggccctttt | |
| RU5 | accacagatgcaccccacta | tgaggcaggatgattgcttg | |
| RU6 | ccacctgagtctcggtctcc | gcaccagcgtccagaatatg | |
| UBA1 Ex1 | ttccgagctccgcgcaaact | ttcctgccaaagatgagctgaagc | |
| Intron(In) 1−1 | ggaggctcaggactgcgagac | ggcgcctcgtcagctcagga | |
| In1−2 | gggatgaggcctcctacattgtt | cctcatacccattattcaggcttac | |
| In1−3 | ctccatcgcaaggtaagttgctg | gggatgagccttagaacattgctggc | |
| In1−4 | gctgggattacaggcgtcatct | ccgcctcaggaacaaggg | |
| In1−5 | gcagcaataggcgggcgt | gccacacacccccattcat | |
| UBA1 Ex1′ | ttgtgtcggcggctcggctgtaag | ccgccaactcctcaaggagccgaa | |
| In1′−1 | acatgctagccgcgctcag | ggaagatgctagggccaggg | |
| In1′−2 | tgcagtcaggggttttcagctt | cctggacgacatagtgaaacaattctg | |
| In1′−3 | caccctcagagcagctgtgatacc | gaggctgtgagggtggatgg | |
| In1′−4 | cccagaaagcttgacatgctcc | gggtttgagggaggagtcttggag | |
| In1′−5 | cctcaggtcagtgagttctgaccacag | ctgactgacctggggtagagatctc | |
| In1′−6 | tcagtgtgggaggacttgtgtgaa | ctggtggtgggggtgactg | |
| In1′−7 | cggttttcccctcccaaagc | gagtgcaggagggaagtgaggc | |
| UBA1 Ex2 | atgtccagctcgccgctgtccaag | gcacttcggacaacacggactg | |
| XIST + 1 | ccttcagttcttaaagcgctgcaa | ggtgatttttttaaagaaatacgccat | |
| XIST + 780 | tgggttgttgcactctctgg | ctgcctgacctgctatcatcc | |
| PGK1 + 98 | gcacgtcggcagtcggct | cctcataacgacccgcttccc | |
| PGK1 + 700 | acttgctggcctccaatctc | agaaaccgtgttggcaagtg | |
| RBM10 + 67 | gtaggcggcagtgagtttcc | tcctccaactctcccagctc | |
| RBM10 + 600 | ccacttattgggtgggaagg | tttccatggatgagcgtcag | |
| H19 ICR | ccgagaaaatagccattgcctacagt | catgttcctttgagtcctgggtgta | |
| Eif2 | ggttggaggccagtatgaga | cggcaacatgctgttaagac | |
| KIAA0522 | agccagctgggataaggaat | cctggggtcttcacctaaca | |
| Bisulfite sequencing | RBM10 Ex24 | gggtagggaaggatagagtgttg | aactccacctccacccctcta |
| RU2 to RU3 | ggttttttgggtttgggtatagtgagttg | aaaatttcactcttatcccccaaac | |
| RU5 | agtttgttaaagtgatgggattatagg | ccaaactaaactaaaactcctaaactcaaac | |
| RU6 to UBA1 Ex1 | gggtgtgtttttttatgtggaggt | caccctcctccaaatactatc | |
| In1−1 | ggtattgtggaggtgttaggtttttgtg | caaaacgccccttcaacacgtccc | |
| In1−4 | gttgtggaaattgaggtatttgttaaag | cacccaaaactaccctacccaac | |
| UBA1 Ex1′ | gtttttgggggtggggtttaag | aaatccactccccctccctactc |
Chromatin immunoprecipitation using native chromatin
We used mouse monoclonal antibodies specific to different H3 modifications (30) and commercial antibodies including di + tri-methyl H3K4 (H3K4me2 + 3; Abcam; ab6000), di-methyl H3K9 (H3K9me2; Abcam; ab1220), tri-methyl H3K27 (H3K27me3; Abcam; ab6002), tri-methyl H4K20 (H4K20me3; Abcam; ab9053). Control mouse IgG (Jackson Immunoresearch) was also used.
To profile the histone modifications, native chromatin was prepared and used for chromatin immunoprecipitation (ChIP) assay as described previously (15,31) with slight modifications. Briefly, ∼15 µg of native chromatin (∼2–4 nucleosomes) was incubated with 15–20 µl hybridoma culture sup or 10–20 µg IgG for 15 h at 4°C in ChIP buffer [25 mM Tris–HCl (pH 7.6), 500 mM NaCl, 5 mM EDTA and Protease inhibitor cocktail (Roche; Complete, EDTA-free)]. After further incubation for 2 h with 80 µl Dynabeads M-280 Sheep anti-mouse IgG (for mouse IgG; Dynal), or 50 µl Dynabeads Protein G (for rabbit IgG; Dynal), chromatin-antibody complexes (‘bound’ fraction) were separated from antibody-free chromatin (‘unbound’ fraction) using a magnetic stand (Dynal). After rinsing three times with ChIP buffer, bound-chromatin was eluted from the beads using ChIP buffer containing 1% SDS. At least three independent ChIP experiments were performed for each antibody.
ChIP using cross-linked chromatin
To analyze the binding of CTCF and RNA polymerase with the chromatin, we used another ChIP method using cross-linked chromatin as described before (30,32). Sonicated chromatin (∼300–1500 bp; ∼15 µg) was incubated with mouse monoclonal anti-CTCF (BD transduction Laboratories, 612148; 15 µg), anti-RNA polymerase, B8-1 (33) (10 µg), 8WG16 (Covance; 10 µl) and H14 (Covance; 10 µl), or normal mouse IgG (Jackson Immunoresearch; 5 µg) for 15 h at 4°C in RIPA buffer [50 mM Tris–Cl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate and Protease inhibitor cocktail (Complete, EDTA-free; Roche)]. After further incubation for 2 h with 80 µl Dynabeads M-280 Sheep anti-mouse IgG (Dynal), or anti-mouse IgM (Dynal) for H14, immunoprecipitates were collected, washed with RIPA buffer containing 150 mM and 500 mM NaCl, and resuspended in 200 µl of 10 mM Tris–HCl (pH 8.0), 5 mM EDTA, 300 mM NaCl and 0.5% SDS. The chromatin-bound beads (‘bound’ fraction) and the input fraction were incubated at 65°C for 12 h to reverse the cross-linking.
Quantitative analysis of ChIP samples
DNA was extracted from the input, ‘bound’ and ‘unbound’ fractions with phenol/chloroform, precipitated with ethanol, and re-suspended in 60 µl TE [10 mM Tris–HCl (pH 7.6) and 1 mM EDTA]. The DNA sample (1 µl) was used for quantitative PCR with QuantiTect SYBR Green PCR (Qiagen) using an Opticon2 (BioRad). Each PCR was run in triplicate to control PCR variation. Primers used here are listed in Table 1.
DNA methylation assay
Bisulfite treatment was carried out using EZ DNA methylation kit (ZYMO RESEARCH) according to the manufacturer’s instruction, and the completion of reaction was confirmed using control samples containing non- and fully-methylated templates. To amplify bisulfite-treated genomic DNA, primers for PCR amplification are designed to contain degenerate nucleotides (C to T) except CpG dinucleotide sequence (Table 1). PCR products were cloned into Topo-TA vector (Invitrogen) and the nucleotide sequence was determined using a PRISM3100-Avant (ABI).
RESULTS
Status of human X chromosomes in hybrid cell lines resembles that in diploid human cells
To analyze active and inactive X chromosomes (Xa and Xi) separately, we used human–mouse hybrid cell lines, A9 (7149)-5 [hereafter described as HX; (27)] and CF150 [CF; (28)] harboring active and inactive human X-chromosome, respectively. These were generated by microcell-mediated chromosome transfer into mouse A9 cells.
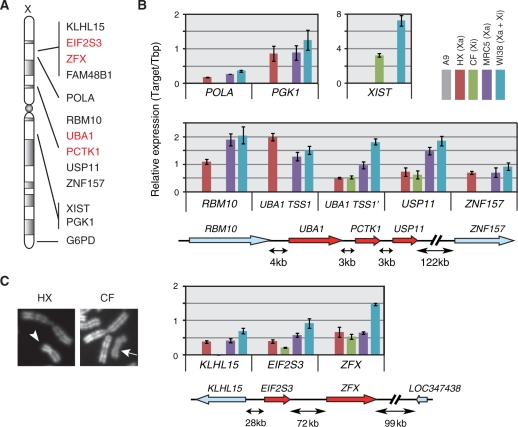
We first examined whether the active or inactive status of human X chromosomes was maintained in HX and CF cells. By quantitative RT-PCR, the expression of several human X-linked genes was examined in A9 (parent mouse cells; a negative control), HX and CF, and compared with that in diploid human male and female cells (i.e. MRC5 harboring Xa and WI38 harboring both Xa and Xi, respectively) (Figure 1A and B). The expression of typical human X-linked genes that are repressed on Xi (i.e. POLA, PGK1 and G6PD) was detected in cells harboring human Xa, such as HX, MRC5 and WI38 (Figure 1B, top; data not shown). In contrast, XIST was expressed in cells harboring human Xi (CF and WI38; Figure 1B, top). To evaluate the replication timing of human X chromosomes in hybrid cells, the cytogenetic examination using R-banding (29) was employed. In chromosome spreads from HX cells, human X exhibited a typical banding pattern of active X (Xa) due to its asynchronous replication, whereas all human X in CF spreads appeared as homogenous late replication staining, characteristic of the Xi (Figure 1C). These results indicated that human Xa and Xi were stably maintained in hybrid cell lines HX and CF, respectively (27,28).
Figure 1.
The status of human X chromosomes in the human–mouse hybrid cell lines. (A) Schematic human X-chromosome map with the inactivated and escape genes examined in this study. (B) Quantitative RT–PCR analysis of human X-linked genes in different cell lines. Total RNA was prepared from A9 (parental mouse cell line for HX and CF: without human X), HX (mouse line harboring active human X), CF (mouse line harboring inactive human X), MRC5 (46, XY; male diploid human cells harboring active X) and WI38 (46, XX; female diploid human cells harboring active and inactive Xs). The levels of transcripts were analyzed using real-time PCR, and expressed as relative amounts to human TBP (for MRC5 and WI38) or mouse Tbp (for A9, HX and CF). (C) Representative X chromosome banding patterns. After the incubation with BrdU for 6 h to label late replicating DNA, cells were treated with colcemid for chromosome spreading and R-banding. A typical human active X chromosome is observed in HX cell spread (left panel; arrowhead). In CF cell spread, human inactive X chromosome shows homogenous dimmer staining (right panel; arrow).
To explore the chromatin boundary between inactivated and escape genes, we next validated the expression of three previously reported escape genes (2,7,34,35), UBA1 (ubiquitin-like modifier activating enzyme 1; also known as UBE1) and PCTK1 (PCTAIRE protein kinase 1), EIF2S3 (eukaryotic translation initiation factor 2, subunit 3 gamma) and their flanking genes (Figure 1A and B). UBA1 consists of two transcription start sites (TSSs) with the alternative non-coding first exons. Hereafter, the first TSS is numbered +1 and called UBA1 TSS1, and the second, alternative TSS (at +3036) called UBA1 TSS1′; accordingly, the first exons from TSS1 and TSS1′ are called Ex1 and Ex1′, respectively. Quantitative RT-PCR analysis showed that the transcripts from Ex1 were detected in HX, MRC5 and WI38, and those from Ex1′ in all cells harboring human X including CF (Figure 1B, middle). The relative amount of UBA1 Ex1′ transcripts was roughly 2-fold in WI38 compared with MRC5, whereas that of Ex1 was comparable in the two. These results suggest that TSS1′, but not TSS1, escaped inactivation in UBA1 gene in WI38 and CF.
In the same locus, transcripts from 3′ flanking genes to UBA1 (i.e. PCTK1 and USP11) were detected in all cell lines including CF harboring Xi (Figure 1B, middle), confirming previous data showing these genes escape from silencing as a cluster in other cell lines (7,34). In contrast, the expression of RBM10, located 4 kb upstream of UBA1, was detected only in the cells containing Xa (Figure 1B, middle), like other typical X-linked genes that undergo X-inactivation.
Another previously known escape gene EIF2S3 was transcribed from both human Xa and Xi in hybrid cells (Figure 1B, bottom), as anticipated (7,35). Among the genes near EIF2S3 (Figure 1A), KLHL15 (Kelch-like 15) located 28 kb upstream EIF2S3 was silenced in Xi (Figure 1B, bottom). In contrast, zinc finger protein, X-linked (ZFX), which is located 72 kb downstream EIF2S3 and known to be an escape gene (7), was transcribed in all cells (Figure 1C). We did not obtain any evidence for the expression of LOC347438/FAM48B1 (family with sequence similarity 48, member B1), a single exon gene located further 99 kb downstream, in any cell lines tested here (data not shown).
These results are entirely consistent with the previous data obtained by different hybrid cells (7,34), supporting the notion that clusters of genes spanning tens of kilobases escape X-inactivation constituting escape domains, and that chromatin boundaries between inactivated and escape regions should be present in the intergenic intervals.
Histone modification profile in RBM10-UBA1 intergenic region
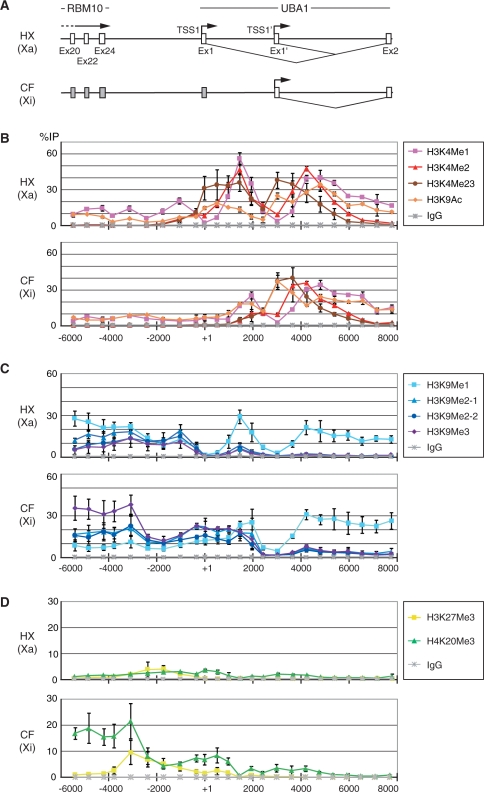
As the inactivated RBM10 and escape gene UBA1 are only ∼4 kb apart, we focused on this region to investigate the boundary between silenced and active chromatin using hybrid cells. By using ChIP, we profiled histone modifications characteristic to euchromatin and heterochromatin along a 14 kb region covering the last exon of RBM10 through the second exon of UBA1 (Figure 2). Histone H3 K9 acetylation and K4 methylation are in general regarded as euchromatic modifications associated with the transcriptional activation (12,36–38). ChIP of di + tri-methyl H3K4 (H3K4me2 + 3) showed that these modifications were highly enriched around TSS1 and TSS1′ on Xa (HX), and around TSS1′ on Xi (CF) (Figure 2B), in good agreement with their transcription status (Figure 1B). As H3K4me2 distributed downstream H3K4me2 + 3, H3K4me3 appeared to be enriched more proximal to the TSSs. H3K4me1 was observed further downstream. H3K9ac was also enriched at active TSSs and downstream. Thus, all modifications characteristic to active chromatin were found at active TSSs and downstream but not in the intergenic region between the 3′-end of RBM10 and UBA1 TSS1. These results suggest that the active markers are simply correlated with transcription activities and may not be used as particular marks at the chromatin boundary in this locus.
Figure 2.
Histone modification profiles in the ∼12 kb region encompassing RBM10 exon 20 to UBA1 exon 2. ChIP was performed using antibodies against different modifications on histone H3K4, H3K9, H3K27 and H4K20. The enrichment of each region in immunoprecipitates was evaluated using real-time PCR and expressed as the percentage to the input (%IP). The mean values of three independent ChIP experiments are plotted with the standard deviations. (A) Schematic representation of the locus in HX (harboring Xa) and CF (harboring Xi). (B) ChIP with histone H3K4me1, me2, me2+3 and H3K9ac. (C) ChIP with histone H3K9me1, me2 and me3. (D) ChIP with histone H3K27me3 and H4K20me3.
H3K9me2 and H3K9me3 were reported as key modifications for heterochromatin formation and maintenance (12). Whereas H3K9me3 was observed little throughout the intergenic region of RBM10-UBA1 locus on Xa, the same modification was highly enriched in inactivated RBM10 last exons through the proximal downstream region on Xi (Figure 2C). The level of H3K9me3 was remarkably diminished at the intergenic region between RBM10 and UBA1 (∼2 kb upstream UBA1 TSS1) and disappeared at active TSS1′ on Xi (Figure 2B). ChIP assay using two independent specific antibodies indicated that H3K9me2 showed similar distribution to H3K9me3. H3K9me1 was broadly detected from RBM10 to UBA1 except at around TSSs (Figure 2C). The level of monomethylation was relatively high in RBM10 transcribed region on Xa. As H3K9me1 was also enriched in UBA1 transcribed region on Xa and Xi, this modification seems to be correlated with active chromatin, in good agreement with recent genome-wide analysis (38).
We further examined other modifications associated with repressed chromatin such as H3K27me3 and H4K20me3 (21,39). H4K20me3 was highly enriched in the inactivated RBM10 to intergenic region on Xi similarly to H3K9me3 (Figure 2D). In contrast, enrichment of H3K27me3 was restricted to the transition point of H3K9me3 and H4K20me3 on Xi (Figure 2D). From these results, the heterochromatic feature of H3K9me3 and H4K20me3 enrichment in the intergenic proximal of RBM10 appears to be disrupted at ∼2 kb upstream of the UBA1 TSS1, and a chromatin boundary is established at this region.
Little transcription around the RBM10-UBA1 chromatin boundary
Non-coding transcripts are often found in locus control regions that regulate transcription of structural genes (40–44) and siRNA could participate in establishing heterochromatin (45–48). Therefore, we investigated whether we could detect transcripts from the RBM10-UBA1 intergenic region using strand-specific quantitative RT-PCR (Figure 3A and B).
Figure 3.
The presence of transcripts and RNA polymerase II on RBM10-UBA1 region. (A) Schematic representation of the locus in HX (harboring Xa) and CF (harboring Xi). (B) Relative amounts of sense and antisense transcripts measured by quantitative RT-PCR. (C) RNA polymerase II association. ChIP was performed using antibodies directed against Rpb8 (a shared subunit of RNA polymerase I, II and III; B8), unphosphorylated form of Pol II (8WG), and Ser5-phosphorylated form of Pol II (H14). The enrichment is expressed as the percentage to the input (%IP); the mean values from three independent experiments are plotted with the standard deviations.
RBM10 mRNA was clearly detected along the last several exons by priming RT with the reverse primer in HX harboring Xa, but not in CF harboring Xi (Figure 3B). In RBM10 proximal downstream regions (encompassing to the regions designated RU1 to RU3, from −3808 to −1595), sense transcripts primed with the reverse primers were also detected in HX (Figure 3B). These transcripts were likely to be read-through from RBM10 transcription unit, as the exon sequences were amplified from cDNA primed with the reverse primers in the intergenic regions (RU1-3) (data not shown). Antisense transcripts in these RBM10 proximal regions were, in contrast, undetectable in both cells under the similar experimental conditions. Transcripts from either direction were not detected in RU4 region (from −1722 to −1595) in both tested cells (Figure 3B). Subtle antisense transcripts, primed with the forward primers, were observed in RU5 (from −1010 to −915) and RU6 (from −308 to −223) regions through UBA1 gene on both Xa and Xi. These results suggest that RBM10 read-through and UBA1 antisense transcripts are terminated before approaching to RU4 region. We also detected weak signals of UBA1 sense transcripts in RU6 region in cells harboring Xa (HX). In UBA1 gene, the sense transcripts were clearly detected in the first exons (Ex1 and Ex1′, starting from TSS1 and TSS1′, respectively) and exon 2 (Ex2) in HX, whereas the transcripts were only detected in Ex1′ and Ex2, but not in Ex1, in CF (Figure 3B), consistent with the ChIP data with methylated H3K4 and H3K9ac associating with active TSSs (Figure 2).
To refine these results, we also used ChIP assay to investigate the presence of RNA polymerases in RBM10-UBA1 locus by using three monoclonal antibodies recognizing different forms (Figure 3C). B8 reacts with RPB8, a shared subunit of RNA polymerases I, II and III (33), and so chromatin fragments bound by any polymerase could be precipitated. 8WG16 and H14 recognize unphosphorylated and Ser5-phosphorylated forms of the C-terminal domain in the largest subunit of RNA polymerase II, respectively (49,50). While the pre-initiation complex on a promoter contains unphosphorylated form, Ser5 is phosphorylated during the initiation of transcription or when polymerases slow down at the posing and termination sites (51,52). ChIP with these antibodies revealed that RNA polymerase II was enriched around active RBM10 TSSs (Figure 3C). Ser5-phosphorylated form (recognized by H14) was subtly detected from the last exons of RBM10 to the proximal intergenic region (RU1 and in particular RU2, but not RU3), exclusively on Xa, which is probably associated with the termination of read-through transcripts. As polymerases were not detected in the intergenic region on Xi (Figure 3C), any specific transcripts, or transcription as such, does not appear to be crucial for the formation and maintenance of the chromatin boundary in this region.
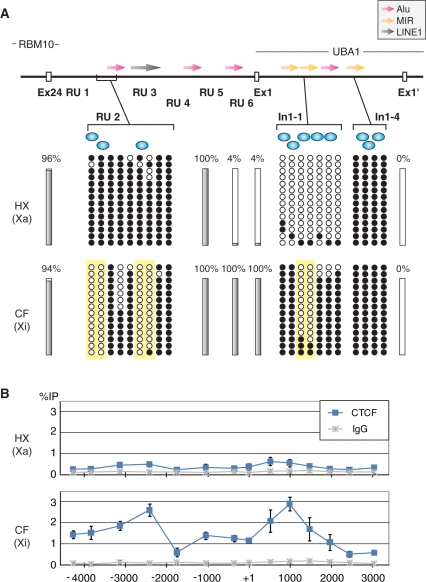
CpG hypomethylation and CTCF binding around the chromatin boundary on Xi
We next investigated DNA methylation at the RBM10-UBA1 locus (Figure 4A). CpG dinucleotides on Xa were almost fully methylated from the last exon of RBM10 through the intergenic regions (RU2 and RU5), and hypomethylated near TSS1 (RU6) and exon 1 of UBA1. CpG hypomethylation was also found in the 5′ region of the first intron (In1−1) and around TSS1′, but CpGs at the 3′ region of the first intron (In1−4) were fully methylated. These results suggest that CpG hypomethylation is limited around active TSSs including promoter and enhancer elements on Xa. In contrast, while most CpG sites were fully methylated on Xi other than around TSS1′, some CpGs in RU2 and In1−1 regions were remarkably protected from the methylation (Figure 4A). Such Xi-specific CpG hypomethylation might be involved in the chromatin boundary formation.
Figure 4.
DNA methylation status and CTCF binding at the chromatin boundary and flanking region. (A) The methylation status of the regional and individual CpG dinucleotides in RBM10-UBA1 region. Top: The gene structure is schematically represented with the positions of repeat sequences (Alu, red; MIR, yellow; and LINE1, grey) and putative CTCF binding sites (blue circle). Middle and bottom: DNA methylation status in HX and CF cells. Solid bars represent the methylation status of regions where no particular site exhibits a distinct pattern among several CpG sites. The percentages of methylation levels are indicated. Small circles represent the individual methylation status; methylated and unmethylated CpG sites are shown as filled and open circles, respectively. The inactive X-specific hypomethylated area is highlighted by yellow background. (B) ChIP with anti-CTCF. The enrichment is expressed as the percentage to the input (%IP); the mean values from three independent experiments are plotted with the standard deviations.
CTCF is known to be a methylation-sensitive DNA binding protein with chromatin insulator and enhancer blocking function (53–55). As CTCF also binds to 5′ regions of escape genes like Jarid1c and Eif2s3 (26), we investigated CTCF binding in RBM10-UBA1 locus. By homology search with the previously reported CTCF binding motif CCGCNNGGNGGCAG (56), several potential CTCF binding sites were found in RU1, RU2, In1−1 and In1−4, close to the hypomethylated CpG sites (Figure 4A). We then examined the CTCF binding in these regions using cross-linked ChIP (Figure 4B). In both HX and CF cells, the positive-control loci, including mouse H19 ICR [imprinting control region; (57)] and human EIF2S3 promoter (26), were enriched in anti-CTCF immunoprecipitates (data not shown). CTCF was distributed in the entire RBM10-UBA1 intergenic region on Xi, with peaks at RU2–RU3 and In1−1–In1−2, in contrast to Xa, where little CTCF binding is observed (Figure 4B). Thus, the CpG hypomethylation and CTCF binding around RU2–RU3 region appear to be correlated with the chromatin boundary formation.
DISCUSSION
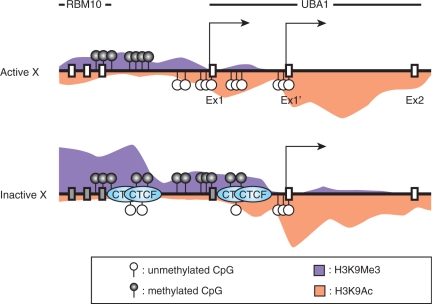
A number of genes on human Xi chromosome are expressed, or escaped from the silencing, even after the whole chromosome is inactivated in somatic cells. To distinguish the Xa and Xi alleles, F1 hybrid mice from different parent strains are often used (26); however, the analysis is restricted to the regions containing small nucleotide polymorphisms on mouse X chromosome. Inter-species hybrid cell lines containing only either active or inactive human X chromosome allow higher resolution analysis without allelic examination (7,26). We here used human–mouse hybrid cells to investigate the chromatin boundary between inactivated RBM10 and escape UBA1 genes on human X chromosome. As summarized in Figure 5, the TSS1 was silenced on Xi, whereas the TSS1 and the downstream alternative transcription start site (TSS1′) were both used on Xa. By detailed profiling of the histone modifications and transcription status using ChIP and real-time PCR at ∼500 bp intervals across the intergenic region, a boundary between inactive and active chromatin was found around the middle of the intergenic region (∼2 kb upstream the UBA1 TSS1) on Xi. Importantly, similar histone modification profiles were observed in diploid human fibroblasts. In male cells harboring only Xa (MRC5), the distributions of H3K4me1-3 and H3K9me1-3 were similar to those in HX (Supplementary Figure S1). In female cells harboring Xa and Xi (WI38), their distributions appeared as the mixture of HX and CF (Supplementary Figure S1). These results support the idea that the epigenetic status of X chromosomes that was established in human cells is maintained in the hybrid cells, as seen by others (7,26). We also found CTCF binding and hypomethylated CpG in this chromatin boundary region specifically on Xi; in contrast, H3K9 acetylation and RNA polymerases were hardly detected (Figure 5).
Figure 5.
Schematic representation of RBM10-UBA1 region. The enrichment of histone modifications (H3K9Ac and H3K9me3), CpG methylation status and CTCF binding sites are shown.
Histone modifications around the RBM10-UBA1 boundary on Xi
In the RBM10-UBA1 locus on Xi, H3K9me3 and H4K20me3 were enriched in RBM10 gene body and the proximal downstream region but diminished at the middle in the intergenic region. H3K27me3, a modification enriched in facultative heterochromatin, was distinctively detected around this boundary region. These data are consistent with the mosaic structure of different levels of heterochromatin found on human Xi (58), and possibly correlated with the organization of chromatin boundary formation and maintenance of escape genes.
In contrast to H3K9me3 and H3K9me2, H3K9me1 was broadly distributed throughout the transcribed region other than around active UBA1 TSSs, where H3K9 was heavily acetylated, suggesting that H3K9me1 is distributed broadly in relatively opened chromatin (38). All active TSSs also showed the prominent peak of H3K4me3, associated with those of H3K4me2 and then H3K4me1 shifted to downstream. These data are consistent with the previous genome wide and locus specific data (36,37,59). The monomethylated form may be a neutral state for H3K4 in open chromatin, because unmethylated and higher methylated forms may recruit DNA methyltransferase 3L (60) and transcription machineries (61).
Possible mechanisms of chromatin boundary formation in RBM10-UBA1 locus
CTCF-dependent insulators have been characterized in β-globin and imprinted IGF2/H19 loci (53,54,57,62). Non-coding transcripts and hyper-acetylation of H3/H4 were often associated with these insulators (63,64), suggesting the involvement of transcription in the insulator function. On mouse and human Xi, CTCF binding has also been reported at the 5′ end of escape genes, Jarid1c, Eif2s3x and EIF2S3, adjacent to an inactivated gene (26). Although the boundary between RBM10 and UBA1 seems similar to these insulators in terms of CTCF binding, there are two notable distinct features. First, it does not appear to be a typical LCR/ICR-type insulator, as little RNA polymerase and acetylated H3 were associated around the boundary. Second, the insulator function may not be very strong, because the transcription from TSS1 was repressed on Xi. The presence of two TSSs (i.e. tandem promoter/enhancer elements) may also contribute to maintain the UBA1 transcription activity by suppressing heterochromatin spreading from the upstream inactive region. In fact, chicken β-globin insulator, containing DNaseI-hypersensitive site 4, was not sufficient to intercept the X-inactivation, when GFP reporter gene flanked with the insulator was introduced into X-linked Hprt locus (65). Thus, CTCF-mediated insulators per se may not be sufficient to maintain the transcription activity on Xi due to the global silencing of the whole chromosome. Alternatively, protein complexes assembled on strong promoter/enhancer may also recruit CTCF to block the heterochromatin spreading.
The correlation between CTCF binding and DNA methylation has well been characterized in the insulators of the imprinted genes (54,57) as well as in the enhancer of Tsix (56). In the RBM10-UBA1 intergenic region, the site-specific CpG hypomethylation was also detected exclusively on Xi in the methylated domains on Xa. From the ChIP data, CTCF binding does not appear to be restricted to this hypomethylated region, but the binding was rather spread throughout the intergenic region. It is thus suggested that the CTCF-mediated insulator activity of UBA1 boundaries may be regulated by the specific CpG methylation, but the insulator machinery may progressively spread from the DNA methylation-free cardinal point to their vicinities. In good agreement with this notion, among seven CTCF binding sites in human H19 differential methylation region (DMR), only the sixth site is reported to act as a key regulator for switching between IGF2 and H19 expression, whereas the other sites can be hyper-methylated (66).
Consistent with previous reports (7,34), we here showed that PCTK1 and USP11, located 3 and 6 kb downstream UBA1, respectively, were also escaped in CF cells. As many escape genes are clustered (7), it is likely that the escaping from the inactivation is regulated at chromatin domain levels. Recent studies also suggest that the chromatin insulator activity is correlated with the formation of long-distance chromatin loops by CTCF and cohesin (67,68). During and after X inactivation, Jarid1c gene remains located outside the Xist-RNA accumulated, transcriptionally silent compartment (69), suggesting that the inactivated and escape domains can be separated into different chromatin loops anchored through CTCF and cohesin. If this is the case, the UBA1-PCTK1-USP11 escape domain may be extruded from the inactivated X chromosome body. The 3′ boundary of this escape domain should be laid in the intergenic region between USP11 and ZNF157, which is located ∼122 kb downstream, as ZNF157 and further downstream genes are silenced [Figure 1B and data not shown; (7)]. The large intergenic sequence between USP11 and ZNF157, however, presented a great difficulty to exploring the 3′ boundary of UBA1 escape domain.
Repetitive sequences in human RBM10-UBA1 intergenic region
In human, more than 15% of the X-linked genes escape the inactivation, and escape genes are spread throughout X chromosome with enrichments in PARs (7). Recent bioinformatic analyses revealed that the primary DNA sequence is correlated with the chromatin status on human Xi; Alu repetitive elements and CAG/CGT and GATA repeats are significantly enriched in the escape domain, whereas the long interspersed nuclear element (LINE) and mammalian-wide interspersed repeat (MIR) preferentially distributed in inactivated domains (70,71). The UBA1 boundary region indeed contains an Alu variant, in which the hypomethylated CpG sites were mapped. As mouse Uba1 is not an escape gene (72), the insertion of Alu element between RBM10 and UBA1 may contribute to the establishment of the escape domain in human. Although the downstream boundary region is not identified, fragmented Alu repeat-containing sequences account for ∼45% of the entire 122 kb intergenic region between USP11 and ZNF157, supporting the notion that Alu sequence may disturb the heterochromatin spreading on Xi. Further identification of the boundaries between the inactivated and escape domains may draw more clarified view how those domains are established and maintained.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-aid, the Genome Network Project, and the Special Coordination Funds for Promoting Science and Technology, each from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Funding for open access charge: Grant-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr T.K. Mohandas and Dr M. Oshimura for cell lines, and R. Feil and N. Takagi for valuable comments on the manuscript.
REFERENCES
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Brown CJ, Willard HF. Localization of a gene that escapes inactivation to the X chromosome proximal short arm: implications for X inactivation. Am. J. Hum. Genet. 1990;46:273–279. [PMC free article] [PubMed] [Google Scholar]
- 3.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 4.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 5.Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet. Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- 6.Brown CJ, Greally JM. A stain upon the silence: genes escaping X inactivation. Trends Genet. 2003;19:432–438. doi: 10.1016/S0168-9525(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 7.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 8.D'E;sposito M, Ciccodicola A, Gianfrancesco F, Esposito T, Flagiello L, Mazzarella R, Schlessinger D, D'U;rso M. A synaptobrevin-like gene in the Xq28 pseudoautosomal region undergoes X inactivation. Nat. Genet. 1996;13:227–229. doi: 10.1038/ng0696-227. [DOI] [PubMed] [Google Scholar]
- 9.Disteche CM. Escapees on the X chromosome. Proc. Natl Acad. Sci. USA. 1999;96:14180–14182. doi: 10.1073/pnas.96.25.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashworth A, Rastan S, Lovell-Badge R, Kay G. X-chromosome inactivation may explain the difference in viability of XO humans and mice. Nature. 1991;351:406–408. doi: 10.1038/351406a0. [DOI] [PubMed] [Google Scholar]
- 11.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 13.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 14.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'N;eill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. Embo J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. Embo J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters AH, O'C;arroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 18.Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 19.Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr. Biol. 2002;12:247–251. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- 20.Peters AH, Mermoud JE, O'C;arroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 21.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 22.Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, Avner P, Heard E. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol. Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu. Rev. Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 24.Goodfellow PJ, Mondello C, Darling SM, Pym B, Little P, Goodfellow PN. Absence of methylation of a CpG-rich region at the 5' end of the MIC2 gene on the active X, the inactive X, and the Y chromosome. Proc. Natl Acad. Sci. USA. 1988;85:5605–5609. doi: 10.1073/pnas.85.15.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 26.Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Kugoh H, Mitsuya K, Meguro M, Shigenami K, Schulz TC, Oshimura M. Mouse A9 cells containing single human chromosomes for analysis of genomic imprinting. DNA Res. 1999;6:165–172. doi: 10.1093/dnares/6.3.165. [DOI] [PubMed] [Google Scholar]
- 28.Ellison J, Passage M, Yu LC, Yen P, Mohandas TK, Shapiro L. Directed isolation of human genes that escape X inactivation. Somat. Cell Mol. Genet. 1992;18:259–268. doi: 10.1007/BF01233862. [DOI] [PubMed] [Google Scholar]
- 29.Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- 30.Kimura H, Hayashi-Takanaka Y, Goto Y, Takizawa N, Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- 31.Gregory RI, Randall TE, Johnson CA, Khosla S, Hatada I, O'N;eill LP, Turner BM, Feil R. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell Biol. 2001;21:5426–5436. doi: 10.1128/MCB.21.16.5426-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 33.Jones E, Kimura H, Vigneron M, Wang Z, Roeder RG, Cook PR. Isolation and characterization of monoclonal antibodies directed against subunits of human RNA polymerases I, II, and III. Exp. Cell Res. 2000;254:163–172. doi: 10.1006/excr.1999.4739. [DOI] [PubMed] [Google Scholar]
- 34.Carrel L, Clemson CM, Dunn JM, Miller AP, Hunt PA, Lawrence JB, Willard HF. X inactivation analysis and DNA methylation studies of the ubiquitin activating enzyme E1 and PCTAIRE-1 genes in human and mouse. Hum. Mol. Genet. 1996;5:391–401. doi: 10.1093/hmg/5.3.391. [DOI] [PubMed] [Google Scholar]
- 35.Ehrmann IE, Ellis PS, Mazeyrat S, Duthie S, Brockdorff N, Mattei MG, Gavin MA, Affara NA, Brown GM, Simpson E, et al. Characterization of genes encoding translation initiation factor eIF-2gamma in mouse and human: sex chromosome localization, escape from X-inactivation and evolution. Hum. Mol. Genet. 1998;7:1725–1737. doi: 10.1093/hmg/7.11.1725. [DOI] [PubMed] [Google Scholar]
- 36.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalla R, Zhang MQ. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomic. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schotta G, Lachner M, Peters AH, Jenuwein T. The indexing potential of histone lysine methylation. Novartis Found. Symp. 2004;259:22–37. discussion 37–47, 163–169. [PubMed] [Google Scholar]
- 40.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 41.Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum. Mol. Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- 42.Zwart R, Sleutels F, Wutz A, Schinkel AH, Barlow DP. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001;15:2361–2366. doi: 10.1101/gad.206201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling J, Pi W, Yu X, Bengra C, Long Q, Jin H, Seyfang A, Tuan D. The ERV-9 LTR enhancer is not blocked by the HS5 insulator and synthesizes through the HS5 site non-coding, long RNAs that regulate LTR enhancer function. Nucleic Acids Res. 2003;31:4582–4596. doi: 10.1093/nar/gkg646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling J, Baibakov B, Pi W, Emerson BM, Tuan D. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Biol. 2005;350:883–896. doi: 10.1016/j.jmb.2005.05.039. [DOI] [PubMed] [Google Scholar]
- 45.Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 46.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 47.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 48.Schramke V, Allshire R. Those interfering little RNAs! Silencing and eliminating chromatin. Curr. Opin. Genet. Dev. 2004;14:174–180. doi: 10.1016/j.gde.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 50.Patturajan M, Wei X, Berezney R, Corden JL. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol. Cell Biol. 1998;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warren SL, Landolfi AS, Curtis C, Morrow JS. Cytostellin: a novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J. Cell Sci. 1992;103(Pt 2):381–388. doi: 10.1242/jcs.103.2.381. [DOI] [PubMed] [Google Scholar]
- 52.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 53.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 54.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 55.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 56.Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295:345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 57.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 58.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl Acad. Sci. USA. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol. Cell Biol. 2007;27:1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken beta-globin domain. Embo J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 64.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Ciavatta D, Kalantry S, Magnuson T, Smithies O. A DNA insulator prevents repression of a targeted X-linked transgene but not its random or imprinted X inactivation. Proc. Natl Acad. Sci. USA. 2006;103:9958–9963. doi: 10.1073/pnas.0603754103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takai D, Gonzales FA, Tsai YC, Thayer MJ, Jones PA. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum. Mol. Genet. 2001;10:2619–2626. doi: 10.1093/hmg/10.23.2619. [DOI] [PubMed] [Google Scholar]
- 67.Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 68.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 69.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McNeil JA, Smith KP, Hall LL, Lawrence JB. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 2006;16:477–484. doi: 10.1101/gr.4627606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Willard HF, Mukherjee S, Furey TS. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput. Biol. 2006;2:e113. doi: 10.1371/journal.pcbi.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Disteche CM, Zacksenhaus E, Adler DA, Bressler SL, Keitz BT, Chapman VM. Mapping and expression of the ubiquitin-activating enzyme E1 (Ube1) gene in the mouse. Mamm. Genome. 1992;3:156–161. doi: 10.1007/BF00352460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.