Abstract
19F NMR spectroscopy has proved to be a valuable tool to monitor functionally important conformational transitions of nucleic acids. Here, we present a systematic investigation on the application of 5-fluoro pyrimidines to probe DNA and RNA secondary structures. Oligonucleotides with the propensity to adapt secondary structure equilibria were chosen as model systems and analyzed by 1D 19F and 1H NMR spectroscopy. A comparison with the unmodified analogs revealed that the equilibrium characteristics of the bistable DNA and RNA oligonucleotides were hardly affected upon fluorine substitution at C5 of pyrimidines. This observation was in accordance with UV spectroscopic melting experiments which demonstrated that single 5-fluoro substitutions in double helices lead to comparable thermodynamic stabilities. Thus, 5-fluoro pyrimidine labeling of DNA and RNA can be reliably applied for NMR based nucleic acid secondary structure evaluation. Furthermore, we developed a facile synthetic route towards 5-fluoro cytidine phosphoramidites that enables their convenient site-specific incorporation into oligonucleotides by solid-phase synthesis.
INTRODUCTION
The use of fluorine labels in context with NMR spectroscopy represents an emerging tool to study structure and function of nucleic acids (1–3). In contrast to 1H NMR spectroscopy, the problem of resonance degeneracy is practically absent for 19F NMR spectroscopy. Together with the option to site-specifically incorporate fluorine labels into DNA and RNA and thus to avoid time-consuming signal assignment procedures, these advantages can be exploited to monitor conformational changes and folding of nucleic acids or to investigate binding processes in small molecule/nucleic acid or enzyme/nucleic acid complexes. This was demonstrated with short oligonucleotides (4–17) but also with transfer RNAs (18,19), ribozymes (20), RNA aptamers (21), or with substrates of DNA methyltransferases (22), to mention a few examples.
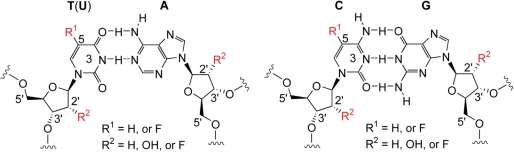
Concerning the position of labeling, fluorine modifications at both the ribose and the nucleobase unit come into consideration (Figure 1). The majority of 19F NMR studies on nucleic acids have involved 5-fluoro uracil so far which is easily accessible as nucleoside phosphoramidite either for solid phase synthesis of DNA or RNA (6,23), or as nucleoside triphosphate derivative for enzymatic RNA synthesis by T7 polymerase (18,19,24,25). Significantly less studies deal with 5-fluoro cytosine in nucleic acids (22,24–27). A comprehensive and comparative study on the usage of both 5-fluoro uracil and 5-fluoro cytosine as potential labels for NMR based DNA and RNA secondary structure probing (28) is lacking to date and represents a major goal here. This includes a critical assessment of the impact of these fluorine modifications on DNA and RNA base pairing properties. For such an undertaking, we considered short bistable RNA and DNA oligonucleotides to be the most appropriate systems (29–34). Bistable nucleic acids are highly sensitive with respect to single nucleotide mutations or chemical modifications since they directly reflect the interference by a change of the secondary structure equilibrium position. We previously took advantage of this sensitivity to explore the influence of nucleobase methylations and to control switching of secondary structures for a series of RNAs (30,31). Others extended the latter concept by using photolabile chemical modifications to study photochemically induced RNA folding by real time NMR spectroscopy (33,35,36) and other biophysical methods (37–41). In this sense, the use of short bistable oligonucleotides will directly reveal the potential influence of 5-fluoro pyrimidine labeling on secondary structure formation—if there is any.
Figure 1.
Chemical structure of Watson–Crick base pairs in DNA and RNA with potential positions for non-invasive fluoro labeling as indicated. While ribose 2′-fluoro labels were evaluated for RNA secondary structure probing previously in our laboratory (42), the present study is focused on a comprehensive comparison of 5-fluoro pyrimidine labeling for both DNA and RNA structure probing by 19F NMR spectroscopy.
MATERIALS AND METHODS
Synthesis of 5-fluoro cytidine phosphoramidites (5 and 9)
General
1H, 13C and 31P NMR spectra were recorded on a Bruker DRX 300 MHz, Avance II+ 600 MHz or Varian Unity 500 MHz instrument. The chemical shifts are referenced to the residual proton signal of the deuterated solvents: CDCl3 (7.26 ppm), d6-DMSO (2.49 ppm) for 1H NMR spectra; CDCl3 (77.0 ppm) or d6-DMSO (39.5 ppm) for 13C NMR spectra. 31P-shifts are relative to external 85% phosphoric acid. 1H- and 13C-assignments were based on COSY and HSQC experiments. UV-spectra were recorded on a Varian Cary 100 spectrophotometer. Analytical thin-layer chromatography (TLC) was carried out on silica 60F-254 plates. Flash column chromatography was carried out on silica gel 60 (230–400 mesh). All reactions were carried out under argon atmosphere. Chemical reagents and solvents were purchased from commercial suppliers and used without further purification. Organic solvents for reactions were dried overnight over freshly activated molecular sieves (4 Å).
N4-acetyl-5-fluoro cytidine (2)
A suspension of 5-fluoro cytidine 1 (300 mg, 1.149 mmol) in anhydrous ethanol (12 ml) was treated with acetic anhydride (1.7 ml, 13.78 mmol) and stirred for 14 h at room temperature. After evaporation of the solvent, the product was coevaporated three times with diethylether and dried under high vacuum. The crude product was then suspended in 5 ml diethylether, filtrated and washed thoroughly with ether. Yield: 346 mg of 2 as white powder (99%). TLC (CH2Cl2/CH3OH, 9/1): Rf = 0.17; 1H NMR (300 MHz, DMSO): δ 2.20 (s, 3H, COCH3); 3.59 [m, 1H, H1–C(5′)]; 3.75 [m, 1H, H2–C(5′)]; 3.89 [m, 1H, H–C(4′)]; 3.97 [m, 2H, H–C(3′) + H–C(2′)]; 4.98 [m, 1H, HO–C(2′) or HO–C(3′)]; 5.29 [m, 1H, HO–C(5′)]; 5.49 [m, 1H, HO–C(3′) or HO–C(2′)]; 5.67 [s, 1H, H–C(1′)]; 8.65 [d, J = 6.6 Hz, 1H, H–C(6)]; 10.52 [s, br, 1H, HN–C(4)] ppm; 13C NMR (75 MHz, DMSO): δ 25.24 (COCH3); 60.00 [C(5′)]; 68.81, 75.11 [C(3′), C(2′)]; 84.67 [C(4′)]; 90.92 [C(1′)]; 130.39; 130.85; 135.99; 139.21; 153.03; 153.91; 170.12 (COCH3) ppm; UV/Vis (MeOH): λ (ε) = 260 (5100) nm (mol−1 dm3 cm−1); ESI-MS (m/z): [M + H]+ calculated for C11H14FN3O6, 304.25; found 304.09.
N4-acetyl-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine (3)
To a suspension of N4-acetyl-5-fluoro cytidine 2 (121 mg, 0.383 mmol) in pyridine (1.5 ml) was added 4,4′-dimethoxytritylchloride (143 mg, 0.421 mmol) in three portions over a period of 1 h. The reaction mixture was stirred for 14 h at room temperature, evaporated and coevaporated with toluene and dichloromethane. The crude product was purified by column chromatography on SiO2 (CH2Cl2/CH3OH, 99/1–97/3 v/v). Yield: 144 mg of 3 as white foam (60%). TLC (CH2Cl2/CH3OH, 9/1): Rf = 0.50; 1H NMR (300 MHz, CDCl3): δ 2.64 (s, 3H, COCH3); 3.34 [dd, J = 2.4, 11.1 Hz, 1H, H1–C(5′)]; 3.40 [dd, J = 2.4, 11.1 Hz, 1H, H2–C(5′)]; 3.58 [s, br, 1H, HO–C(3′)]; 3.78, 3.79 (2s, 6H, 2× OCH3); 4.40 [m, 1H, H–C(4′)]; 4.48 [m, 1H, H–C(3′)]; 4.53 [m, 1H, H–C(2′)]; 5.62 [s, br, 1H, HO–C(2′)]; 5.79 [d, J = 3.9 Hz, 1H, H–C(1′)]; 6.82 [m, 4H, H–C(ar)]; 7.25 [m, 9H, H–C(ar)]; 8.02 [s, br, 1H, HN–C(4)]; 8.11 [d, J = 5.7 Hz, 1H, H–C(6)] ppm; 13C NMR (75 MHz, CDCl3): δ 26.12 (COCH3); 55.18 (2× OCH3); 62.88 [C(5′)]; 72.00 [C(3′)]; 77.04 [C(2′)]; 86.14 [C(4′)]; 87.18; 93.39 [C(1′)]; 113.27, 113.32 [C(ar)]; 127.03 [C(ar)]; 127.79, 127.97 [C(ar)]; 128.43, 128.88; 129.79, 129.91 [C(ar)]; 134.90, 134.28 [C(ar)]; 144.04 [C(ar)]; 149.64; 152.60; 152.75; 154.09; 158.63; 158.68; 170.87 (COCH3) ppm; UV/Vis (MeOH): λ (ε) = 260 (5400) nm (mol−1 dm3 cm−1); ESI-MS (m/z): [M + H]+ calculated for C32H32FN3O8, 606.62; found 606.06.
N4-acetyl-2′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine (4a) and N4-acetyl-3′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine (4b)
Compound 3 (565 mg, 0.933 mmol) and silver nitrate (285 mg, 1.679 mmol) was stirred in THF (3.9 ml) and pyridine (0.4 ml) overnight at room temperature. Then, tert.-butyldimethylchlorosilane (253 mg, 1.679 mmol) was added and stirring was continued for 1 h. The resulting white suspension was filtered, the solvents were evaporated and the residue was coevaporated with dichloromethane. The crude product was purified by column chromatography on SiO2 (hexane/ethylacetate, 6/4 – 5/5 v/v). Yield: 325 mg of 4a (48%) and 95 mg of 4b (14%) as white foam. TLC (hexane/ethylacetate, 3/7): Rf (4a) = 0.50, Rf (4b) = 0.24; 1H NMR (300 MHz, CDCl3): 4a: δ 0.23 (s, 3H, SiCH3); 0.34 (s, 3H, SiCH3); 0.96 [s, 9H, SiC(CH3)3]; 2.41 [d, J = 9.3 Hz, 1H, HO–C(3′)]; 2.68 (s, br, 3H, COCH3); 3.57 [m, 2H, H2-C(5′)]; 3.80 [s, 6H, 2× OCH3]; 4.12 [m, 1H, H–C(4′)]; 4.40 [m, 1H, H–C(2′)]; 4.45 [m, 1H, H–C(3′)]; 5.77 [s, 1H, H–C(1′)]; 6.85 [d, J = 8.7 Hz, 4H, H–C(ar)]; 7.23–7.44 [m, 9H, H–C(ar)]; 7.81 [s, br, 1H, HN–C(4)]; 8.38 [d, J = 4.5 Hz, 1H, H–C(6)] ppm; 4b: δ 0.00 (s, 3H, SiCH3); 0.09 (s, 3H, SiCH3); 0.87 [s, 9H, SiC(CH3)3]; 2.71 (s, br, 3H, COCH3); 3.13 [s, br, 1H, HO–C(2′)]; 3.33 [dd, J = 3.0, 9.0 Hz 1H, H1–C(5′)]; 3.58 [m, 1H, H2–C(5′)]; 3.82 (s, 6H, 2× OCH3); 4.18 [m, 1H, H–C(4′)]; 4.22 [m, 1H, H–C(2′)]; 4.37 [m, 1H, H–C(3′)]; 5.96 [d, J = 3.0 Hz, 1H, H–C(1′)]; 6.86 [d, J = 9.0 Hz, 4H, H–C(ar)]; 7.25–7.40 [m, 9H, H–C(ar)]; 7.58 [s, br, 1H, HN–C(4)]; 8.25 [m, 1H, H–C(6)] ppm; 13C NMR (75 MHz, CDCl3): 4a: δ − 5.23, −4.19 (2× SiCH3); 18.22 [(SiC(CH3)3]; 25.98 [SiC(CH3)3]; 26.43 (COCH3); 55.37 (2× OCH3); 61.32 [C(5′)]; 69.22 [C(3′)]; 67.57 [C(2′)]; 83.45 [C(4′)]; 87.32; 91.09 [C(1′)]; 113.47, 133.51 [C(ar)]; 127.21 [C(ar)]; 128.12, 128.16 [C(ar)]; 128.46; 130.13, 130.19 [C(ar)]; 135.23, 135.48, 144.43 [C(ar)]; 152.66; 152.81; 158.83; 158.87; 171.64 (COCH3) ppm; UV/Vis (MeOH): 4a: λ (ε) = 260 (5500) nm (mol−1 dm3 cm−1); ESI-MS (m/z): 4a: [M + H]+ calculated for C38H46FN3O8Si, 720.88; found 720.21; 4b: [M + H]+ calculated for C38H46FN3O8Si, 720.88; found 720.23.
N4-acetyl-2′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine 3′-(2-cyanoethyl)-N,N-diisopropylphosphoramidite (5)
Compound 4a (251 mg, 0.349 mmol) was dissolved in a mixture of N-ethyldiisopropylamine (597 µl, 3.487 mmol) and 1-methylimidazole (138 µl, 1.743 mmol) in dichloromethane (5.4 ml). After 15 min at room temperature, (2-cyanoethyl)-N,N-diisopropyl-chlorophosphoramidite (166 mg, 0.697 mmol) was added slowly and the solution was stirred at room temperature for 2.5 h. Then, the solvent was evaporated, the residue was coevaporated with dichloromethane and dried on high vacuum. The crude product was purified by column chromatography on SiO2 [hexane/ethylacetate, 7/3 – 5/5 v/v (+0.5% NEt3)]. Yield: 186 mg of 5 (mixture of diastereomers) as white foam (58%). TLC (hexane/ethylacetate, 3/7): Rf = 0.57, 0.45; 1H NMR (600 MHz, CDCl3): δ 0.16, 0.28 (2s, 12H, 4× SiCH3); 0.91, 0.92 (2s, 18H, 2× [SiC(CH3)3]; 1.00–1.16 {m, 24H, 2× [(CH3)2CH]2N}; 2.42, 2.60 (2m, 4H, 2× CH2CN); 2.68 (s, br, 6H, 2× COCH3); 3.48–3.90 {m, 12H, 2× [(CH3)2CH]2N, 2× H2–C(5′), 2× POCH2}; 3.80 (s, 12H, 4× OCH3); 4.34–4.37 [m, 4H, 2× H–C(3′) + 2× H–C(4′)]; 4.47 [m, 2H, 2× H–C(2′)]; 4.67, 4.75 [2m, 2H, 2× H–C(1′)]; 6.81–6.85 [m, 8H, H–C(ar)]; 7.23–7.47 [m, 18H, H–C(ar)]; 8.37, 8.42 [2m, 2H, 2× H–C(6)] ppm; 31P NMR (121 MHz, CDCl3): δ 149.2, 151.5 ppm; UV/Vis (MeOH): λ (ε) = 260 (5700) nm (mol−1 dm3 cm−1); ESI-MS (m/z): [M + H]+ calculated for C47H63FN5O9PSi, 921.10; found 920.24.
N4-benzoyl-5-fluoro cytidine (6)
A suspension of 5-fluoro cytidine 1 (100 mg, 0.383 mmol) in anhydrous ethanol (4 ml) was treated with benzoic anhydride (1.04 g, 4.594 mmol) and stirred for 14 h at room temperature. After evaporation of the solvent, the product was coevaporated three times with diethylether and dried under high vacuum. The crude product was then suspended in 2 ml diethylether, filtrated and washed thoroughly with ether. Yield: 134 mg of 6 as white powder (99%). TLC (CH2Cl2/CH3OH, 90/10): Rf = 0.40; 1H NMR (300 MHz, DMSO): δ 3.63 [m, 1H, H1–C(5′)]; 3.74 [m, 1H, H2–C(5′)]; 3.91 [m, 1H, H–C(4′)]; 4.04 [m, 2H, H–C(3′) + H–C(2′)]; 5.05 [m, 1H, HO–C(2′) or HO–C(3′)]; 5.32 [m, 1H, HO–C(5′)]; 5.51 [m, 1H, HO–C(3′) or HO–C(2′)]; 5.74 [s, 1H, H–C(1′)]; 7.51 [m, 2H, H–C(ar)]; 7.62 [m, 1H, H–C(ar)]; 7.99 [m, 2H, H–C(ar)]; 8.61 [m, 1H, H–C(6)]; 11–12.5 [s, br, 1H, HN–C(4)] ppm; 13C NMR (75 MHz, DMSO): δ 60.30 [C(5′)]; 69.21, 74.90 [C(3′), C(2′)]; 85.00 [C(4′)]; 90.37 [C(1′)]; 129.11–129.85 [C(ar), C(6)]; 133.47 [C(ar)]; 137.59; 140.73 ppm; UV/Vis (MeOH): λ (ε) = 260 (1300) nm (mol−1 dm3 cm−1); ESI-MS (m/z): [M + H]+ calculated for C15H16FN3O6, 366.32; found 366.03.
N4-Benzoyl-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine (7)
To a suspension of N4-benzoyl-5-fluoro cytidine 6 (247 mg, 0.676 mmol) in pyridine (2.6 ml) was added 4,4′-dimethoxytritylchloride (252 mg, 0.744 mmol) in three portions over a period of 1 h. The reaction mixture was stirred for 14 h at room temperature, evaporated and coevaporated with toluene and dichloromethane. The crude product was purified by column chromatography on SiO2 (CH2Cl2/CH3OH, 99/1 – 97/3 v/v). Yield: 251 mg of 7 as white foam (56%). TLC (CH2Cl2/CH3OH, 9/1): Rf = 0.60; 1H NMR (300 MHz, CDCl3): δ 1.85 [s, br, 1H, HO–C(2′)]; 3.42 [m, 2H, H2–C(5′)]; 3.61 [s, br, 1H, HO–C(3′)]; 3.76, 3.77 (2s, 6H, 2× OCH3); 4.37 [m, 1H, H–C(4′)]; 4.48 [m, 1H, H–C(3′)]; 4.57 [m, 1H, H–C(2′)]; 5.85 [d, J = 3.6 Hz, 1H, H–C(1′)]; 6.84 [m, 4H, H–C(ar)]; 7.19–7.43 [m, 9H, H–C(ar)]; 7.48 [m, 2H, H–C(ar)]; 7.57 [m, 1H, H–C(ar)]; 8.07 [m, 1H, H–C(6)]; 8.16 [m, 2H, H–C(ar)] ppm; 13C NMR (75 MHz, CDCl3): δ 55.20 (2× OCH3); 62.89 [C(5′)]; 71.76 [C(3′)]; 76.68 [C(2′)]; 85.71 [C(4′)]; 87.40; 92.39 [C(1′)]; 113.33, 113.36 [C(ar)]; 127.08 [C(ar)]; 127.88, 128.03 [C(ar)]; 128.48 [C(ar)]; 129.83, 129.98 [C(ar)]; 133.13 [C(ar)]; 135.02, 135.37 [C(ar)]; 144.08 [C(ar)]; 158.67, 158.70 [C(ar)] ppm; UV/Vis (MeOH): λ (ε) = 260 (2200) nm (mol−1 dm3 cm−1); ESI-MS (m/z): [M + H]+ calculated for C37H34FN3O8, 668.69; found 668.15.
N4-benzoyl-2′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine (8a) and N4-benzoyl-3′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine (8b)
Compound 7 (251 mg, 0.376 mmol) and silver nitrate (115 mg, 0.677 mmol) was stirred in THF (1.5 ml) and pyridine (0.15 ml) overnight at room temperature. Then, tert.-butyldimethylchlorosilane (102 mg, 0.677 mmol) was added and stirring was continued for 1 h. The resulting white suspension was filtered, the solvents were evaporated and the residue was coevaporated with dichloromethane. The crude product was purified by column chromatography on SiO2 (hexane/ethylacetate, 8/2 – 7/3 v/v). Yield: 171 mg of 8a (58%) and 21 mg of 8b (7%) as white foam. TLC (hexane/ethylacetate, 7/3): Rf (8a) = 0.50, Rf (8b) = 0.26; 1H NMR (300 MHz, CDCl3): 8a: δ 0.20 (s, 3H, SiCH3); 0.21 (s, 3H, SiCH3); 0.96 [s, 9H, SiC(CH3)3]; 2.64 [m, 1H, HO–C(3′)]; 3.52 [m, 2H, H2–C(5′)]; 3.82 (s, 6H, 2× OCH3); 4.18 [m, 1H, H–C(4′)]; 4.39 [m, 1H, H–C(3′)]; 4.46 [m, 1H, H–C(2′)]; 5.98 [d, J = 2.7 Hz, 1H, H–C(1′)]; 6.88 [d, J = 8.1 Hz, 4H, H–C(ar)]; 7.23 – 7.49 [m, 11H, H–C(ar)]; 7.57 [m, 1H, H–C(ar)]; 8.11 [d, J = 5.7 Hz, 1H, H–C(6)]; 8.29 [d, J = 7.2 Hz, 2H, H–C(ar)]; 13.04 [s, br, 1H, HN–C(4)] ppm; 8b: δ 0.00 (s, 3H, SiCH3); 0.09 (s, 3H, SiCH3); 0.88 [s, 9H, SiC(CH3)3]; 2.84 [m, 1H, HO–C(2′)]; 3.36 [dd, J = 2.7, 11.1 Hz, 1H, H1–C(5′)]; 3.51 [dd, J = 2.1, 11.1 Hz, 1H, H2–C(5′)]; 3.80 (s, 6H, 2× OCH3); 4.10 [m, 1H, H–C(4′)]; 4.25 [m, 1H, H–C(2′)]; 4.33 [m, 1H, H–C(3′)]; 5.98 [d, J = 3.0 Hz, 1H, H–C(1′)]; 6.86 [d, J = 8.7 Hz, 4H, H–C(ar)]; 7.25–7.48 [m, 11H, H–C(ar)]; 7.55 [m, 1H, H–C(ar)]; 8.00 [d, J = 5.7 Hz, 1H, H-C(6)]; 8.28 [d, J = 7.5 Hz, 2H, H–C(ar)]; 13.01 [s, br, 1H, HN–C(4)] ppm; 13C NMR [75 MHz, CDCl3]: 8a: δ − 5.02, −4.50 (2× SiCH3); 18.17 [SiC(CH3)3]; 25.82 [SiC(CH3)3]; 55.41 (2× OCH3); 62.68 [C(5′)]; 70.93 [C(3′)]; 76.59 [C(2′)]; 84.07 [C(4′)]; 87.54; 89.35 [C(1′)]; 113.55, 133.58 [C(ar)]; 125.68; 127.33 [C(ar)]; 127.92, 127.98, 128.09, 128.24, 128.45, 129.28, 130.18, 133.17 [C(ar)]; 135.12; 135.39; 144.36; 152.60; 152.85; 158.91, 158.93 [C(ar)] ppm; UV/Vis (MeOH): 8a: λ (ε) = 260 (2900) nm (mol−1 dm3 cm−1); ESI-MS (m/z): 8a: [M + H]+ calculated for C43H48FN3O8Si, 782.95; found 782.16; 8b: [M + H]+ calculated for C43H48FN3O8Si, 782.95; found 782.21.
N4-benzoyl-2′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro cytidine 3′-(2-cyanoethyl)-N,N-diisopropylphosphoramidite (9)
Compound 8a (125 mg, 0.160 mmol) was dissolved in a mixture of N-ethyldiisopropylamine (185 µl, 1.60 mmol) and 1-methylimidazole (37 µl, 0.800 mmol) in dry dichloromethane (2.5 ml). After 15 min at room temperature, (2-cyanoethyl)-N,N-diisopropyl-chlorophosphoramidite (74 mg, 0.320 mmol) was added slowly and the solution was stirred at room temperature for 2.5 h. Then, the solvent was evaporated, the residue was coevaporated with dichloromethane and dried on high vacuum. The crude product was purified by column chromatography on SiO2 [hexane/ethylacetate, 7/3 v/v (+0.5% NEt3)]. Yield: 90 mg of 9 (mixture of diastereomers) as white foam (57%). TLC (hexane/ethylacetate, 7/3): Rf = 0.47; 1H NMR (600 MHz, CDCl3): δ 0.13, 0.14, 0.15, 0.16 (4s, 12H, 4× SiCH3); 0.92, 0.93 [2s, 18H, 2× SiC(CH3)3]; 1.05–1.26 {m, 24H, 2× [(CH3)2CH]2N}; 2.40, 2.64 (2m, 4H, 2× CH2CN); 3.45 [m, 4H, 2× H2–C(5′)]; 3.57–3.61 {m, 5H, 2× [(CH3)2CH]2N + 1× POCH(1)dia1}; 3.73 [m, 1H, 1× POCH(1)dia2]; 3.798, 3.804 (2s, 12H, 4× OCH3); 3.84 [m, 1H, 1× POCH(2)dia1]; 3.95 [m, 1H, POCH(2)dia2]; 4.26–4.35 [m, 4H, 2× H–C(3′) and H–C(4′)]; 4.46, 4.54 [2m, 2H, 2× H–C(2′)]; 5.91, 6.00 [2d, J = 4.2 Hz, 2H, 2× H–C(1′)]; 6.84–6.87 [m, 8H, H–C(ar)]; 7.23–7.53 [m, 22H, H–C(ar)]; 7.54 [m, 2H, H–C(ar)]; 8.07, 8.14 [2d, J = 6.0 Hz, 2H, 2× H–C(6)]; 8.28 [d, J = 7.8 Hz, 4H, H–C(ar)] ppm; 31P NMR (121 MHz, CDCl3): δ 150.4, 150.9 ppm; UV/Vis (MeOH): λ (ε) = 260 (2400) nm (mol−1 dm3 cm−1); ESI-MS (m/z): [M + H]+ calculated for C52H65FN5O9PSi, 983.17; found 982.12.
Solid-phase synthesis of oligonucleotides
2′-O-TOM standard nucleoside phosphoramidites were obtained from GlenResearch or ChemGenes. 2′-O-tert.-Butyldimethylsilyl-5′-O-(4,4′-dimethoxytrityl)-5-fluoro uridine phosphoramidite was purchased from ChemGenes, 5′-O-(4,4′-dimethoxytrityl)-5-fluoro 2′-deoxyuridine phosphoramidite and 5′-O-(4,4′-dimethoxytrityl)-5-fluoro-O4-(2,4,6-trimethylphenyl) 2′-deoxyuridine phosphoramidite were obtained from GlenResearch. All solid supports for DNA and RNA synthesis were purchased from GE Healthcare (Custom Primer Supports: riboC Ac 80, riboA 80, riboG 80, riboU 80; dA 80s, dC 80s, dG 80s, T 80s). All oligonucleotides were synthesized on Pharmacia instruments (Gene Assembler Plus) following DNA/RNA standard methods; detritylation (2.0 min): dichloroacetic acid/1,2-dichloroethane (4/96); coupling (3.0 min): phosphoramidites/acetonitrile (0.1 M × 120 µl) were activated by benzylthiotetrazole/acetonitrile (0.35 M × 360 µl); capping (3 × 0.4 min): A: Ac2O/sym-collidine/acetonitrile (20/30/50), B: 4-(dimethylamino)pyridine/acetonitrile (0.5 M), A/B = 1/1; oxidation (1.0 min): I2 (10 mM) in acetonitrile/sym-collidine/H2O (10/1/5). Solutions of standard amidites, tetrazole solutions and acetonitrile were dried over activated molecular sieves overnight. Solutions of 5-fluoro pyrimidine modified phosphoramidites were only dried for 4–6 h over molecular sieves before consumption.
Deprotection of oligonucleotides
DNA oligonucleotides were deprotected by treatment with ammonia in water (32%) and ethanol (final volume 1 ml, aqueous ammonia/ethanol 3/1, v/v) at 55°C for 16 h. The solution was evaporated to dryness and the crude oligonucleotide was dissolved in 1.0 ml of water. RNA oligonucleotides (including those containing N4-acetyl protected C5F) were deprotected by using MeNH2 in EtOH (8 M, 0.60 ml) and MeNH2 in H2O (40%, 0.60 ml) at room temperature for 5–6 h. After complete evaporation of the solution, the 2′-O-TOM protecting groups were removed by treatment with tetrabutylammonium fluoride trihydrate (TBAF•3H2O) in THF (1 M, 0.95 ml) for at least 12 h at 37°C. The reaction was quenched by addition of triethylammonium acetate (TEAA) (1 M, pH 7.0, 0.95 ml). The volume of the solution was reduced to 1 ml and the solution was loaded on a GE Healthcare HiPrep 26/10 Desalting column (2.6 × 10 cm; Sephadex G25). The crude RNA was eluted with H2O, evaporated to dryness and dissolved in 1.0 ml of water.
Analysis and purification of oligonucleotides
Analysis of crude oligonucleotides after deprotection was performed by anion-exchange chromatography on a Dionex DNAPac100 column (4 × 250 mm) at 80°C. Flow rate: 1 ml min−1; eluant A: 25 mM Tris–HCl (pH 8.0), 6 M urea; eluant B: 25 mM Tris–HCl (pH 8.0), 0.5 M NaClO4, 6 M urea; gradient: 0–40% B in A within 30 min (<20 nt) or 0–60% B in A within 45 min (>20 nt); UV-detection at 265 nm. Crude RNA products (trityl-off) were purified on a semipreparative Dionex DNAPac100 column (9 × 250 mm). Flow rate: 2 ml min−1; gradient: Δ5–10% B in A within 20 min. Fractions containing oligonucleotide were loaded on a C18 SepPak cartridge (waters/millipore), washed with 0.1–0.2 M (Et3NH)+ HCO3- and H2O, eluted with H2O/CH3CN and lyophilized to dryness. The purified oligonucleotides were characterized by mass spectrometry on a Finnigan LCQ Advantage MAX ion trap instrumentation connected to an Amersham Ettan micro LC system (negative-ion mode with a potential of −4 kV applied to the spray needle). LC: Sample (250 pmol of oligonucleotide dissolved in 20 µl of 20 mM EDTA solution; average injection volume: 10–20 µl); column (Amersham µRPC C2/C18; 2.1 × 100 mm) at 21°C; flow rate: 100 µl min−1; eluant A: 8.6 mM TEA, 100 mM 1,1,1,3,3,3-hexafluoro-2-propanol in H2O (pH 8.0); eluant B: methanol; gradient: 0–100% B in A within 30 min; UV detection at 254 nm.
NMR spectroscopy
19F NMR spectra with 1H-decoupling were recorded at a frequency of 564.7 MHz on a Bruker Avance II + 600 MHz NMR spectrometer equipped with a 5 mm QNP probe. Typical experimental parameters were chosen as follows: spectral width 5.647 kHz, 19F excitation pulse 12.4 μs, acquisition time 1.0 s, relaxation delay 2 s, number of scans 2 K, proton decoupling using Waltz-16 with γB1 = 1 kHz. Prior to Fourier transformation all time domain data was processed with an exponential window function using a line broadening factor of 2 Hz. 19F-resonances were referenced relative to external CCl3F. Sample preparation: oligonucleotides (triethylammonium salts) together with the adequate amount of sodium arsenate buffer were lyophilized to dryness, and subsequently dissolved in H2O/D2O (9/1, v/v, 500 μl). All samples were heated to 90°C for 2 min, then cooled to room temperature and equilibrated for one hour before measurements.
RESULTS AND DISCUSSION
Synthesis of 5-fluoro pyrimidine containing DNA and RNA
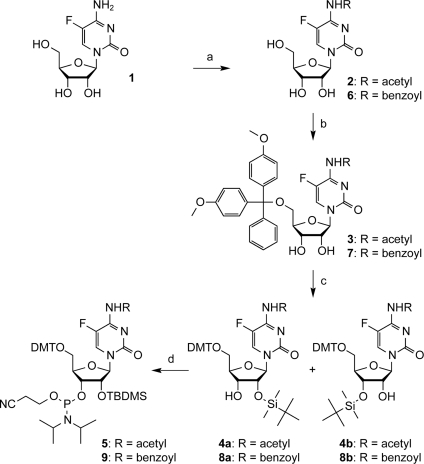
All oligonucleotides investigated here were chemically synthesized by solid-phase synthesis. For 5-fluoro pyrimidine (dU5F or dC5F) modified DNA, we used the commercially available building blocks of either 5′-O-(4,4′-dimethoxytrityl)-5-fluoro 2′-O-deoxyuridine 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite or 5′-O-(4,4′-dimethoxytrityl)-5-fluoro-O4-(2,4,6-trimethylphenyl) 2′-deoxyuridine 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite (26). The latter was converted into 5-fluoro 2′-deoxycytidine after oligonucleotide synthesis by ammonium hydroxide used for the routine deprotection steps. For 5-fluoro uridine (U5F) modified RNA, we applied commercially available 2′-O-(tert.-butyldimethylsilyl)-5′-O-(4,4′-dimethoxytrityl)-5-fluoro uridine 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite (6,23). For the preparation of 5-fluoro cytidine (C5F) modified RNAs, we elaborated a short synthetic route towards 5-fluoro cytidine phosphoramidites equipped for solid-phase synthesis (Scheme 1). Up to now, 5-fluoro cytidine containing RNAs have been provided mostly by enzymatic methods, producing uniformly 5-fluoro cytidine labeled derivatives (24,25), or by chemical solid-phase synthesis using a convertible 5-fluoro-O4-methyl uridine nucleoside (6). Ammonolysis has been required to convert the uridine precursor into the corresponding cytidine nucleoside after completed strand assembly (NH3/MeOH, r.t., 80 h) (6). The severe limitations of ammonolysis for the deprotection of larger chemically synthesized RNA strands prompted us to develop a synthetic route for the straightforward incorporation of 5-fluoro cytidine into RNA.
Scheme 1.
Synthesis of 5-fluoro cytidine phosphoramidites 5 and 9; (a) 12 eq. acetic anhydride (for 2) or benzoic anhydride (for 6) in anhydrous ethanol, room temperature, 14 h, 99% (2) and 99% (6); (b) 1.1 eq. 4,4′-dimethoxytrityl chloride in pyridine, room temperature, 14 h, 60% (3) and 56% (7); (c) 1.8 eq. silver nitrate, 1.8 eq. tert.-butyldimethylchlorosilane in tetrahydrofurane/pyridine, room temperature, 15 h, 48% (4a) and 58% (8a) (yields after separation from 4b and 8b, respectively); (d) 2 eq. (2-cyanoethyl)-N,N-diisopropylchlorophosphoramidite, 10 eq. N-ethyldiisopropylamine, 5 eq. 1-methylimidazole in dichloromethane, room temperature, 2.5 h, 58% (5) and 57% (9).
Facile synthesis of a 5-fluoro cytidine phosphoramidite derivative and incorporation into RNA
The synthesis started with commercially available 5-fluoro cytidine 1 using acetic anhydride in anhydrous ethanol for acetylation at C(4)–NH2 to obtain compound 2 (Scheme 1). In analogy, the usage of benzoic anhydride resulted in derivative 6. After regioselective protection of the 5′-OH with the 4,4′-dimethoxytrityl group under standard conditions to yield compounds 3 and 7, respectively, the 2′ hydroxy group was reacted with tert.-butyldimethylchlorosilane in the presence of silver nitrate. The resulting derivatives 4a and 8a, respectively, were finally converted into phosphoramidite 5 and 9 by reaction with (2-cyanoethyl)-N,N-diisopropylchlorophosphoramidite in the presence of 1-methylimidazole and N-ethyldiisopropylamine in dichloromethane. This route provided compound 5 in a 17% overall yield and compound 9 in a 14% overall yield in four steps with three chromatographic purifications. In total, 0.7 g of 5 and 0.4 g of 9 were prepared in the course of this study. These building blocks were incorporated into RNA by solid-phase synthesis with >98% coupling efficiencies. Importantly, deprotection of RNA containing N4-acetyl-5-fluoro cytidine was accomplished under standard conditions using methylamine in H2O/EtOH whereas deprotection of the N4-benzoyl protected counterpart required treatment with aqueous ammonia prior to application of methylamine to circumvent potential substitution of the phenyl carboxamide moiety at C(4) by methylamine. Because of this, we preferred the preparation of 5-fluoro cytidine modified RNAs using the N4-acetyl protection concept.
The molecular weights of all 5-fluoro substituted DNA and RNA oligonucleotides synthesized in this study were confirmed by LC-ESI mass spectrometry (Table 1).
Table 1.
Selection of chemically synthesized 5-fluoro uracil and 5-fluoro cytosine modified DNA and RNA oligonucleotidesa
| No. | Sequencea | Length | Isolated yield |
Molecular weight |
||
|---|---|---|---|---|---|---|
| (nt) | OD 260 nm | (nmol) | Calculated (amu) | Foundb (amu) | ||
| S2 | 5′-d(TCG TAC CGG AAG GTA CGA ACC dU5FTC CG)-3′ | 26 | 88 | 320 | 7960.1 | 7958.8 |
| S2b | 5′-d(CCG GAA GGT ACG AAC CdU5FT CCG)-3′ | 21 | 78 | 350 | 6420.1 | 6419.1 |
| S4 | 5′-r(GAC CGG AAG GUC CGC CU5FU CC)-3′ | 20 | 68 | 320 | 6374.8 | 6374.9 |
| S4b | 5′-r(CGG AAG GUC CGC CU5FU CC)-3′ | 17 | 86 | 500 | 5395.2 | 5394.8 |
| S5 | 5′-r(GAC CGG AAG GUC CGC CU5FU5F CC)-3′ | 20 | 66 | 323 | 6392.8 | 6393.1 |
| S5b | 5′-r(CGG AAG GUC CGC CU5FU5F CC)-3′ | 17 | 27 | 161 | 5413.2 | 5412.8 |
| S6 | 5′-d(TCG TAC CGG AAG GTA CGA ACdC5F TTC CG)-3′ | 26 | 150 | 550 | 7974.1 | 7973.6 |
| S6b | 5′-d(C CGG AAG GTA CGA ACdC5F TTC CG)-3′ | 21 | 93 | 420 | 6434.1 | 6433.0 |
| S7 | 5′-r(GAC CGG AAG GUC CGC C5FUU CC)-3′ | 20 | 64 | 310 | 6374.8 | 6373.5 |
| S7b | 5′-r(CGG AAG GUC CGC C5FUU CC)-3′ | 17 | 64 | 370 | 5395.2 | 5394.7 |
adU5F, 5-fluoro 2′-deoxy uridine; dC5F, 5-fluoro 2′-deoxy cytidine; U5F, 5-fluoro uridine; C5F, 5-fluoro cytidine.
bLC-ESI MS.
Bistable nucleic acids
Bistable nucleic acids exist in dynamic equilibria between two defined secondary structures (29). We have previously shown this behavior for a series of rationally designed oligoribonucleotides (29–31) and we also demonstrated the applicability of 2′-fluoro ribose labeling to probe the secondary structures of bistable RNA (42). The intention of the present study is on the one hand, to introduce bistable DNA oligonucleotides and on the other hand, to assess a fluoro label that can be used in equivalent manner for both RNA and DNA secondary structure probing. The previously applied 2′-fluoro ribose label does not fulfill this requirement since this modification favors ribose C3′-endo over C2′-endo conformation and therefore supports structural integrity of RNA only (11). For these reasons, we focused on the assessment of 5-fluoro pyrimidine labeling in the present comparative study.
Rational design of bistable oligonucleotides
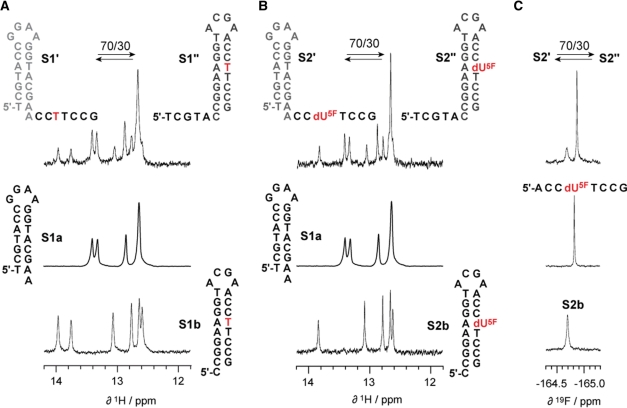
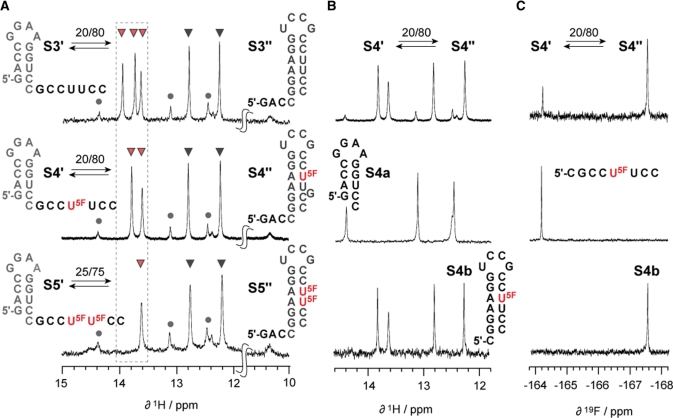
Following the sequence design of two haipins that compete for the same internal sequence partition, we demonstrated that a 26 nt DNA with the sequence of 5′-TCG TAC CGG AAG GTA CGA ACC TTC CG-3′ S1 exhibited a 70:30 (estimated accuracy ±10) structure equilibrium between fold S1′ and S1″ (Figure 2A). This was verified by comparative imino proton 1H NMR spectroscopy with a set of shorter reference oligonucleotides (S1a and S1b) that were corresponding to either of the two potential folds. To the best of our knowledge, this sequence is the first example of a short DNA with pronounced bistable secondary structure characteristics. For the bistable RNA system, we relied on a previously introduced 20 nt RNA, 5′-GAC CGG AAG GUC CGC CUU CC-3′ S3 that exhibited a 20:80 equilibrium between fold S3′ and S3″ under the buffer conditions used (Figure 3A, top) (30).
Figure 2.
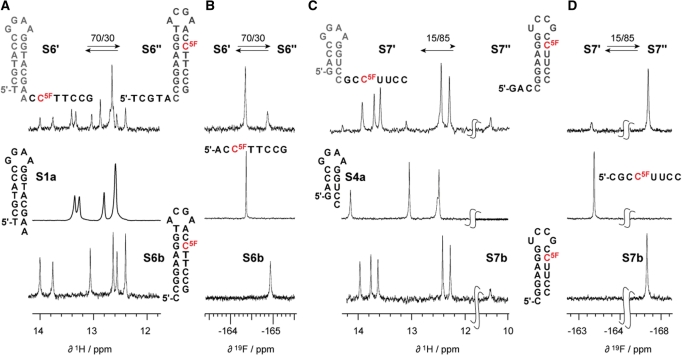
Bistable 26 nt DNA oligonucleotide. (A) Imino proton 1H NMR spectra of bistable DNA S1 (existing in a dynamic equilibrium of secondary structures S1′ and S1″) and of reference hairpins S1a and S1b. (B) Same for the 5-fluoro 2′-deoxyuridine modified bistable DNA S2 (S2' and S2″) and the corresponding reference hairpins S1a and S2b. (C) 1H decoupled 19F NMR spectra of S2 and the corresponding reference hairpin S2b and reference single strand 5′-d[ACC(dU5F)TCCG]. Conditions: 0.3 mM oligonucleotide, 25 mM sodium arsenate buffer, H2O/D2O = 9/1, pH 7.0, 300 K.
Figure 3.
Bistable 20 nt RNA oligonucleotide. (A) Comparison of imino proton 1H NMR spectra of bistable RNA S3 (existing in a dynamic equilibrium of secondary structures S3′ and S3″) and the single and double 5-fluoro uridine modified derivatives S4 and S5, respectively. (B) Imino proton 1H NMR spectra of S4 and of reference hairpins S4a and S4b. (C) 1H decoupled 19F NMR spectra of S4 and the corresponding reference hairpin S4b and reference single strand 5′-r(CGCC(U5F)UCC). Conditions: 0.3 mM oligonucleotide, 25 mM sodium arsenate buffer, H2O/D2O = 9/1, pH 7.0, 300 K.
Bistable DNA and RNA with 5-fluoro uracil labels
Alternative secondary structures of the same RNA sequence can be distinguished by use of 19F NMR spectroscopy if a distinct 2′-F modified nucleoside resides within the double helix of 1-fold while it is part of a single-stranded region within the alternative fold; we demonstrated this aspect previously (42). However, since 2′-F modified nucleosides are incompatible with the structural integrity of DNA as outlined above, we considered that the chemical environment of a fluorine atom attached at C5 of 2′-deoxyuridine should also be different for the two cases of double helical versus single-stranded arrangements. We therefore expected different chemical shift values of the corresponding 19F NMR resonances. In this sense, we replaced thymidine by 5-fluoro 2′-deoxyuridine in position 22 of S1 to yield S2 and analyzed the respective modified bistable DNA (Figure 2B). Indeed, the equilibrium position of S2, analyzed by comparative imino proton 1H NMR spectroscopy exhibited a 70:30 ratio between S2′ and S2″ and was well comparable to that of the unmodified DNA S1. Also, the proton-decoupled 19F NMR spectrum displayed two individual resonances (separated by ∼0.35 ppm) corresponding to the two secondary structures (Figure 2C). The ratio determined on basis of the 19F NMR signals was 70:30 and again in good agreement with the value determined from the 1H NMR spectrum. Together, this indicated that the influence of a single 5-fluoro uracil substituent on the DNA structure equilibrium was minor and within the range of accuracy. Interestingly, the 19F NMR resonance of 5-fluoro uracil showed an increased line width when it was involved in base pairing compared to the situation when it was located in an unpaired strand region. Moreover, we stress that the 1H NMR spectra of the bistable DNA S2 as well as the hairpin reference S2b were lacking the N(3)–H imino proton resonances of the respective dA:dU5F base pairs at 300 K (compare Figure 2A, top to Figure 2B, top; and Figure 2A, bottom to Figure 2B, bottom). All other imino protons showed similar chemical shift values as the ones observed for the unmodified DNA counterparts, S1 and S1b. Importantly, we observed the same behavior for the 20 nt bistable RNA S3 when one or even two A:U base pairs were replaced by A:U5F to yield S4 and S5, respectively (Figure 3A). Again, at 300 K, the 1H NMR imino proton resonance pattern of S4 and S5 was lacking one or two signals, respectively, that were attributed to 5-fluoro substituted Watson–Crick base pairs while all other imino protons provided chemical shift values comparable to that of the unmodified RNA counterpart. Our finding fits to a report by James and coworkers who showed in a temperature dependent series of 1H NMR spectra for the self-complementary duplex of [d(GGAAT(dU5F)CC)]2 that the dU5F N(3)–H resonance is extremely broad at low temperatures and becomes non-observable at room temperature (5). Also Hennig et al., observed for a uniformly 5-fluoro uridine labeled HIV TAR hairpin significant line broadening of 5-fluoro uridine N(3)–H imino protons together with a downfield chemical shift change between 0.6 and 0.7 ppm (24). The line broadening most likely reflects an increased exchange rate of dA:dU5F and A:U5F imino protons and relates to the considerable electronegativity of the 5-fluorine atom that lowers the apparent pKa of uridine N(3)–H from 9.18 ± 0.02 (unmodified uridine) to a value of 7.55 ± 0.02 (5-fluoro uridine) (43).
With respect to the structure equilibrium of the bistable RNA S3 (S3′:S3″ = 20:80), a single 5-fluoro uridine substitution, as in S4, did not change the equilibrium ratio of 20:80 between S4′ and S4″, determined by 1H NMR and 19F NMR spectroscopy in independent manner (Figure 3B and C). The chemical shift difference between the 19F resonances assigned for the RNA folds of S4′ and S4″ was 3.5 ppm, and therefore remarkably larger compared to the chemical shift difference of 0.35 ppm observed for the 19F resonances of the two DNA folds S2′ and S2″. This most likely reflects the different stacking patterns of Watson–Crick base pairs within A- and B-form (Figure 4 and Supplementary Data) and moreover, the much more pronounced difference in the chemical environment of a 5-fluoro pyrimidine atom that resides within an RNA duplex compared to its RNA single strand than the respective environments encountered for DNA.
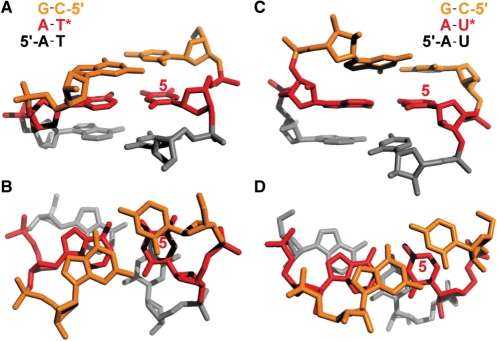
Figure 4.
Three-dimensional model for the chemical environment of A:T and A:U base pairs in S1″ and S3″ assuming standard A- and B-form, respectively. The position of fluoro labeling is indicated by an asterisk or the number 5 in red color. (A) DNA duplex with sequence as indicated (side view). (B) Same as (A) (top view). (C) RNA duplex with sequence as indicated (side view). (D) Same as (C) (top view). For stereo views see Supplementary Data.
Taken together, 5-fluoro uracil represents a non-invasive label for structural probing by 19F NMR spectroscopy of both, bistable DNA and RNA. This rationale holds true even for double labeling; the bistable RNA S5 which contained two successive 5-fluoro uridine replacements revealed only a marginally shifted equilibrium position for the two secondary structures of S5′ and S5″ (Figure 3C, bottom).
Bistable DNA and RNA with 5-fluoro cytosine labels
The bistable DNA and RNA sequences, S1 and S3, were labeled with single 5-fluoro cytosines and—as their uracil counterparts—turned out to be non-invasive labels. The dC5F at position 21 of DNA S6 resulted in an equilibrium position between S6′ and S6″ of 70:30 (Figure 5A) and was unchanged compared with the unmodified counterpart DNA S1 (Figure 2A). Also for the C5F modified RNA S7, the determined equilibrium position of 15:85 between S7′ and S7″ (Figure 5B) revealed that the structure equilibrium was—within the limits of accuracy—unaffected compared with the non-modified counterpart S3 (Figure 3A).
Figure 5.
5-Fluoro cytidine modified bistable DNA and RNA oligonucleotides, S6 and S7. (A) Imino proton 1H NMR spectra of bistable DNA S6 (existing in a dynamic equilibrium of secondary structures S6′ and S6″) and of reference hairpins S1a and S6b. (B) 1H decoupled 19F NMR spectra of S6 and the corresponding reference hairpin S6b and reference single strand 5′-ACC5FTTCCG. (C) Same as (A) for the 5-fluoro cytidine modified bistable RNA S7 (S7′ and S7″) and the corresponding reference hairpins S4a and S7b. (D) Same as (A) for S7 and the corresponding reference hairpin S7b and reference single strand 5′-CGCC5FUUCC. Conditions: 0.3 mM oligonucleotide, 25 mM sodium arsenate buffer, D2O, pH 7.0, 300 K; (for spectra of 5-fluoro cytidine labeled oligonucleotides in H2O/D2O = 9/1 see Supplementary Data).
For protonated 5-fluoro cytidine, a significantly lower pKa value of pKa[N(3)–H+] = 2.3 was reported when compared to their unmodified counterpart (pKa[N(3)–H+] = 4.2) (44). However, opposed to the significantly broad shape of the imino proton resonances for dA:dU5F and A:U5F base pairs, the imino protons of dG:dC5F and G:C5F base pairs displayed the typical linewidth for N–H•••N signals at 300 K. This observation accords with the higher thermodynamic stability of G:C versus A:U base pairs and therefore decreased N–H•••N exchange rates. In consent with a report by Henning et al. (24), the guanine N(1)–H imino proton experienced a slight shift upfield when involved in base pairing with C5F.
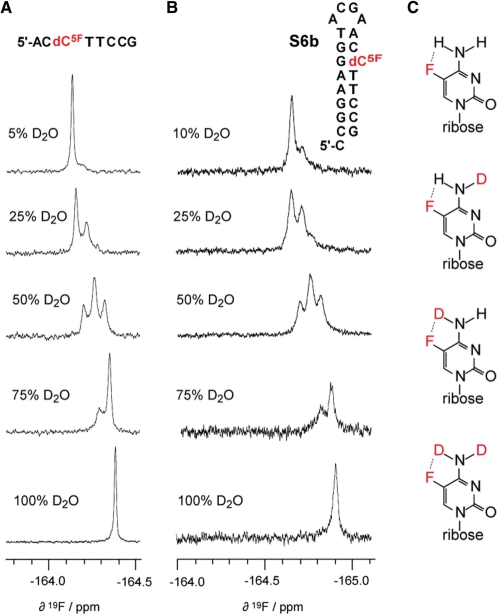
For the 5-fluoro cytosine DNA and RNA series, we observed a pronounced solvent induced isotope shift effect. Figure 6 depicts the typical signal pattern of a dC5F modified single-stranded DNA, 5′-AC(dC5F)TTCCG, (Figures 5A and 6A) and of the hairpin S6b (dC5F involved in base pairing, Figure 6B) when the ratio of H2O:D2O was gradually altered. Also for C5F modified RNAs we observed this effect (see Supporting Information). Solvent induced shifts of fluorine resonances have been previously reported for small organic molecules as well as fluorine labeled biomolecules (45). In the case of 5-fluoro cytidine modified oligonucleotides, the appearance of additional signals cannot only be attributed to solvent exposure of the fluorine nucleus but additionally stems from the isotopic replacement of exchangeable protons within the exocyclic amino group of the nucleobase (Figure 6C). A potential intramolecular hydrogen bond between C(5)–F•••H–N(C4) may be the cause for this pronounced effect and would be consistent with the lack of this phenomenon in the 5-fluoro uracil series.
Figure 6.
Solvent-induced isotope shift observed for 5-fluoro cytidine modified oligonucleotides. (A) 19F NMR spectra of 5′-ACC5FTTCCG recorded in varying mixtures of H2O/D2O. (B) Same for DNA hairpin S6b. Conditions: 0.3 mM oligonucleotide, 25 mM sodium arsenate buffer, pH 7.0, 300 K. (C) Deuterium exchange at the exocyclic amino group of 5-fluoro cytidine can result in four structures, causing individually shifted 19F resonance signals that are differently populated at different H2O/D2O ratios.
Influence of 5-fluoro pyrimidine modifications on thermodynamic stability
Since 5-fluoro modifications left the equilibrium position of bistable DNA and RNA unchanged, we expected their impact on thermodynamic base pairing stability to be minor. To provide evidence for this assumption, we recorded UV melting curves of DNA hairpin references S1b (unmodified), S2b (dU5F) and S6b (C5F) (Supplementary Data). In aqueous buffer system of 150 mM NaCl and 10 mM phosphate buffer, at pH 7.0 and at 2 µM DNA concentration, their Tm values varied only within 72.5 ± 1.0°C and their thermodynamic stabilities were in the small range of ΔG° = 7.1 ± 0.5 kcal mol−1. The Tm values of the RNA hairpin references S3b (unmodified), S4b (U5F), S5b (U5FU5F) and S7b (C5F) also varied only within one degree, 75.5 ± 1.0°C, and their thermodynamic stabilities were within the range of ΔG° = 7.1 ± 0.3 kcal mol−1. In extrapolation, these thermal and thermodynamic data suggest that the stabilities of the two competing hairpin structures in bistable oligonucleotides are minimally affected by the 5-fluoro pyrimidine labels.
Pseudoknot formation of a riboswitch aptamer
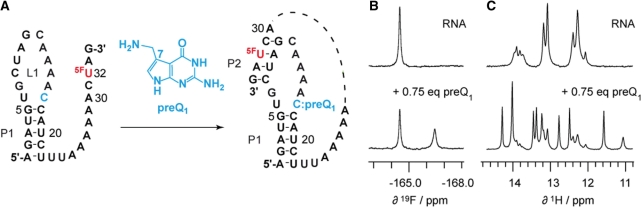
To demonstrate the value of 5-fluoro pyrimidine labeling for biologically relevant RNAs, we monitored the formation of a riboswitch aptamer/ligand complex (Figure 7). Based on the recently published X-ray and NMR structures of a 7-aminomethyl-7-deazaguanine (preQ1) sensing mRNA aptamer domain (46–49), we expected that fluorine labeling of U32 in the single-stranded nucleotide 3′-overhang (…C(5F–U32)AG-3′) would allow to follow base pairing with four complementary nucleotides in the loop (double-helix formation). Indeed, a significant shift of the fluorine resonance from 164.4 to 166.5 ppm was observed upon ligand addition corresponding to the free versus ligand-bound RNA (Figure 7). Ongoing experiments focus on the verification of ligand-induced secondary structure rearrangements within the expression platform of the full-length preQ1 riboswitch domain containing single fluorine labels.
Figure 7.
PreQ1 sensitive riboswitch aptamer. (A) RNA secondary structure model for ligand-induced folding (49). The 5-fluoro uridine label is positioned in a single-stranded region in the free RNA whereas it is located in a double-helical segment in the ligand-bound form. (B) 19F NMR spectra before and after addition of preQ1. (C) Corresponding 1H NMR spectra. Increase of ligand concentration results in higher population of the aptamer/ligand complex (data not shown). Conditions: 0.6 mM oligonucleotide, 25 mM sodium arsenate buffer, H2O/D2O = 9/1, pH 6.5, 298 K.
CONCLUSIONS
With the present study, we demonstrated the reliability of 5-fluoro labeling at pyrimidine nucleobases for secondary structure probing by 19F NMR spectroscopy. For bistable DNA and RNAs, the impact of single 5-fluoro pyrimidine substitutions on the equilibrium position was within the accuracy of determination. Watson–Crick base pairing properties were preserved corresponding to the observation that thermodynamic stabilities remained unchanged for the modified versus non-modified double helices. 5-fluoro pyrimidine modifications hence turned out to be non-invasive spin label options for secondary structure evaluation by 19F NMR spectroscopy. This was not expected in such strict manner, since the pKa values of the N(3) imino proton of 5-fluoro uracil and of N(3) protonated 5-fluoro cytosine are decreased by the order of two. A change of pKa values could potentially affect base pair formation, in particular base pairing strength because of higher proton mobility and higher proton exchange rates with the solvent, and consequently could affect the RNA structure equilibrium. However, our study showed that this concern did not come true.
In comparison with RNA 2′-F ribose labeling that we explored previously in our laboratory (42), 5-fluoro labeling is superior in one aspect: the modification does not directly affect ribose puckering and is therefore applicable for both DNA and RNA. However, we recall that 2′-F ribose represents an ideal label if constitutionally equivalent labeling of all four standard nucleosides is required (42). We also recall that 2′-F modified pyrimidine nucleosides shifted the equilibrium position of bistable RNAs by ∼25% (42) which can be regarded as disadvantage compared to 5-F pyrimidine labeling. Only 2′-F modified purine nucleosides left the RNA equilibrium position unchanged as observed here for the 5-fluoro pyrimidine series (42).
RNA with site-selective 5-F pyrimidine labels are conveniently accessible by chemical solid-phase synthesis for a size up to ∼65 nucleotides. Larger RNAs with site-specific labels in nmol quantities that are required for various types of biophysical experiments are accessible by combined approaches using chemical synthesis and enzymatic ligation methods as we have recently exemplified for adenine and TPP riboswitches containing fluorescent dyes (50–53). We consider the advantage of site-specific 5-F pyrimidine labeling over fully 5-F pyrimidine labeled RNA [which are efficiently prepared biochemically (24,25)] in the avoidance of constitutional assignments of 19F resonances and also in the avoidance of signal overlaps that are likely to be encountered in the fully labeled counterparts. We are confident that the use of RNAs labeled with single fluorine probes is a requirement to assist facile verification of RNA secondary structure models and we are currently working along this line.
Taken together, we consider this study as an important step on the way to identify reliable fluoro labels for RNA structure probing by NMR spectroscopy, with the long-term aim to contribute to the elucidation of folding pathways of biologically relevant, mid-sized RNA (up to ∼200 nt), such as riboswitches (49–52).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding by the Austrian Science Fund FWF (I317-N17 to R.M.; SFB F17 to R.K.) and the Ministry of Science and Research bm:wf (GenAU project ‘Non-coding RNAs’, P0726-012-012 to R.M.) is acknowledged. Funding for open access charge: Austrian Science Fund FWF.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Cobb SL, Murphy CD. F-19 NMR applications in chemical biology. J. Fluorine Chem. 2009;130:132–143. [Google Scholar]
- 2.Dalvit C. Ligand- and substrate-based F-19 NMR screening: principles and applications to drug discovery. Prog. Nucl. Magnet. Reson. Spectr. 2007;51:243–271. [Google Scholar]
- 3.Kreutz C, Micura R. Investigations on fluorine-labeled ribonucleic acids by 19F NMR spectroscopy. In: Herdewijn P, editor. Modified Nucleotides in Biochemistry, Biotechnology and Medicine. New York: Wiley-VCH; 2008. pp. 3–22. [Google Scholar]
- 4.Gmeiner WH, Pon RT, Lown JW. Synthesis, annealing properties, F-19 NMR characterization, and detection limits of a trifluorothymidine-labeled antisense oligodeoxyribonucleotide 21-mer. J. Org. Chem. 1991;56:3602–3608. [Google Scholar]
- 5.Stolarski R, Egna W, James TL. Solution structure of the EcoRI DNA octamer containing 5-fluorouracil via restrained molecular dynamics using distance and torsion angle constraints extracted from NMR spectral simulations. Biochemistry. 1992;31:7027–7042. doi: 10.1021/bi00146a003. [DOI] [PubMed] [Google Scholar]
- 6.Fisher A, Gdaniec Z, Biala E, Lozynski M, Milecki J, Adamiak RW. F-19 NMR of RNA. The structural and chemical aspects of 5-fluoro-cytidine and -uridine labelling of oligoribonucleotides. Nucleosides Nucleotides. 1996;15:477–488. [Google Scholar]
- 7.Sahasrabudhe PV, Gmeiner WH. Solution structures of 5-fluorouracil-substituted RNA duplexes containing G-U wobble base pairs. Biochemistry. 1997;36:5981–5591. doi: 10.1021/bi9700577. [DOI] [PubMed] [Google Scholar]
- 8.Reif B, Wittmann V, Schwalbe H, Griesinger C, Wörner K, Jahn-Hoffman K, Engels J, Bermel W. Structural comparison of oligoribonucleotides and their 2′-deoxy-2′-fluoro analogs by heteronuclear NMR spectroscopy. Helv. Chim. Acta. 1997;80:1952–1971. [Google Scholar]
- 9.Pfaff DA, Clarke KM, Parr TA, Cole JM, Geierstanger BH, Tahmassebi DC, Dwyer TJ. Solution structure of a DNA duplex containing a guanine-difluorotoluene pair: a wobble pair without hydrogen bonding? J. Am. Chem. Soc. 2008;130:4869–4878. doi: 10.1021/ja7103608. [DOI] [PubMed] [Google Scholar]
- 10.Arnold JRP, Fisher J. Structural equilibria in RNA as revealed by F-19 NMR. J. Biomol. Struct. & Dyn. 2000;17:843–856. doi: 10.1080/07391102.2000.10506573. [DOI] [PubMed] [Google Scholar]
- 11.Luy B, Marino JP. Measurement and application of H-1-F-19 dipolar couplings in the structure determination of 2′-fluorolabeled RNA. J. Biomol. NMR. 2001;20:39–47. doi: 10.1023/a:1011210307947. [DOI] [PubMed] [Google Scholar]
- 12.Olejniczak M, Gdaniec Z, Fischer A, Grabarkiewicz T, Bielecki L, Adamiak RW. The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucleic Acids Res. 2002;30:4241–4249. doi: 10.1093/nar/gkf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott LG, Geierstanger BH, Williamson JR, Hennig M. Enzymatic synthesis and F-19 NMR studies of 2-fluoroadenine-substituted RNA. J. Am. Chem. Soc. 2004;126:11776–11777. doi: 10.1021/ja047556x. [DOI] [PubMed] [Google Scholar]
- 14.Olsen GL, Edwards TE, Deka P, Varani G, Sigurdsson STh, Drobny GP. Monitoring tat peptide binding to TAR RNA by solid-state P-31-F-19 REDOR NMR. Nucleic Acids Res. 2005;33:3447–3454. doi: 10.1093/nar/gki626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barhate NB, Barhate RN, Cekan P, Drobny G, Sigurdsson STh. A nonafluoro nucleoside as a sensitive F-19 NMR probe of nucleic acid conformation. Org. Lett. 2008;10:2745–2747. doi: 10.1021/ol800872a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seela F, Xu K. DNA with stable fluorinated dA and dG substitutes: syntheses, base pairing and F-19-NMR spectra of 7-fluoro-7-deaza-2′-deoxyadenosine and 7-fluoro-7-deaza-2′-deoxyguanosine. Org. Biomol. Chem. 2008;6:3552–3560. doi: 10.1039/b806145a. [DOI] [PubMed] [Google Scholar]
- 17.Graber D, Moroder H, Micura R. F-19 NMR Spectroscopy for the Analysis of RNA Secondary Structure Populations. J. Am. Chem. Soc. 2008;130:17230–17231. doi: 10.1021/ja806716s. [DOI] [PubMed] [Google Scholar]
- 18.Chu WC, Horowitz J. F-19 NMR of 5-fluorouracil-substituted transfer-RNA transcribed invitro—resonance assignment of fluorouracile-guanine base-pairs. Nucleic Acids Res. 1989;17:7241–7252. doi: 10.1093/nar/17.18.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu WC, Liu J, Horowitz J. Localization of the major ethidium bromide binding site on tRNA. Nucleic Acids Res. 1997;25:3944–3949. doi: 10.1093/nar/25.19.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammann C, Norman DG, Lilley D. Dissection of the ion-induced folding of the hammerhead ribozyme using F-19 NMR. Proc. Nat. Acad. Sci. 2001;98:5503–5508. doi: 10.1073/pnas.091097498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreutz C, Kählig H, Konrat R, Micura R. A general approach for the identification of site-specific RNA binders by F-19 NMR spectroscopy: Proof of concept. Angew. Chem. Int. Ed. Engl. 2006;45:3450–3453. doi: 10.1002/anie.200504174. [DOI] [PubMed] [Google Scholar]
- 22.Klimasauskas S, Szyperski T, Serva S, Wüthrich K. Dynamic modes of the flipped-out cytosine during HhaI methyltransferase-DNA interactions in solution. EMBO J. 1998;17:317–324. doi: 10.1093/emboj/17.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gmeiner WH, Sahasrabudhe PV, Pon RT. Synthesis of 5′-O-(4,4′-dimethoxytrityl)-2′-O-(tert.-butyldimethylsilyl)-5-fluorouridine 3′-(cyanoethyl N,N-Diisopropylphosphoramidite) and its use in the synthesis of RNA. J. Org. Chem. 1994;59:5779–5783. [Google Scholar]
- 24.Hennig M, Scott LG, Sperling E, Bermel W, Williamson JR. Synthesis of 5-fluoropyrimidine Nucleotides as sensitive NMR probes of RNA structure. J. Am. Chem. Soc. 2007;129:14911–14921. doi: 10.1021/ja073825i. [DOI] [PubMed] [Google Scholar]
- 25.Hennig M, Munzarová ML, Bermel W, Scott LG, Sklenár V, Williamson JR. Measurement of long-range H-1-F-19 scalar coupling constants and their glycosidic torsion dependence in 5-fluoropyrimidine-substituted RNA. J. Am. Chem. Soc. 2006;128:5851–5858. doi: 10.1021/ja060165t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacMillan AM, Chen L, Verdine GL. Synthesis of an oligonucleotide suicide substrate for DNA methyltransferases. J. Org. Chem. 1992;57:2989–2991. [Google Scholar]
- 27.Schmidt S, Pein CD, Fritz HJ, Cech D. Chemical synthesis of 2′-deoxyoligonucleotides containing 5-fluoro-2′- deoxycytidine. Nucleic Acids Res. 1992;20:2421–2426. doi: 10.1093/nar/20.10.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hart JM, Kennedy SD, Mathews DH, Turner DH. NMR-assisted prediction of RNA secondary structure: identification of a probable pseudoknot in the coding region of an r2 Retrotransposon. J. Am. Chem. Soc. 2008;130:10233–10239. doi: 10.1021/ja8026696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Höbartner C, Micura R. Bistable secondary structures of small RNAs and their structural probing by comparative imino proton NMR spectroscopy. J. Mol. Biol. 2003;325:421–431. doi: 10.1016/s0022-2836(02)01243-3. [DOI] [PubMed] [Google Scholar]
- 30.Höbartner C, Ebert MO, Jaun B, Micura R. RNA two-state conformation equilibria and the effect of nucleobase methylation. Angew. Chem. Int. Ed. 2002;41:605–609. [Google Scholar]
- 31.Höbartner C, Mittendorfer H, Breuker K, Micura R. Triggering of RNA secondary structures by a functionalized nucleobase. Angew. Chem. Int. Ed. 2004;43:3922–3925. doi: 10.1002/anie.200460068. [DOI] [PubMed] [Google Scholar]
- 32.Micura R, Höbartner C. On secondary structure rearrangements and equilibria of small RNAs. ChemBioChem. 2003;4:984–990. doi: 10.1002/cbic.200300664. [DOI] [PubMed] [Google Scholar]
- 33.Fürtig B, Wenter P, Reymond L, Richter C, Pitsch S, Schwalbe H. Conformational dynamics of bistable RNAs studied by time-resolved NMR spectroscopy. J. Am. Chem. Soc. 2007;129:16222–16229. doi: 10.1021/ja076739r. [DOI] [PubMed] [Google Scholar]
- 34.Fürtig B, Buck J, Manoharan V, Bermel W, Jäschke A, Wenter P, Pitsch S, Schwalbe H. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;27:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 35.Wenter P, Fürtig B, Hainard A, Schwalbe H, Pitsch S. Kinetics of photoinduced RNA refolding by real-time NMR spectroscopy. Angew. Chem. Int. Ed. 2005;44:2600–2603. doi: 10.1002/anie.200462724. [DOI] [PubMed] [Google Scholar]
- 36.Wenter P, Fürtig B, Hainard A, Schwalbe H, Pitsch S. A caged uridine for the selective preparation of an RNA fold and determination of its refolding kinetics by real-time NMR. ChemBioChem. 2006;7:417–420. doi: 10.1002/cbic.200500468. [DOI] [PubMed] [Google Scholar]
- 37.Höbartner C, Silverman SK. Modulation of RNA tertiary folding by incorporation of caged nucleotides. Angew. Chem. Int. Ed. 2005;44:7305–7309. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]
- 38.Mayer G, Heckel A. Biologically active molecules with a “light switch”. Angew. Chem. Int. Ed. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 39.Heckel A, Buff MCR, Raddatz MSL, Müller J, Pötzsch B, Mayer G. An anticoagulant with light-triggered antidote activity. Angew. Chem. Int. Ed. 2006;45:6748–6750. doi: 10.1002/anie.200602346. [DOI] [PubMed] [Google Scholar]
- 40.Mikat V, Heckel A. Light-dependent RNA interference with nucleobase-caged siRNAs. RNA. 2007;13:2341–2347. doi: 10.1261/rna.753407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer G, Müller J, Mack T, Freitag DF, Höver T, Pötzsch B, Heckel A. Differential regulation of protein subdomain activity with caged bivalent ligands. ChemBioChem. 2009;10:654–657. doi: 10.1002/cbic.200800814. [DOI] [PubMed] [Google Scholar]
- 42.Kreutz C, Kählig HP, Konrat R, Micura R. Ribose 2′-F labeling: a simple tool for the characterization of RNA secondary structure equilibria by F-19 NMR spectroscopy. J. Am. Chem. Soc. 2005;127:11558–11559. doi: 10.1021/ja052844u. [DOI] [PubMed] [Google Scholar]
- 43.Knobloch B, Linert W, Sigel H. Metal ion-binding properties of (N3)-deprotonated uridine, thymidine, and related pyrimidine nucleosides in aqueous solution. Proc. Natl Acad. Sci. USA. 2005;102:7459–7464. doi: 10.1073/pnas.0501446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oyerle AK, Strobel SA. Biochemical detection of cytidine protonation within RNA. J. Am. Chem. Soc. 2000;122:10259–10267. [Google Scholar]
- 45.Hansen PE. Isotope effects on chemical shifts of proteins and peptides. Magn. Reson. Chem. 2000;38:1–10. [Google Scholar]
- 46.Klein DJ, Edwards TE, Ferré-D'A;maré AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat. Struct. Mol. Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J. Biol. Chem. 2009;284:11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang M, Peterson R, Feigon J. Structural Insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol. Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 49.Rieder U, Lang K, Kreutz C, Polacek N, Micura R. Evidence for pseudoknot formation of class I preQ1 riboswitch aptamers. ChemBioChem. 2009;10:1141–1144. doi: 10.1002/cbic.200900155. [DOI] [PubMed] [Google Scholar]
- 50.Lang K, Micura R. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat. Protoc. 2008;3:1457–1466. doi: 10.1038/nprot.2008.135. [DOI] [PubMed] [Google Scholar]
- 51.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. ChemBioChem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 53.Rieder R, Höbartner C, Micura R. Enzymatic ligation strategies for the preparation of purine riboswitches with site-specific chemical modifications. Methods Mol. Biol. 2009;540:15–24. doi: 10.1007/978-1-59745-558-9_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.