Abstract
One critical step in RNA interference (RNAi) experiments is to design small interfering RNAs (siRNAs) that can greatly reduce the expression of the target transcripts, but not of other unintended targets. Although various statistical and computational approaches have been attempted, this remains a challenge facing RNAi researchers. Here, we present a new experimentally validated method for siRNA design. By analyzing public siRNA data and focusing on hyperfunctional siRNAs, we identified a set of sequence features as potency selection criteria to build an siRNA design algorithm with support vector machines. Additional bioinformatics filters were also included in the algorithm to increase RNAi specificity by reducing potential sequence cross-hybridization or microRNA-like effects. Independent validation experiments were performed, which indicated that the newly designed siRNAs have significantly improved performance, and worked effectively even at low concentrations. Furthermore, our cell-based studies demonstrated that the siRNA off-target effects were significantly reduced when the siRNAs were delivered into cells at the 3 nM concentration compared to 30 nM. Thus, the capability of our new design program to select highly potent siRNAs also renders increased RNAi specificity because these siRNAs can be used at a much lower concentration. The siRNA design web server is available at http://www5.appliedbiosystems.com/tools/siDesign/.
INTRODUCTION
RNA interference (RNAi) is a naturally occurring mechanism for messenger RNA (mRNA) degradation in animals and plants (1–3). RNAi has been widely used to study gene functions by targeted cleavage of mRNA transcripts. Because of its convenience as well as its low cost, RNAi-based gene expression knockdown has become one of the most rapidly adopted molecular biology techniques in recent years. One popular way to initiate RNAi-induced mRNA degradation is through the introduction of chemically synthesized small interfering RNAs (siRNAs) into cells.
Over the past few years, there have been extensive studies on designing siRNAs with high mRNA knockdown efficiency (4). Randomly selected siRNA sequences were screened to identify features that are relevant to siRNA potency. One feature, for example, is that the 5′-end of the siRNA guide strand should have lower thermodynamic stability compared to the 3′-end because the guide strand of an siRNA duplex must be preferentially taken up by the RNA-induced silencing complex for effective mRNA degradation (5,6). Additionally, the base composition at certain positions in an siRNA also plays an important role in determining the siRNA potency (7,8). A high propensity of secondary structure in the guide siRNA strand may prevent its binding to the mRNA target site and reduce siRNA silencing efficacy (9,10). In addition, the availability of the mRNA target binding sites by the RNA-induced silencing complex may also be important for siRNA potency (11–13).
Multiple statistical and computational models have been proposed in recent years to design functional siRNA. For example, Reynolds et al. (7) have developed an siRNA design model by empirically summarizing relevant selection features. More recently, by analyzing over 2000 randomly selected siRNA sequences, Huesken et al. (8) have developed a neural network model to predict siRNA potency. There have been other siRNA prediction models using various machine learning techniques (14–21). Despite intense research efforts on siRNA design, there is still significant room for algorithmic improvement by optimizing the computational feature selection and modeling process. More importantly, few of the existing design algorithms have been experimentally validated, which limits their practical applications.
Here, we present an experimentally validated siRNA design algorithm built with support vector machines (SVMs) to predict hyperfunctional siRNAs. This algorithm employs a new feature selection process, and combines both feature filtering and modeling processes. Comparative analysis indicates that our new algorithm has significantly improved performance over the existing algorithm trained with the same data set. Also importantly, our new algorithm has been rigorously validated experimentally for its ability to select hyperfunctional siRNAs that function effectively even at low concentrations. The high efficacy of these siRNAs at low concentrations makes it possible to reduce RNAi off-target effects by using a much reduced amount of siRNAs, as demonstrated in our cell-based screening studies.
MATERIALS AND METHODS
Data retrieval
An siRNA data set was analyzed in our study for algorithm training and testing (8). This data set contains the sequences and knockdown data for over 2000 siRNA sequences randomly selected from the transcript sequence positions. The RefSeq sequences were downloaded from the NCBI ftp site (22).
Computational tools and data analysis
The SVM package LIBSVM was used to construct our siRNA prediction models (http://www.csie.ntu.edu.tw/∼cjlin/libsvm/). For SVM analysis, a Radial Basis Function was used for kernel transformation. Grid search was performed to identify the optimal parameters (C = 128 and γ = 0.002) for the Radial Basis Function kernel using the recommended protocol by LIBSVM. In the Huesken study, the performance of BIOPREDsi was evaluated using an independent data set [Figure 2c in ref. (8)]. In our algorithm comparison analysis, this testing data set was used to evaluate the performance of BIOPREDsi by determining the percentage of selected testing siRNAs that were at least 80 or 90% as efficient as the positive control. Only the siRNAs with the highest BIOPREDsi scores (top 10% among all testing siRNAs) were selected to represent the performance of BIOPREDsi.
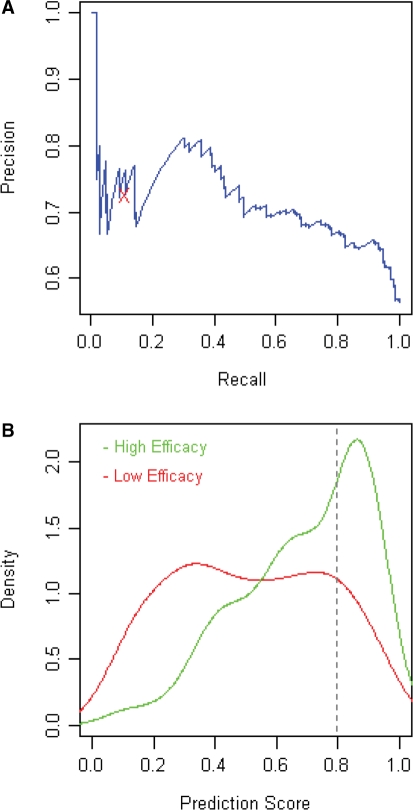
Figure 2.
Evaluation of the predictive power of the design model. (A) Precision–recall analysis to evaluate the prediction performance. The ‘X’ label represents the prediction performance using the threshold SVM prediction score (0.8). (B) The score distributions for high-efficacy and low-efficacy siRNAs. High efficacy was defined to be at least 90% as efficient as the positive control. All other siRNAs in the data set are defined as low-efficacy siRNAs.
RNA secondary structure stability, represented by the ΔG value, was calculated with RNAFold (23). Statistical computing was performed with the R package (http://www.r-project.org/). The statistical significance (P-value) for the selected biochemical features was calculated with Student’s t-test, chi-square test or hypergeometric test.
The web server for siRNA design using our new design algorithm is available at http://www5.appliedbiosystems.com/tools/siDesign/
siRNA validation by reverse transcription and real-time polymerase chain reaction
Two validation experiments were included in this study. For the first validation experiment, all siRNAs were synthesized by Ambion (Austin, TX, USA). HeLa cells were reverse transfected with siRNAs in triplicate using siPORT™ NeoFX™ Transfection Agent. Forty-eight hours post-transfection, RNA was isolated using the MagMAX-96 Total RNA Isolation Kit, then reverse transcribed using the RETROscript® First Strand cDNA Synthesis Kit (Ambion). Real-time polymerase chain reaction (PCR) was performed on an Applied Biosystems 7900HT Fast Real-Time PCR System using TaqMan® Gene Expression Assays, followed by relative quantitation using the ΔΔCt method with 18S ribosomal RNA (rRNA) as the endogenous control. Silencer Negative Control #1 siRNA-treated cells were included for knockdown efficiency analysis. Knockdown results were represented as the average of median percent mRNA remaining compared to Silencer Negative Control #1 siRNA-treated samples from two duplicated experiments.
For the second validation experiment, the performance of siRNA products designed using three different algorithms was directly compared. Ten gene targets were tested side by side for mRNA knockdown efficacy in HeLa cells. One hundred siRNAs selected for these genes were ordered from Ambion (designed with our new algorithm), Qiagen [designed with the BIOPREDsi algorithm (8)] or Dharmacon [designed with the Reynolds algorithm (7)]. All siRNAs were transfected at 3 and 30 nM final concentrations using Lipofectamine™ RNAiMax siRNA transfection agent (Invitrogen) according to the manufacturer’s recommendations. After 48 h, mRNA levels were analyzed using the TaqMan® Gene Expression Cell-to-Ct™ kit and the TaqMan® gene expression assays (Applied Biosystems).
Cell-based mitosis assays
siRNAs were transfected in the U2-OS osteosarcoma cell line at both 3 and 30 nM concentrations. Mitotic cells were identified using immunofluorescence detection of phosphorylated histone H3 as the antigen marker. Immunofluorescence signals were collected using automated image acquisition of assay plates on the ImageXpress Micro automated fluorescence microscope (Molecular Devices, Toronto, Canada). Images were analyzed using the MetaMorph image analysis software package. Nine positions in each 96-well sample were collected, and the data were averaged across three biological replicates and compared to similarly treated negative siRNA control samples (scrambled non-targeting siRNA). Following imaging analysis, data are evaluated according to the following protocol: (i) Index calculation: for each site within a well, the number of mitotic cells was normalized against the total number of nuclei and the area of non-cell background was subtracted from the total image area to calculate the area occupied by cells, and then normalized against the number of nuclei. (ii) Sample well average calculation: the average of all sites within a well was calculated. Additionally, all sites of all negative control wells per plate were averaged. (iii) Normalization: samples in each plate were normalized against the average of negative siRNA control wells. (iv) Triplicate average calculation: the average of the plate triplicates was calculated for each treatment and readout.
RESULTS
Identification of sequence features relevant to siRNA efficacy
Huesken et al. (8) have analyzed over 2000 randomly selected siRNA sequences for their target knockdown efficiency as determined by fluorescence reporter assays. Based on this training data set, a neural network model was developed, which was shown to have a robust performance in selecting siRNAs with high efficacy (8). This same data set was analyzed in our study for the development of a new improved siRNA selection algorithm. As a major deviation from the existing algorithms, we did not use all random siRNA sequences in the data set for the identification of features relevant to siRNA potency. Instead, we compared the 200 most potent siRNAs and the 200 least potent siRNAs to identify features that characterized the hyperfunctional siRNAs. Our design goal was to confidently pick the most potent siRNAs while rejecting other siRNAs including the ones with moderate knockdown activity. Since SVM is a binary classification process, we reasoned that by comparing the ‘best’ and the ‘worst’ siRNA sequences directly, relevant predictive features for hyperfunctional siRNAs can be more sensitively identified. In contrast, if all siRNA sequences in the data set were used, the selection process would be biased toward the siRNAs with moderate efficacy because these siRNAs significantly outnumbered the hyperfunctional siRNAs. Below is a summary of the selection features that were significantly associated with hyperfunctional siRNAs. The complete set of features used for SVM training is listed in Supplementary Table S2.
Base composition
The base composition of each of the 21 positions in an siRNA guide strand sequence was analyzed and the significant patterns were identified by the hypergeometric test (Supplementary Table S1). The analysis results in general agreed well with the previous studies (7,8). For example, for potent siRNA sequences, the first position of the guide strand was overwhelmingly A or U, and A was overrepresented at position 10.
Strand bias
Previous studies demonstrate the importance of differential end stability in predicting functional siRNAs using small sets of siRNAs (5,6). Here, we analyzed the Huesken data set and the same conclusion was observed. In general, the guide strands of highly potent siRNAs had significantly less stable 5′-ends compared to the guide strands of less potent or non-functional siRNAs (P = 1.9E-06, Student’s t-test).
GC content and siRNA binding stability
siRNAs with very high GC content are less likely to be functional (7). This conclusion was confirmed by analyzing the Huesken data set (P = 4.0E-13, Student’s t-test). Thermodynamic properties may play an important role in siRNA functionality. One important thermodynamic feature is the stability of target transcript binding by the siRNA. This is an overlapping feature to the GC content feature because siRNAs with high GC content tend to bind tightly to the target sites. However, the binding free energy was calculated with the nearest neighbor method (24), and thus was considered as a more accurate measurement of binding stability than the GC content. A large number of non-functional siRNAs bind extremely tightly to their target sites (P = 3.8E-14, Student’s t-test).
The secondary structures of the siRNAs and their target sites
An siRNA is less likely to be functional if its mRNA target site is inaccessible due to secondary structure formation (11). The local secondary structures for 81 nucleotides around the target sites were calculated with RNAFold (23). Compared to non-functional siRNAs, highly potent siRNAs tend to target regions with less potential for stable secondary structures (P = 1.2E-11, Student’s t-test). We also considered the secondary structures of the siRNAs. Our analysis showed that none of the functional siRNAs have highly stable secondary structures (P = 6.2E-12, Student’s t-test). One previous study suggested that a functional siRNA is more likely to have an accessible 3′-end in its guide strand so that the guide strand can easily enter into the RNA-induced silencing complex (25). Thus, in our study we also calculated the end accessibility of the siRNAs (i.e. to examine whether the 3′-end is freely exposed) as represented by binary values.
An SVM model to predict siRNA potency by integrating multiple sequence features
An SVM-based siRNA design algorithm was developed by integrating the sequence features described above. The program workflow is presented in Figure 1. The LIBSVM package was used for algorithm training and testing.
Figure 1.
The workflow for functional siRNA prediction with a new design algorithm.
Similar to the feature selection process, only the most and the least potent siRNAs were used to build SVM models. Although there were 2430 siRNA sequences in the original data set, only ∼300 sequences with >95 or <65% knockdown efficiencies were used for model training. After manually examining the incorrect predictions by the SVM, some of the failures were identified to be related to a few extreme values in several training features (e.g. extremely stable secondary structure of the siRNA). This posed a challenge to SVM modeling, as these extreme values did not seem to be properly modeled by the SVM due to their rare frequency in the training data. To address this issue, the data set was first filtered to remove sequences with extreme feature values before the SVM training and testing process. These sequence filters are listed in Table 1. Most functional siRNAs were retained after excluding the siRNAs with extreme feature values. Therefore, the improvement of the prediction performance did not lead to a significant increase of the false negative rate. For example, one important filter is the exclusion of sequences with C or G at the 5′-end. Most functional siRNAs (85%) contain A or T at this position. Thus, by excluding sequences with C or G at the 5′-end, half of the 2430 siRNAs from the original data set were excluded from our SVM analysis, while most functional siRNAs were still kept. Because the 5′-end base feature was so significant at identifying potent sequences, it became a dominating factor in the SVM model and overshadowed other less obvious but important features. Thus, by focusing on a subset of siRNA sequences all starting with A or T, other relevant features were more likely to be properly represented in the SVM model. In developing our final design algorithm, training data filtration was performed before SVM modeling. The combination of both the filtering step and the SVM modeling process led to improved prediction performance, resulting in 77% prediction specificity as described in detail below.
Table 1.
Prediction features that were filtered before the SVM modeling process
| Filtered features | Value range |
|---|---|
| 5′-end base | A or U |
| GC content (%) | 25–55 |
| Strand bias (ΔG) (kcal/mol) | −3.5 to 6.0 |
| Duplex binding (ΔG) (kcal/mol) | ≥ −39.0 |
| siRNA folding (ΔG) (kcal/mol) | ≥ −5.0 |
| Target site folding (ΔG) (kcal/mol) | ≥ −30.0 |
The Huesken data set was randomly partitioned, with 90% of the siRNAs as the training data and 10% of the siRNAs as the testing data. The data partition process was repeated three times so that three distinct training and testing groups were generated. Three SVM models were generated based on the three training groups. Each data partition led to an SVM model with a similar performance when applied to the testing data. A testing siRNA was considered to be potent if it was at least 90% efficient in knockdown as compared to the positive control. On average, the fraction of correctly predicted potent siRNAs in the testing set was 77.0%, which was much higher than the fraction of all potent siRNAs in the testing set (29.5%).
Precision–recall curves were drawn to evaluate the prediction performance of our new prediction model. Precision–recall curves are commonly used in machine learning to address binary decision problems, and they are a sensitive way to examine the prediction precision (correctly predicted positives divided by total predicted positives) in relation to the recall rate (correctly predicted positives divided by total actual positive training samples). In most cases for siRNA design, only a few functional siRNAs need to be selected for each gene target. Therefore, precision–recall analysis is an especially useful technique to examine the prediction performance for the few top siRNA candidates. As shown in Figure 2A, the precision rate is 70–80% when the recall rate is <40%. The calculated P-values by the SVM model were used as the algorithm prediction scores. The score distributions for the functional and non-functional siRNAs were compared to evaluate the differentiating power of the scoring system (Figure 2B). High-efficacy siRNAs were defined to be at least 90% as efficient as the positive control, and all other siRNAs were defined to be low-efficacy siRNAs. The high-efficacy siRNA scores were significantly separated from the low-efficacy siRNA scores, with a major mode at ∼0.8 (Figure 2B). Therefore, the score 0.8 was chosen as the cutoff threshold for the prediction of functional siRNAs.
The new model performance was compared to the BIOPREDsi algorithm (8). The BIOPREDsi algorithm (a neural network model) was developed by analyzing the same training data set as used in our study. A testing data set was also presented to evaluate the performance of BIOPREDsi in ref. (8); however, the details of this data set are not publicly available. Thus, the algorithms cannot be directly compared using the same testing data set. In our comparative analysis, the performance of our model was evaluated with three randomly partitioned testing data sets as described earlier. These training data sets had not been used in the model training process and thus represent independent data. The average SVM model performance for predicting the knockdown efficacy of the three testing data sets was presented in Table 2. Our new model was shown to have significantly improved performance over the Huesken algorithm in predicting hyperfunctional siRNAs that were at least 90% as efficient as the positive control (77 and 47% correct predictions for the new model and BIOPREDsi, respectively).
Table 2.
Performance comparison of different siRNA design algorithms
| 80% of control | 90% of control | |
|---|---|---|
| New SVM model (%) | 95 | 77 |
| BIOPREDsi (%) | 69 | 47 |
| Random siRNA (%) | 46 | 30 |
Two knockdown thresholds, 80 or 90% efficacy of the positive control, were chosen in the algorithm comparative analysis. The percentages of the siRNAs that rendered at least the threshold knockdown efficacy were presented here. For the BIOPREDsi algorithm, the prediction performance was estimated by analyzing the siRNAs with the highest prediction scores (top 10%, see ‘Materials and Methods’ for details).
Bioinformatics filters for siRNA specificity
The mechanisms for siRNA off-targeting are not well understood at present. There might be multiple mechanisms that lead to unintended siRNA knockdown. It is possible that off-targeting would occur if there is significant sequence cross-hybridization between an siRNA oligo and its unintended target transcript. To address this concern, multiple sequence-based filters were implemented in our new design algorithm to reduce potential sequence cross-reactivity. It is known that low sequence complexity contributes to cross-hybridization (26). Thus, transcript sequences were evaluated for sequence complexity, and low-complexity regions were identified by the DUST program and avoided when selecting siRNA sequences (27).
Mismatches can reduce oligo priming stability and sometimes even a single mismatch within a long stretch of the nucleotide duplex could have a significant destabilizing effect (28,29). Thus, continuous base pairing contributes significantly to nucleotide duplex stability, potentially leading to cross-hybridization. Expression profiling studies also suggest that contiguous base pairing contributes to non-specific siRNA targeting (30). In our siRNA sequence selection process, we employed an algorithm that we previously developed to quickly identify stretches of 15 contiguous bases that match perfectly to unintended transcripts in the transcriptome (31). In brief, redundant 15-mer sequences were identified using a computational hashing technique with two 10-mer sequences as the basic hash keys. Every possible 15-mer in an siRNA sequence was tested against all transcript sequences in the transcriptome. An siRNA candidate would be excluded from further consideration if a repetitive 15-mer was present in its sequence. As a further step to reduce cross-reactivity, BLAST search was performed against all known sequences in the transcriptome and an siRNA candidate would be discarded if the BLAST score was >30 [threshold value adopted based on our previous study (31)].
Another possible mechanism for unintended siRNA off-targeting could be the so-called ‘microRNA effect’, i.e. an siRNA could behave like a microRNA (miRNA) and target multiple unintended transcripts (32,33). The miRNA targeting process is not well understood to date. However, many studies indicate that the miRNA seed regions (positions 2–8) are usually required to match perfectly to the target transcript. Thus, if an siRNA behaves like a miRNA, its seed sequence typically pairs perfectly to the non-specific target transcripts. As a seed sequence only contains seven nucleotides, it is not possible to select such a short sequence that is unique in the whole transcriptome. To partly address this potential ‘miRNA-like’ effect, we focused on comparing the siRNA seed sequences with the seeds from all known miRNAs. We hypothesized that if an siRNA shares the same seed sequence with an endogenous miRNA, then this siRNA would have similar functions to the miRNA by targeting a similar group of transcripts. To this end, the seed sequence of an siRNA candidate was examined and an siRNA candidate would be discarded if its seed sequence was shared by any known miRNA.
Experimental validation of siRNA efficacy
During the SVM modeling process, the performance of our siRNA selection algorithm was computationally validated using published data sets. However, the best way to determine true model performance is to test the predictions with new independent experimental data. To this end, a double-blind randomized experiment was designed to evaluate model performance. Similar design strategies are widely used in clinical trials for unbiased evaluation of competing treatment plans. Our new design algorithm was compared to two other methods. One method was based on a linear statistical model after analyzing an in-house training data set, and had been used to design genome-wide Silencer® siRNA products at Ambion; the other method selected the siRNAs from random positions in the transcript sequences using a software random function.
To reduce potential variations from gene-specific effects, we selected 14 genes covering a wide expression spectrum in HeLa cells. Ten siRNAs for each gene (three from the new model, three from the previous algorithm and four from randomly selected positions) were evaluated at two different concentrations, 3 and 30 nM. To reduce potential experimental bias, the identity of all these siRNAs was blinded from the researchers performing the validation experiments, including the siRNA synthesis, transfection, and reverse transcription (RT) and real-time PCR validation of the remaining mRNA expression level. The siRNA identity was only revealed at the final data analysis stage. To further reduce the experimental variation, all 140 siRNAs were synthesized in one batch. Each siRNA was transfected in triplicate into the cells and real-time RT–PCR was performed in duplicate for each transfection. The entire validation experiment was repeated twice on different days. Thus, the efficacy of each siRNA was evaluated using 12 knockdown measurements. The siRNAs were tested sequentially according to the Random Index, which randomly determined the order of the siRNAs from the three algorithms for each gene (Supplementary Table S3). The siRNA sequences included in this validation experiment as well as the knockdown efficacy at 3 and 30 nM are summarized in Supplementary Table S3. The 3 nM siRNA transfection result is also presented in Figure 3A. A similar result was obtained for the 30 nM siRNA transfection experiment. Compared to the randomly designed siRNAs, the algorithm used to design Silencer® siRNAs was a major improvement at picking functional siRNAs. More than 80% of the siRNAs picked by this algorithm reduced the mRNA level by at least 80% at both 30 and 3 nM concentrations, which was in agreement with our previous observations. More interestingly, siRNAs designed with our new model rendered even higher efficacy. All 42 siRNAs selected by the new model led to at least 80% reduction of gene expression at both 30 and 3 nM concentrations. Most of these siRNAs reduced the gene expression level by >90% even at a low concentration (3 nM).
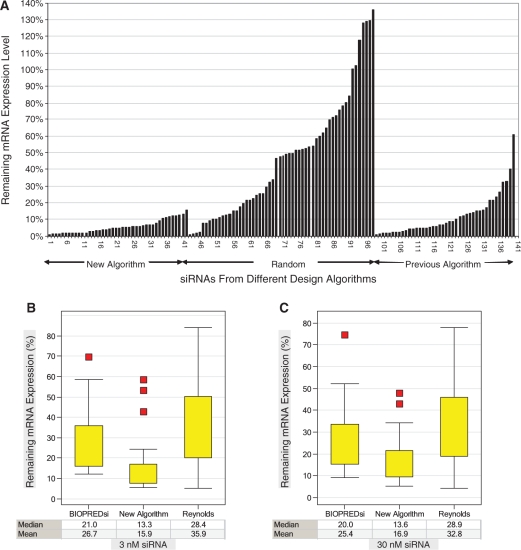
Figure 3.
Experimental validation of the new design algorithm. (A) Three siRNA design algorithms were compared in this experiment. Fourteen genes were selected. For each gene, three siRNAs were picked by the new algorithm, three were picked by our previous algorithm and four were picked by a random selection process. The remaining mRNA expression level was normalized to the negative control siRNA. All siRNAs were transfected into HeLa cells at 3 nM in triplicate in a double-blind randomized experiment for testing the knockdown efficiency. The siRNAs from each algorithm were sorted by their knockdown efficiency. (B and C) A second validation experiment to compare three widely used design algorithms. siRNAs for 10 genes were designed with three algorithms. For each gene, three siRNAs were picked by the new algorithm, three were picked by the BIOPREDsi algorithm and four were picked by the Reynolds algorithm. The remaining mRNA expression level was normalized to the negative control siRNA. All siRNAs were transfected into HeLa cells either at 3 nM (B) or 30 nM (C) concentrations.
To further validate the performance of our new design algorithm, a separate RNAi knockdown experiment was performed to directly compare our newly designed siRNAs with the siRNAs designed with two widely used algorithms. In this experiment, 100 siRNAs designed for 10 genes were compared. Ten siRNAs for each gene, including three designed by the new algorithm, three by BIOPREDsi (8) and four by the Reynolds algorithm (7), were analyzed for gene knockdown efficacy at two concentrations, 30 and 3 nM. The gene IDs and knockdown data are summarized in Supplementary Table S4. As shown in Figure 3B and C, comparative analysis indicates that our new algorithm had significantly improved performance at both siRNA concentrations compared to the other algorithms (P = 4.1E-05 and 0.0001, respectively, when compared with the Reynolds algorithm at 3 or 30 nM with Student’s t-tests; P = 0.004 and 0.01, respectively, when compared with BIOPREDsi at 3 or 30 nM). Interestingly, none of the 10 siRNAs designed for LDLR (including three from our new algorithm) could reduce the mRNA expression to <30% at either 3 or 30 nM, indicating that a small number of genes might be recalcitrant to RNAi-based gene expression knockdown.
The effect of siRNA concentration on gene knockdown
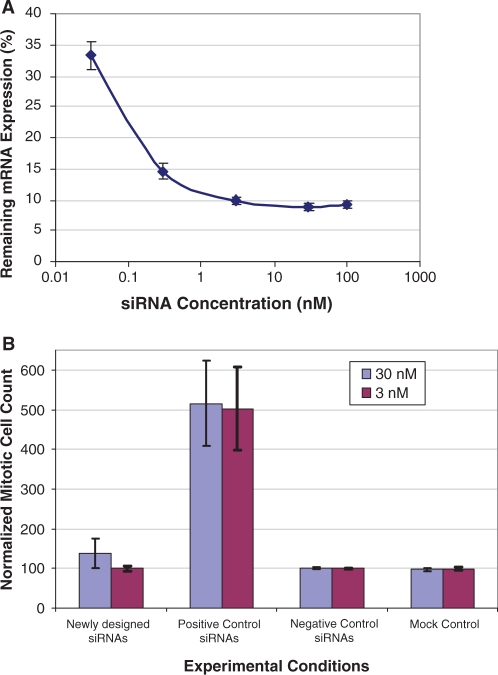
The efficacy of the siRNAs selected by the new model was further evaluated at five siRNA concentrations. For this experiment, 73 siRNAs were designed to target 10 genes. These siRNAs effectively suppressed the expression of the target transcripts when introduced at high concentrations as shown in Figure 4A. Interestingly, the knockdown potency was essentially unchanged when the siRNA concentration was decreased to 3 nM. The siRNA potency decreased dramatically when the concentration was further decreased to 0.03 nM. Thus, RNAi experiments can be performed using a few namomolar of our newly designed siRNAs without loss of knockdown efficiency.
Figure 4.
Dose effect on siRNA efficacy and specificity. (A) Experimental evaluation of the siRNA potency at different concentrations. Seventy-three siRNAs targeting 10 genes were analyzed. Each siRNA was transfected into HeLa cells at five concentrations, 0.03, 0.3, 3, 30 and 100 nM. The siRNA knockdown readings at each concentration were averaged and presented in the graph. The error bars represent the standard error of the mean. (B) Cell-based mitosis assays to evaluate the siRNA off-target effects. Fourteen newly designed siRNAs, targeting three genes unrelated to mitosis, were analyzed. Each siRNA was transfected at both 3 and 30 nM, and potential unintended effects on mitosis were determined. Eleven siRNAs targeting WEE1 and PLK1 were included as positive controls. The data were normalized to three negative control siRNAs. In the mock control transfection experiment, no RNA duplex was added.
Reduced off-target effects with hyperfunctional siRNAs at a low concentration
Since the siRNAs designed by our new algorithm rendered ample gene knockdown even at low concentrations (Figure 4A), we further characterized these siRNAs to determine whether a decreased siRNA concentration leads to significantly reduced off-target effects.
High-content cell microscopy assays were performed to evaluate the impact of siRNA concentration on off-target effects. Fourteen siRNAs designed for three genes (FDFT1, LDLR and SC5DL) were tested in this study. These genes are mainly involved in cholesterol biosynthesis. FDFT1, LDLR and SC5DL have been extensively characterized by various researchers and have not been observed to have direct impacts on cell cycle control or cellular mitosis. Thus, we hypothesized that any impacts on cellular mitosis that were observed following transfection of siRNA designed to target one of these genes could be the consequence of off-targeting. The mitosis screening assay data were normalized to three negative control siRNAs included in each plate. As positive controls, 11 siRNAs were also included in the screening experiment targeting genes WEE1 and PLK1, which are known to be directly involved in the mitosis process (34,35). As expected, the knockdown of either WEE1 or PLK1 led to a significant increase in the number of cells in mitosis (Figure 4B). This cellular phenotype from the positive control siRNAs was not affected by the amount of siRNAs introduced into the cells, indicating that the positive control siRNAs worked effectively at 3 nM. As to the 14 siRNAs targeting genes unrelated to mitosis, they behaved similarly to the negative control siRNAs or mock transfection controls (no RNA duplex added) when transfected at 3 nM. Little change in the number of mitotic cells was observed at this siRNA concentration. In contrast, when present at a higher concentration (30 nM), three of these siRNAs led to measurable cellular mitosis phenotype, resulting in an increased sample variability as measured by the SEM readings presented in Figure 4B (7.5 and 36.6 for 3 and 30 nM siRNAs, respectively).
DISCUSSION
Techniques to evaluate the performance of siRNA design algorithms
Correlation analysis and Receiver Operator Characteristic (ROC) analysis are two popular approaches in machine learning studies. However, both analyses have major limitations when applied to siRNA design. For siRNA design, usually only a few functional siRNAs need to be selected for each gene from potentially hundreds of siRNA candidates. A typical correlation analysis is to calculate the correlation coefficients for all the siRNAs in the sample space in relation to their knockdown efficiency. Although it is interesting to use a design algorithm to identify many siRNAs with moderate activity, it is much more important practically to identify a few hyperfunctional siRNAs with high precision. Therefore, the prediction precision is a more important measurement than the correlation coefficient. Another issue in siRNA design is that the vast majority of random siRNAs are not functional, and thus the data set is highly skewed toward many non-functional sequences. As a result, the false positive rate is very low in most cases even though the absolute number of false positives may be unacceptably high. In the ROC analysis, this observation is translated into a steep curve with the ROC area close to 1 in most cases. Thus, the ROC curves do not have much differentiating power at comparing different algorithms for the selection of only a few top candidates. Considering these potential limitations, we adopted a new strategy to evaluate siRNA prediction accuracy by performing precision–recall analysis. Precision–recall curves are commonly used in machine learning, and they are especially suitable for studies with a large percentage of negative samples.
siRNA efficacy and specificity
The capability of our new design algorithm for picking hyperfunctional siRNAs with high accuracy has important implications to address the siRNA specificity issue. One major issue in RNAi research is that many unintended genes may also be targeted by an siRNA, leading to widespread off-target effects. The siRNA off-target effect is one of the most challenging issues facing RNAi researchers (4,30,32,36). To date, bioinformatics design has failed to identify siRNA sequences that would specifically target the genes of interest. Although multiple specificity filters were implemented in our new algorithm, the off-target issue is not likely to be completely addressed by bioinformatics design alone. One possible experimental approach to reduce the off-target effects is to use much reduced amounts of siRNAs for gene knockdown. Unfortunately, the knockdown efficiency would decrease drastically at low siRNA concentrations for most commercially available siRNA products. At present, most manufacturers recommend using 30–100 nM siRNA in RNAi experiments, which inevitably leads to significant off-target effects. The high efficacy of our newly designed siRNAs offers a unique opportunity to address this issue experimentally. Our validation results indicate that effective gene expression knockdown can be achieved with a small amount of siRNA. Thus, by decreasing the siRNA concentration, the off-target effects could be significantly reduced as demonstrated in our cell-based mitosis assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Life Technologies.
Conflicts of interest statement. XH.W., R.K.V., L.B., S.M., T.J.S. are employees of Life Technologies, which may potentially benefit from the publication of this work.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Diana Batten, Sarah LaMartina, Tera Schaller, Josquin Holmes and Daniel Williams for technical assistance. We thank Andrew Walsh, Christoph Sachse, Michael Hannus, Christophe Echeverri from Cenix for the development of the mitosis assays and useful discussions. We also thank John Burns, Hugh Pasika and Tian Zhang for reviewing the manuscript.
REFERENCES
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Denli AM, Hannon GJ. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 2003;28:196–201. doi: 10.1016/S0968-0004(03)00058-6. [DOI] [PubMed] [Google Scholar]
- 3.Sontheimer EJ. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- 4.Pei Y, Tuschl T. On the art of identifying effective and specific siRNAs. Nat. Methods. 2006;3:670–676. doi: 10.1038/nmeth911. [DOI] [PubMed] [Google Scholar]
- 5.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 8.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Design of a genome-wide siRNA library using an artificial neural network. Nat. Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 9.Patzel V, Rutz S, Dietrich I, Koberle C, Scheffold A, Kaufmann SH. Design of siRNAs producing unstructured guide-RNAs results in improved RNA interference efficiency. Nat. Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 10.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, Hofacker IL. The impact of target site accessibility on the design of effective siRNAs. Nat. Biotechnol. 2008;26:578–583. doi: 10.1038/nbt1404. [DOI] [PubMed] [Google Scholar]
- 12.Schubert S, Grunweller A, Erdmann VA, Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J. Mol. Biol. 2005;348:883–893. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Heale BS, Soifer HS, Bowers C, Rossi JJ. siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res. 2005;33:e30. doi: 10.1093/nar/gni026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagla B, Aulner N, Kelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O, et al. Sequence characteristics of functional siRNAs. RNA. 2005;11:864–872. doi: 10.1261/rna.7275905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia P, Shi T, Cai Y, Li Y. Demonstration of two novel methods for predicting functional siRNA efficiency. BMC Bioinformatics. 2006;7:271. doi: 10.1186/1471-2105-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saetrom P. Predicting the efficacy of short oligonucleotides in antisense and RNAi experiments with boosted genetic programming. Bioinformatics. 2004;20:3055–3063. doi: 10.1093/bioinformatics/bth364. [DOI] [PubMed] [Google Scholar]
- 17.Shabalina SA, Spiridonov AN, Ogurtsov AY. Computational models with thermodynamic and composition features improve siRNA design. BMC Bioinformatics. 2006;7:65. doi: 10.1186/1471-2105-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teramoto R, Aoki M, Kimura T, Kanaoka M. Prediction of siRNA functionality using generalized string kernel and support vector machine. FEBS Lett. 2005;579:2878–2882. doi: 10.1016/j.febslet.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 19.Peek AS. Improving model predictions for RNA interference activities that use support vector machine regression by combining and filtering features. BMC Bioinformatics. 2007;8:182. doi: 10.1186/1471-2105-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladunga I. More complete gene silencing by fewer siRNAs: transparent optimized design and biophysical signature. Nucleic Acids Res. 2007;35:433–440. doi: 10.1093/nar/gkl1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–647. doi: 10.1093/nar/gkm920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xia T, SantaLucia J., Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 25.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 27.Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput. Appl. Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- 28.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, Lefkowitz SM, Ziman M, Schelter JM, Meyer MR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 29.Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Seed B. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics. 2003;19:796–802. doi: 10.1093/bioinformatics/btg086. [DOI] [PubMed] [Google Scholar]
- 32.Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl Acad. Sci. USA. 2004;101:4419–4424. doi: 10.1073/pnas.0307700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cogswell JP, Brown CE, Bisi JE, Neill SD. Dominant-negative polo-like kinase 1 induces mitotic catastrophe independent of cdc25C function. Cell Growth Differ. 2000;11:615–623. [PubMed] [Google Scholar]
- 36.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.