Abstract
The Rev protein of HIV-1 actively shuttles between nucleus and cytoplasm and mediates the export of unspliced retroviral RNAs. The localization of shuttling proteins such as Rev is controlled by the relative rates of nuclear import and export. To study nuclear export in isolation, we generated cell lines expressing a green fluorescent protein-labeled chimeric protein consisting of HIV-1 Rev and a hormone-inducible nuclear localization sequence. Steroid removal switches off import thus allowing direct visualization of the Rev export pathway in living cells. After digitonin permeabilization of these cells, we found that a functional nuclear export sequence (NES), ATP, and fractionated cytosol were sufficient for nuclear export in vitro. Nuclear pore-specific lectins and leptomycin B were potent export inhibitors. Nuclear export was not inhibited by antagonists of calcium metabolism that block nuclear import. These data further suggest that nuclear pores do not functionally close when luminal calcium stores are depleted. The distinct requirements for nuclear import and export argue that these competing processes may be regulated independently. This system should have wide applicability for the analysis of nuclear import and export.
The bidirectional exchange of macromolecules between the nucleus and cytoplasm occurs through the nuclear pore complex (1). Generally, the cut-off between facilitated transport and diffusion across the nuclear pore complex is 40–50 kDa. However, some smaller molecules such as histone H1 also are transported actively (2). Active transport is a highly regulated process requiring specific targeting sequences, receptors, regulatory proteins, and energy (reviewed in refs. 3 and 4). Nuclear import also requires the small GTPase Ran, p10, and GTP hydrolysis (5, 6). In addition, alternate pathways for nuclear import exist (7, 8). Although many of the requirements of import have been identified, the exact mechanism of translocation across the pore is poorly understood.
Nuclear export, like import, is also initiated by a specific sequence. One of the first nuclear export sequences (NESs) was identified in the HIV-1 transactivating protein Rev (9). This hydrophobic sequence, LPPLERLTLD, is characterized by critical leucine residues; removal of one of these residues inactivates this sequence and prevents export (10). Similar NESs have been identified in several other shuttling proteins (11–19). When the NES is placed on a heterologous protein, it is sufficient to initiate export. An excess amount of the Rev NES peptide competes efficiently with some RNA species including 5S rRNA and spliceosomal U snRNAs for export out of the nucleus (9, 20). These competition studies indicate that Rev utilizes an established nuclear export pathway within the cell.
NES-containing proteins are exported in an active and saturable manner, indicating that the process is receptor-mediated. However, identification of export receptors has proven more difficult than the isolation of import receptors (21–23). Recently, leptomycin B was shown to inhibit Rev export in living cells and the authors suggested that the target of leptomycin B was involved directly in the export process (24). CRM1, a target of leptomycin B, was shown subsequently to interact with NES-containing proteins. Recombinant CRM1 injected into Xenopus laevis oocytes enhanced the export of Rev and overcame the inhibition by leptomycin B (25). In a separate study, CRM1 was essential for the export of protein and mRNA in yeast (26). IκBα also formed a complex with CRM1 in vitro (27). Based on its newly identified function as an export receptor, CRM1 was renamed exportin 1. Exportin 1 exhibits homology to the importin β family of proteins (28). Several yeast proteins that are involved in both import and export also exhibit this homology (29, 30). The importin β family may represent the earliest nuclear transport proteins (31).
Interestingly, Ran-GTP is an essential component of the export receptor complex. Exportin 1 binds cooperatively to Ran-GTP in the presence of NES-containing substrate, and this complex does not form in the presence of Ran-GDP. The export receptor for importin α, CAS, was identified recently and shown to be essential for export in vitro (32). Like exportin 1, this receptor also cooperatively binds importin α and Ran-GTP. Therefore, Ran appears to serve a general, regulatory role in both nuclear export and nuclear import.
Although the signals for nuclear export and a putative export receptor have been identified, the requirements of this interaction have not been studied in the molecular detail that has been possible for nuclear import. The main complication in studying nuclear export has been the lack of an experimental paradigm that would allow discrimination between the import and export processes. Here, we describe a green fluorescent protein (GFP)-labeled, hormone-inducible Rev chimeric protein that allows precise control over the nuclear import and export signals of Rev. The clear advantages of this molecule are that export can be visualized in living cells or semi-intact cells, and the import and export pathways can be examined in isolation by the addition or removal of steroid. We have exploited these approaches to characterize the nuclear export pathway both in living cells and in vitro.
MATERIALS AND METHODS
Expression Vector Construction.
The sequence encoding full-length Rev and the hormone-responsive element from the glucocorticoid receptor (Gr) from pRSV-Rev-Gr (33) was amplified by PCR such that an XhoI site was added to the 5′ and 3′ ends of the fragment. This fragment was then inserted into an engineered XhoI site immediately 3′ of the start codon of GFP in pGreenLantern (Life Technologies, Gaithersburg, MD). This new plasmid, pXRGG, encodes a fusion protein containing full-length Rev, followed by the hormone-responsive element of the rat Gr and GFP. The M10 mutation (10) was introduced into pXRGG via overlap-extension PCR. The plasmid pHRGG is similar to pXRGG; however, a deletion in the 3′ end of the region encoding Gr results in a loss of 58 aa from the hormone-binding region. The protein encoded by this plasmid does not translocate in response to hormone. pHRGG was constructed as described for pXRGG, except the Rev/Gr fragment is flanked by HindIII sites and was inserted into an engineered HindIII site immediately 3′ of the start codon of GFP in pGreenLantern.
Transient Transfections and Isolation of a Stable Cell Line Expressing Rev/Gr/GFP.
HeLa (ATCC, CCL2) cells were seeded at a concentration of 1 × 105 cells per 35-mm dish approximately 24 hr before transfection. Transfections were performed by using Lipofectin Reagent (Life Technologies) according to the manufacturer’s instructions with 1 μg of pXRGG.
Generation of the stable cell line included transfection as described above using pXRGG and pSV2neo at a 20:1 molar ratio. Twenty-four hours after the transfection the cells were placed in DMEM (Life Technologies) containing 10% bovine calf serum (Life Technologies) and 900 μg/ml G418 (Mediatech, Herndon, VA). The positive cell line, RGG2.2, was identified by fluorescence microscopy and isolated by using cloning cylinders.
Import and Export Assay in Living Cells.
Cells expressing Rev/Gr/GFP were seeded on coverslips at a concentration of 8 × 104 cells per well 24 hr before use. Nuclear import was initiated by addition of 1 μM dexamethasone (Calbiochem) for 30 min, or 1 μM corticosterone (Sigma) for 1 hr. After import, monolayers were chilled and washed three times with PBS to remove exogenously added hormone. Cells were incubated in the presence of DMEM without phenol red (Sigma) for indicated times. Unless otherwise stated, all incubations were conducted at 37°C in the presence of 25 mg/ml cycloheximide. Nuclear export was halted by fixing the cells in 3.6% formaldehyde (EM grade 20%, Ladd Research Industries, Burlington, VT) for 30 min at room temperature. After fixation, coverslips were washed with deionized water and mounted on 3 ml of PBS/glycerol (1:10) solution. For quantitation of export, parallel monolayers were permeabilized with 100 μg/ml digitonin (Calbiochem) for 10 min at 4°C and washed with 1 ml PBS before fixation to remove cytosolic Rev/Gr/GFP. Rev/Gr/GFP remaining in the nucleus after digitonin treatment was analyzed by using confocal laser microscopy. Data were normalized to 100% nuclear fluorescence, which was defined as the amount of fluorescence present in cells treated with buffer A alone, and represent the mean ± the SD of triplicate fields.
In Vitro Export and Import Assay.
Cells were treated with steroid as described above, then washed three times with buffer A (8) and permeabilized with 40 μg/ml of digitonin on ice in buffer A. After permeabilization, the coverslips were washed two times in buffer A and placed in a humidified chamber. Each coverslip received 50 μl of complete transport buffer containing buffer A supplemented with 1% BSA, an energy-regeneration system, and untreated rabbit reticulocyte lysate (RRL) (Promega) (8). RRL was fractionated by using a PD-10 column (Pharmacia). Where indicated, 50% RRL was treated with apyrase (20 units) for 15 min at 30°C before the addition of 5 mM AMP-PNP. Nuclear export was quantitated as described above. Nuclear import and thapsigargin and calmidazolium treatments were performed as described (8).
Immunoblot Analysis.
After export in vitro, cells were solubilized with 2× sample buffer (NOVEX) and processed for immunoblot analysis. Proteins were resolved by using 8–16% SDS/PAGE and transferred to nitrocellulose. Rev/Gr/GFP was detected by using GFP polyclonal antisera (CLONTECH), followed by alkaline phosphatase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch), and developed by using BCIP/NBT as per manufacturer’s instructions (Promega).
Fluorescence Microscopy.
Confocal images were obtained by using a Zeiss LSM 410, equipped with a Zeiss Axiovert 100TV microscope. For quantitation, photomultiplier gain and black level were set such that none of the pixels reached saturation in any of the monolayers viewed. The gain settings were held constant within each experiment. The average pixel intensity of the nucleus of each cell in a field was measured by using the Zeiss lsm 410 software. Three separate fields, each containing approximately 50 cells, were measured per condition. Digital images were collected by using a Zeiss Axiovert 100 TV microscope and a Hamamatsu C5810 chilled 3CCD camera (Hamamatsu, Middlesex, NJ).
RESULTS
Rev Nuclear Transport in Living Cells.
The HIV-1 transactivating protein Rev is a nucleolar protein containing both nuclear import and export signals, enabling it to constantly shuttle between the nucleus and cytoplasm. To study the Rev export pathway both in living cells and in vitro, we constructed a hormone-inducible, GFP-labeled chimeric Rev protein (Rev/Gr/GFP) (Fig. 1). This construct contains the full-length Rev sequence and the hormone-responsive element from the glucocorticoid receptor (33) and has a molecular mass of approximately 64 kDa. A stable cell line expressing Rev/Gr/GFP was created to facilitate the quantitation of nuclear transport.
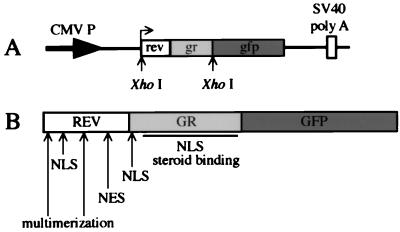
Figure 1.
A diagram of the Rev/Gr/GFP expression plasmid and chimeric protein. (A) pXRGG expression plasmid contains the full-length Rev sequence followed by the sequence encoding the hormone-binding region of Gr. This fragment has been inserted directly after the start codon of the GFP in an engineered XhoI site. (B) The nuclear localization sequence (NLS) is indicated for both the Rev and Gr portion of the chimeric protein. The regions containing the NES and multimerization domain of Rev also are indicated. The steroid-binding region and the second NLS in Gr are shown by the solid bar.
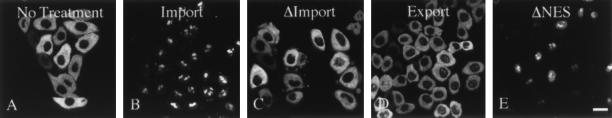
To test whether Rev/Gr/GFP behavior was similar to wild-type Rev, import and export were examined in these cells (Fig. 2). In the absence of steroid, Rev/Gr/GFP was localized to the cytoplasm (Fig. 2A). Translocation to the nucleus and nucleolar localization occurred only with the addition of either the dexamethasone (Fig. 2B) or corticosterone (data not shown). Under similar conditions, a Rev/Gr/GFP mutant containing a deletion in the glucocorticoid-binding domain failed to enter the nucleus in response to steroid (Fig. 2C). Export of Rev/Gr/GFP was initiated by the removal of the steroid, and by 2 hr the chimeric protein was localized again to the cytoplasm (Fig. 2D). Nuclear export required a functional NES, since a Rev/Gr/GFP chimeric protein containing the M10 mutation in the NES failed to export and remained associated with the nucleoli (Fig. 2E). Additionally, both import and export were inhibited at 4°C (data not shown). Therefore, although the steady-state distribution is distinct, the nuclear trafficking of Rev/Gr/GFP is indistinguishable from that of wild-type Rev: both translocate to the nucleus and are targeted to the nucleoli; each require a functional NES for export; and translocation of both is temperature-dependent.
Figure 2.
Nuclear translocation of Rev/Gr/GFP is controlled by steroid. The cell line RGG2.2 that stably expresses Rev/Gr/GFP was used to examine nuclear translocation. Cytoplasmic location of Rev/Gr/GFP before steroid treatment is shown (A). Parallel monolayers of cells were treated with 1 μM dexamethasone for 30 min at 37°C then fixed (B) or washed free of exogenous steroid and incubated for an additional 2 hr at 37°C before fixation (D). Cells transiently expressing Rev/Gr/GFP chimera with a either a nonfunctional NES (E) or a mutation in the hormone-binding region (C) are shown as negative controls. (Bar = 25 μm.)
We obtained additional information by visualizing the nuclear import and export of Rev/Gr/GFP in living cells by time-scan confocal microscopy or video microscopy. Here, we observed that the chimera accumulated around the nuclear membrane before concentration in the nucleolus. Upon removing steroid to initiate export, the nucleoli gradually diminished in intensity with concomitant increase in cytoplasmic concentration; no accumulation was observed at the nuclear periphery (data not shown). These data suggested that, unlike nuclear import of Rev/Gr/GFP, nuclear export of the chimera did not appear to be limited by the rate of movement across the nuclear membrane.
Reconstitution of Rev Nuclear Export in Vitro.
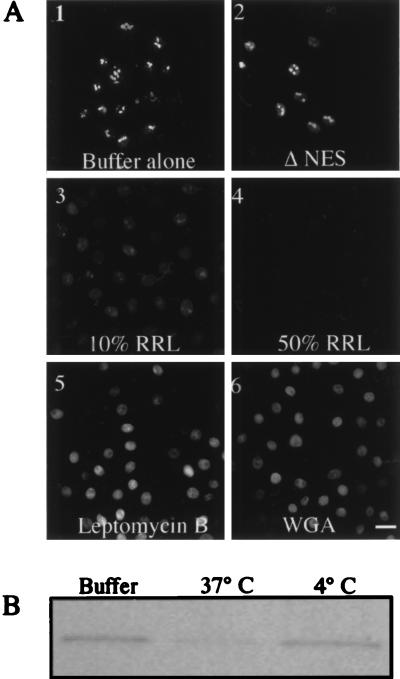
After demonstrating that Rev/Gr/GFP behaved like wild-type Rev in living cells, we developed a system for identifying the requirements of nuclear export in vitro. Rev/Gr/GFP expressing cells first were treated with exogenous steroid to induce import of the chimera. The plasma membrane then was selectively permeabilized with digitonin to release cytosolic components (34). The export pathway was initiated by the removal of steroid and the addition of exogenous factors. This system provides a means to reconstitute export using defined components. Once the Rev/Gr/GFP chimera exports from the nucleus it is soluble in the transport buffer and removed in the washing step; therefore, export is scored as a loss of fluorescence from the nucleus. A range of digitonin concentrations was tested, and 40 μg/ml was found to be optimal as nuclei excluded unconjugated phycoerythrin under these conditions (data not shown). Incubation with transport buffer lacking an ATP-regeneration system and cytoplasmic components failed to initiate export (Fig. 3A-1). The addition of a regeneration system plus nucleotides also did not stimulate export, indicating that additional cytosolic factors are required (data not shown). However, supplementing the transport buffer with an ATP-regeneration system, nucleotides, and 10% RRL stimulated export (Fig. 3A-3). Increasing the amount of RRL to 50% further enhanced the amount of Rev/Gr/GFP exported (Fig. 3A-4). Export was inhibited completely at 4°C (data not shown). Under several of these conditions the export of Rev/Gr/GFP was monitored by immunoblot analysis (Fig. 3B). Leptomycin B, which interacts with exportin 1, inhibited Rev/Gr/GFP export (Fig. 3A-5), as did succinylated wheat germ agglutinin (Fig. 3A-6). Finally, Rev/Gr/GFP containing the M10 mutation in the NES failed to export in this in vitro system (Fig. 3A-2). These data argue that the export of Rev/Gr/GFP is not a result of a nonspecific increase in nuclear membrane permeabilization or of nuclear proteolysis. Instead, export measured in vitro is a result of a specific and active mechanism requiring a functional NES, exogenous cytosolic factors, and an energy regeneration system.
Figure 3.
Reconstitution of nuclear export in vitro. (A) Nuclear export was examined in vitro in RGG2.2 cells by fluorescence microscopy. After incubation with 1 μM corticosterone at 37°C, for 30 min, the cells were washed and permeabilized with 40 μg/ml of digitonin. Permeabilized cells then were incubated either in buffer A (1) or with transport buffer containing 50% RRL (2–6) with and without indicated inhibitors. A-2 represents cells transiently expressing a Rev/Gr/GFP chimera containing the M10 mutation. All cells were visualized by confocal laser microscopy. (Bar = 25 μm.) (B) Immunoblot analysis of nuclear export in vitro. Rev/Gr/GFP was detected in total cellular lysates from cells incubated in buffer A (Buffer) and cells incubated in transport buffer containing 50% RRL at indicated temperatures.
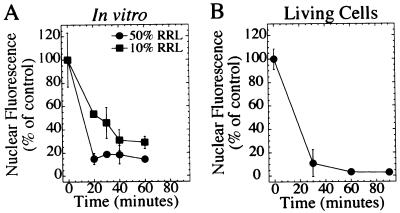
In this in vitro system, the t1/2 for export was approximately 20 min using 10% RRL (Fig. 4A, squares). The export rate was enhanced further by using 50% RRL (Fig. 4A, circles). This rate of export was comparable to that seen in living cells when corticosterone was withdrawn (Fig. 4B). This in vitro export system shares many characteristics seen in vivo: both systems are temperature-sensitive and NES-dependent, and each exhibits a similar export rate. However, this in vitro system allows for isolation of the specific factors required for export.
Figure 4.
The kinetics of nuclear export in vitro is comparable to export in vivo. (A) Digitonin-permeabilized RGG2.2 cells were incubated with export buffer containing either 10% RRL (squares) or 50% RRL (circles) for indicated times. Monolayers incubated in buffer A for 40 min is represented by 0 time. (B) RGG2.2 cells were incubated with 1 μM corticosterone for 30 min. Cells then were washed and incubated in steroid-free medium for indicated times. Before fixation, cells were permeabilized with 100 μg/ml of digitonin to release cytoplasmic Rev/Gr/GFP.
Nuclear Export Requires ATP Hydrolysis.
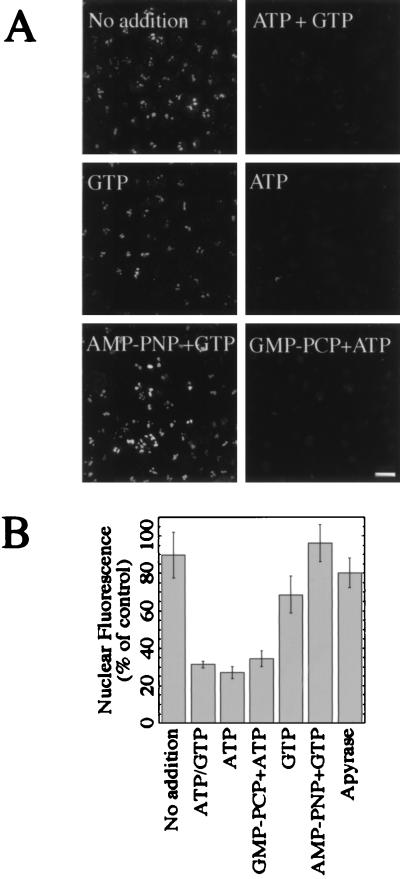
Fractionation of the RRL was performed to identify factors required for nuclear export. Initially, the RRL was fractionated by size-exclusion chromatography to remove small molecules including ions and nucleotides. The fractionated RRL then was used alone or in combination with various nucleotides to stimulate export (Fig. 5). Transport buffer supplemented with the fractionated RRL failed to stimulate export, as indicated by the fluorescent nucleoli (Fig. 5A, No addition). However, supplementing the fractionated RRL with ATP and GTP (1 mM each) restored export (Fig. 5A, ATP + GTP) and resulted in a 70% decrease in fluorescence (Fig. 5B). The addition of GTP alone did not stimulate export (Fig. 5A, GTP). Surprisingly, export was fully restored with the addition of ATP (Fig. 5A, ATP). The amount of export was comparable to that detected when both ATP and GTP were added back (Fig. 5B). A nonhydrolyzable analog of ATP used in combination with GTP did not promote export (Fig. 5A, AMP-PNP + GTP), while addition of GMP-PCP plus ATP did (Fig. 5A, GMP-PCP + ATP). To further test the involvement of ATP, unfractionated RRL was preincubated with apyrase then supplemented with AMP-PNP before it was added to the permeabilized cells (Fig. 5B). This treatment also inhibited export. Taken together, these data argue that the hydrolysis of ATP, but not GTP, is required for nuclear export in vitro.
Figure 5.
ATP hydrolysis is required for nuclear export of Rev/Gr/GFP. (A) Corticosterone-treated, digitonin-permeabilized cells were incubated with transport buffer supplemented with 50% fractionated RRL (No addition) or with fractionated RRL containing added nucleotides (remaining panels) for 30 min at 37°C. The nucleotides ATP and GTP were added at 1 mM, while AMP-PNP and GMP-PCP were used at 5 mM. (Bar = 25 μm.) (B) Nuclear fluorescence shown in A was quantitated by image analysis. The of amount of nuclear fluorescence after incubation with apyrase-treated RRL is also shown.
Nuclear Export Does Not Require Luminal Calcium Stores.
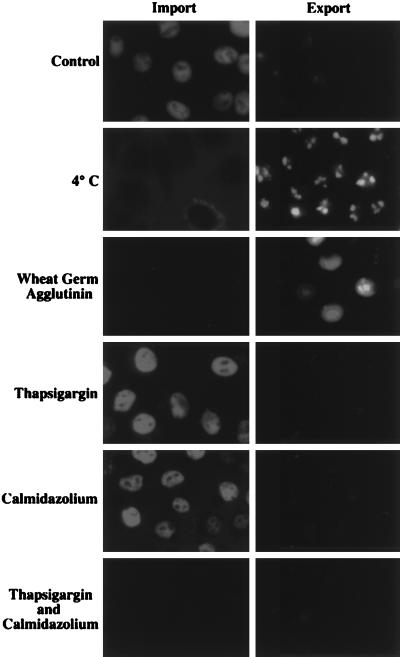
We have found previously that calcium is required for the activation of nuclear protein import (8). Ran/TC4-dependent import is sensitive to the depletion of intracellular calcium stores while calmodulin-dependent import is sensitive to low-cytosolic calcium. We sought to determine whether nuclear export of the Rev protein also required calcium. Export experiments using fractionated rabbit reticulocyte lysate, depleted of small molecules including calcium (Fig. 5), suggested that export is not as sensitive to the depletion of free calcium as import (8). To examine this difference in more detail, thapsigargin was used to deplete calcium from the lumen of the endoplasmic reticulum and calmidazolium was used as a calmodulin antagonist. Cells were assayed in parallel for import and export in vitro under conditions that required active nuclear pores and active transport (Fig. 6). Neither import nor export was sensitive to the inclusion of either thapsigargin and calmidazolium. However, import was blocked by the presence of both thapsigargin and calmidazolium while export was unaffected by the combination of these drugs. Therefore, unlike nuclear import, nuclear export of the Rev protein does not depend on either luminal calcium stores or calmodulin in this system.
Figure 6.
Luminal calcium and calmodulin are not required for nuclear export of Rev/Gr/GFP. Corticosterone-treated RGG2.2 cells (Export) and normal HeLa cells (Import) were digitonin-permeabilized and incubated with transport buffer supplemented with 50% HeLa cytosol for 30 min at 37°C (Control). Import of a phycoerythrin, nuclear localization sequence-conjugate is represented by accumulation of fluorescence in the nuclei, and export of Rev/Gr/GFP is represented as loss of fluorescence from the nuclei. As negative controls, incubations were carried out at 4°C and in the presence of wheat germ agglutinin. Nuclear import and export also were examined in the presence of thapsigargin (10 μM), calmidazolium (8 μM), or both, as described in the Materials and Methods. All images contain at least 8 cells per field, and data are representative of two separate experiments.
DISCUSSION
Direct Visualization of Rev Nuclear Import and Export.
The experimental design presented here provides a way to observe the real-time interaction of Rev with the export machinery and determine its rate-limiting steps. The ability to directly visualize Rev/Gr/GFP export in living cells will become more important as factors involved in export are isolated. Identified factors could be tagged and reintroduced into the Rev/Gr/GFP cell line. Such information has proven invaluable for defining the steps involved in nuclear protein import and should be equally valuable in elucidating the steps involved in nuclear export.
An Assay to Characterize Nuclear Export in Vitro.
Nuclear proteins that shuttle between the nucleus and the cytoplasm contain both import and export sequences, thus complicating the analysis of the nuclear protein export pathway. Extremely clever schemes were devised originally to demonstrate that nuclear proteins rapidly shuttle between cytoplasm and nucleus (35). Some systems have attempted to avoid the complications of separating nuclear import and export by microinjecting synthetic chimeric constructs that contain an NES (9, 25, 36). Still others control export by depletion of essential factors (32). While these types of assays are informative, they are also time consuming and do not lend themselves to biochemical fractionation where high throughput is required. Additionally, when the NES is used out of the context of the native protein, it may be difficult to identify all of the steps in the export pathway. For example, when the NES of Rev is placed on a heterologous protein, this protein is distributed throughout the nucleoplasm, whereas the steady-state localization of full-length Rev is largely nucleolar (37). Since HIV-1 uses Rev to co-opt the host machinery for export of viral mRNAs, this nucleolar accumulation may be an important point of regulation in the Rev nuclear export pathway.
Our system utilizes the full-length Rev molecule to characterize export. Stable expression of the Rev/Gr/GFP chimera allows for easy quantitation and is readily adapted to an in vitro system by digitonin permeabilization and reconstitution with exogenous factors. Also, molecules found to be important for export in vitro can be tested readily in vivo by using the same cell line in combination with molecular genetic approaches and imaging techniques. The semipermeabilized cell system described here already has yielded important new findings regarding the requirements for nuclear export in vitro.
Inhibition of Nuclear Export by Leptomycin B and the Export Receptor Complex.
Leptomycin B inhibits Rev nuclear export in living cells (24). This inhibition can be explained by the ability of leptomycin B to disrupt the export complex in vitro (25, 26). The export complex consists of exportin 1, NES-containing substrate, and Ran-GTP. In our system, leptomycin B effectively inhibited Rev/Gr/GFP export in vitro. Others have shown that leptomycin B prevents NES-containing proteins from binding to exportin 1, thus inhibiting the export of these proteins (27, 38). Leptomycin B inhibition in our system led to the accumulation of Rev in the nucleoplasm. However, it is curious that Rev does not relocalize to the nucleoli when it fails to bind the export receptor. Perhaps Rev is in a different conformational state or has associated with some other factor, such that it is trapped in an intermediate stage of export. Additionally, the chimeric protein did not accumulate at the nuclear pore in the presence of leptomycin B, consistent with exportin 1 facilitating the association of NES-containing proteins to the nuclear pore complex. This is in agreement with published evidence that exportin 1 (CRM1) localizes to the nuclear envelope and can be found in a complex consisting of two other nucleoporins: Nup88 and CAN/Nup214 (28).
Ran-GTP is an essential component of the export complex. Increasing concentrations of an NES-containing protein allow exportin 1 to cooperatively bind Ran-GTP, but not Ran-GDP (25). This phenomenon was also reported for importin α and its export receptor, CAS (32). Additionally, microinjection of a Ran mutant that does not stably bind GTP inhibited the export of an NES-containing protein (36). These studies suggest that hydrolysis of GTP is not a requirement for export. Here we have demonstrated directly that GTP hydrolysis was not required for Rev nuclear export in vitro. Because the regulatory effects of Ran require GTP hydrolysis and Ran is not limiting in the nucleus, its participation in the export complex may represent a way to ensure efficient export of the complex to the cytoplasm (39, 40). Once in the cytoplasm, the complex can be disassembled by GTP hydrolysis because of the presence of RanGAP and RanBP1 (32).
Requirements for Energy and Nucleotides in Nuclear Export.
Using the in vitro system, we have begun to biochemically dissect the energy and nucleotide requirements for nuclear export. Because HIV-1 Rev is a nucleolar protein, nuclear export consists of translocation from the nucleoli to the cytoplasm. This potentially complex process is likely to include many individual steps. Rev nuclear export occurs via an energy-dependent mechanism in living cells, and ATP has been suggested to play a role in the shuttling of other nucleolar proteins (41). Here we found that Rev nuclear export in vitro is due, at least in part, to a requirement for ATP, but not GTP hydrolysis. The absence of ATP, or using a nonhydrolyzable analog of ATP, prevented nuclear export, leaving Rev/Gr/GFP trapped in the nucleolus. However, Rev/Gr/GFP containing the M10 mutation remained nucleolar in the presence of ATP. These data suggest that ATP is necessary, but not sufficient, for release from the nucleoli. It is likely that ATP is required for several steps in the export pathway, including transport across the nuclear pore complex (NPC) as described for other nucleoplasmic proteins (27, 32). The dependence of Rev export on ATP hydrolysis suggests that an ATPase is intrinsic to the export machinery. It will also be of interest to determine whether an ATP-dependent phosphorylation event is involved in the Rev export pathway as demonstrated for other shuttling proteins (42, 43).
Distinct Requirements for Nuclear Import and Export.
Modulating calcium stores in both the lumen of the nuclear envelope and in the cytosol can affect active nuclear import into the nucleus. Specifically, treatment with agents that deplete luminal calcium stores inhibit GTP-dependent nuclear import and diffusion into the nucleus when cytosolic calcium is kept at a low concentration (8, 44, 45). Proteins associated with the nuclear pore that extend into the lumen of nuclear envelope may provide a means to sense these calcium fluxes. The luminal domain of the nucleoporin gp210 contains three putative EF hand domains, suggesting that calcium may modulate the function of the NPC though this protein (44, 46). At elevated cytosolic calcium concentrations, a calmodulin-dependent import pathway is activated that is independent of luminal calcium stores (8). Therefore, to completely block nuclear import at elevated calcium concentrations it is necessary to use agents that deplete luminal calcium and antagonize calmodulin (ref. 8; Fig. 6). In contrast to what has been observed for nuclear import, nuclear export occurred unimpeded when either or both of these pathways were inhibited. These data confirm that a depletion of luminal calcium does not induce nuclear pores to completely shut down transport. Rather, alteration of the luminal and cytosolic calcium concentrations may provide a way to specifically modulate nuclear import while leaving NES-dependent nuclear export unaffected.
Conclusions.
We have described a convenient assay for characterizing nuclear protein export both in living cells and in vitro. This system utilizes the full-length Rev protein, thus ensuring that the export pathway used by this protein is reflected accurately. By replacing the nuclear import and export signals, other pathways may be examined in a similar fashion. Using this system, we have found that the requirements for nuclear export differ substantially from those of import, suggesting different modes of regulation. These data also strongly suggest that nuclear pores do not functionally close upon depletion of calcium stores. This observed asymmetry between import and export may also be reflected in the structural asymmetry of the nuclear pore itself. The ultrastructure of the nucleoplasmic and cytoplasmic surfaces of the nuclear pore is dramatically different. Since the nuclear pore complex mediates both nuclear import and export simultaneously, the independent regulation revealed by our studies may allow a way of coordinating these two competing processes.
Acknowledgments
We thank Tristam Parslow (University of California at San Francisco, CA) for providing pRSV-Rev-Gr, Barbara Wolff (Novard’s Research Institute, Vienna, Austria) for providing leptomycin B, and Jenny Hinshaw (National Institutes of Health, Bethesda, MD) for helpful discussions and critical reading of the manuscript.
ABBREVIATIONS
- NES
nuclear export sequence
- GFP
green fluorescent protein
- Gr
glucocorticoid receptor
- RRL
rabbit reticulocyte lysate
References
- 1.Pante N, Aebi U. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- 2.Breeuwer M, Goldfarb D S. Cell. 1990;60:999–1008. doi: 10.1016/0092-8674(90)90348-i. [DOI] [PubMed] [Google Scholar]
- 3.Gorlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 4.Powers M A, Forbes D J. Cell. 1994;79:931–934. doi: 10.1016/0092-8674(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 5.Melchior F, Paschal B, Evans J, Gerace L. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nehrbass U, Blobel G. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- 7.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 8.Sweitzer T D, Hanover J A. Proc Natl Acad Sci USA. 1996;93:14574–14579. doi: 10.1073/pnas.93.25.14574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 10.Malim M H, Bohnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 11.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 12.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 13.Klemm J D, Beals C R, Crabtree G R. Curr Biol. 1997;7:638–644. doi: 10.1016/s0960-9822(06)00290-9. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M, Gotoh I, Gotoh Y, Nishida E. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 15.Iovine M K, Wente S R. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy R, Wente S R. Nature (London) 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 17.Richards S A, Lounsbury K M, Carey K L, Macara I G. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridell R A, Fischer U, Luhrmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nix D A, Beckerle M C. J Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasquinelli A E, Powers M A, Lund E, Forbes D, Dahlberg J E. Proc Natl Acad Sci USA. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritz C C, Zapp M L, Green M R. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 22.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 23.Stutz F, Neville M, Rosbash M. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 24.Wolff B, Sanglier J J, Wang Y. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 25.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 26.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 27.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 28.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seedorf M, Silver P A. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rout M P, Blobel G, Aitchison J D. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 31.Malik H S, Eickbush T H, Goldfarb D S. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutay U, Bischoff F R, Kostka S, Kraft R, Gorlich D. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- 33.Hope T J, Huang X J, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adam S A, Marr R S, Gerace L. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 36.Richards S A, Carey K L, Macara I G. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 37.Cullen B R, Hauber J, Campbell K, Sodroski J G, Haseltine W A, Rosen C A. J Virol. 1988;62:2498–2501. doi: 10.1128/jvi.62.7.2498-2501.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 39.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 40.Ren M, Drivas G, D’Eustachio P, Rush M G. J Cell Biol. 1993;120:313–323. doi: 10.1083/jcb.120.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu M H, Lam C Y, Yung B Y. Biochem J. 1995;305:987–992. doi: 10.1042/bj3050987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Liu J, DeFranco D B. J Cell Biol. 1997;137:523–538. doi: 10.1083/jcb.137.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beals C R, Sheridan C M, Turck C W, Gardner P, Crabtree G R. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 44.Greber U F, Gerace L. J Cell Biol. 1995;128:5–14. doi: 10.1083/jcb.128.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stehno-Bittel L, Perez-Terzic C, Clapham D E. Science. 1995;270:1835–1838. doi: 10.1126/science.270.5243.1835. [DOI] [PubMed] [Google Scholar]
- 46.Wozniak R W, Bartnik E, Blobel G. J Cell Biol. 1989;108:2083–2092. doi: 10.1083/jcb.108.6.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]