Abstract
The nuclear envelope is a physical barrier between the nucleus and cytoplasm and, as such, separates the mechanisms of transcription from translation. This compartmentalization of eukaryotic cells allows spatial regulation of gene expression; however, it also necessitates a mechanism for transport between the nucleus and cytoplasm. Macromolecular trafficking of protein and RNA occurs exclusively through nuclear pore complexes (NPCs), specialized channels spanning the nuclear envelope. A novel family of NPC proteins, the FG-nucleoporins (FG-Nups), coordinates and potentially regulates NPC translocation. The extensive repeats of phenylalanine-glycine (FG) in each FG-Nup directly bind to shuttling transport receptors moving through the NPC. In addition, FG-Nups are essential components of the nuclear permeability barrier. In this review, we discuss the structural features, cellular functions, and evolutionary conservation of the FG-Nups.
Subcellular compartmentalization of eukaryotic cells into organelles imparts functional and spatial separation of essential cellular processes. Interorganellar communication, however, is required to coordinate activities within the cell. The movement of molecules between the cytoplasm and a given organelle is accomplished by the use of a regulatory transport pore(s) embedded in the organelle membrane. One of the most complex molecular translocons is the nuclear pore complex (NPC), which mediates all traffic of macromolecules in and out of the nucleus.
NPCs are large, selective channels that regulate the nucleocytoplasmic transport of macromolecules but are permeable to the movement of ions, small metabolites, and small proteins by free diffusion. The ability of the NPC to rapidly transport specific macromolecules and coincidently selectively preclude other molecules from entering the nucleus is one of the mysteries of this biological machine. To overcome the permeability barrier, each cargo greater than ∼40 kDa must display a nuclear localization sequence (NLS) or nuclear export sequence (NES). The respective NLS or NES is recognized and bound by a specific transport receptor, of which many exist in eukaryotic cells.
Transport receptors interact with a subset of NPC proteins to mediate translocation and, as such, serve as a molecular bridge between NPC proteins and cargoes to allow efficient nuclear import and export. A unique family of NPC proteins is directly involved, and are designated the FG-nucleoporins (FG-Nups). The FG-Nups are characterized by domains with extensive repeats of phenylalanine-glycine (FG), and these proteins have specific and essential roles in transport through the NPC (discussed below). Recent work has offered many insights into the biophysical nature of the FG-Nups. Structural aspects of interactions between FG-Nups and transport receptors have been resolved, and regulatory roles of FG-Nups in transport, disease, and development have been discovered. Importantly, understanding the structural, functional, and regulatory properties of FG-Nups has provided new insights into a novel paradigm for selective barrier structures in channels and for the mechanism of regulated and efficient nucleocytoplasmic transport. We review here current knowledge regarding the properties and conservation of FG-Nups. We also discuss how these properties are related to biological functions of this protein family and fit into models for the NPC translocation mechanism.
NPC COMPOSITION AND ROBUSTNESS OF TRANSLOCATION
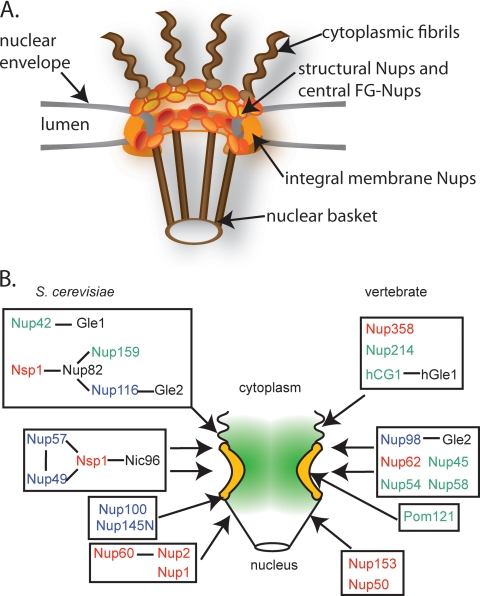
NPCs are assembled from multiple copies of ∼30 different protein components, collectively termed nucleoporins (Nups) (31, 137-139, 159). The NPC proteome includes transmembrane Nups (Poms), which anchor the NPC in the NE, structural Nups, and FG-Nups (159). Recent high-throughput modeling studies predict that the NPC is built from repeating structural modules (3). This repetitive structure is based on the sequential assembly of several copies of each Nup in multiples of eight reflecting the apparent eightfold rotational symmetry of the NPC in the plane perpendicular to the NE (Fig. 1A) (2, 3, 14, 79, 96). Overall, NPCs have an asymmetric shape about the plane of the NE with unordered filaments extending from the cytoplasmic face of the pore. The filaments on the nuclear side of the NPC converge into a basket structure (14, 15, 40, 168). Remarkably, this general structure and many of the components are conserved throughout eukarya (2, 3, 14, 23, 31, 79, 96, 110, 139).
FIG. 1.
FG-Nups are distributed throughout the NPC. (A) Schematic representation of the eightfold radial symmetry of the NPC, showing key aspects of the NPC architecture. (B) Nup subcomplexes and relative NPC substructural localization. Each box represents a biochemically or functional documented subcomplex (from studies summarized by Alber et al. [3]). S. cerevisiae FG- Nups are depicted on the left side; vertebrates are depicted on the right. The FG-Nups (colored text) are found in discrete subcomplexes and substructural locations. This includes Nups containing predominantly FG (green text), GLFG (blue text), and FXFG (red text) repeats. Select structural, non-FG-Nups are shown in black text.
The assembled NPC structure must remain selective while flexing to accommodate cargo-receptor complexes that vary over several orders of magnitude in diameter (159, 178). The vertebrate NPC has been shown to transport signal-bearing gold particles up to 39 nm in diameter (55, 120), as well as to transport similarly large-sized physiological cargoes, including ribosomal subunits (88, 97) and Balbiani ring mRNPs (35). Equally striking is the NPC transport capacity. It is estimated that each of the ∼2,800 NPCs in a HeLa cell transports upwards of 60,000 molecules/min (66). Similarly, Saccharomyces cerevisiae, which has ca. 75 to 150 NPCs per cell (176), is estimated to actively transport 50 to 250 messenger RNAs (mRNAs) transcripts per NPC per min, along with 10 to 20 ribosomal subunits and up to 1,000 transfer RNAs (tRNAs) per pore per min (82). In addition to transporting of all of these distinct types of RNA, NPCs also simultaneously transport large numbers of protein cargoes (65). Thus, trafficking through the NPC is quite robust and efficient, as well as apparently bidirectional (56).
FG-Nups: REPEAT MOTIFS AND TYPES
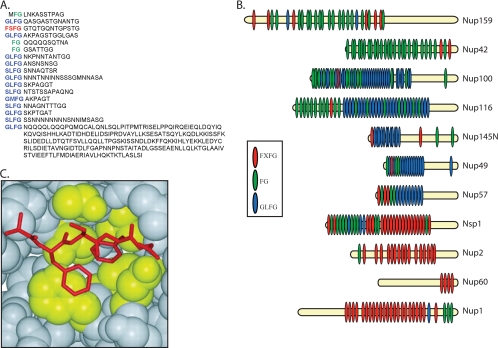
Over the past two decades, multiple studies have indicated that FG-Nups contribute to both the NPC permeability barrier and to the active import and export translocation mechanisms. At the primary amino acid sequence level, FG-Nups have domains with clusters of repeats of Phe-Gly, followed by characteristic spacer sequences (141). The core repeat unit of each FG-repeat is defined as predominantly Phe-Gly (FG), Gly-Leu-Phe-Gly (GLFG), or Phe-any-Phe-Gly (FXFG) (Fig. 2A and B) (141). In addition, the spacer sequences between FG, GLFG, and FXFG repeat types differ slightly in S. cerevisiae (Table 1) and in other organisms (141). Spacer sequences between FXFG repeats are enriched for Ser and Thr and tend to be highly charged; spacers between GLFG repeats are devoid of acidic residues and are enriched for Asn and Gln. Spacers for repeats with an FG core appear to be more degenerate and may have either spacer type. Others have subcategorized FG core repeats further (e.g., PSFG [40a, 121]), but these repeats do not have unique spacer sequences and have also been grouped with the FG class of repeats. Whether the spacer sequences play functional roles in the NPC permeability barrier or active translocation mechanism has not been resolved.
FIG. 2.
Key structural and sequence features of FG-domains in S. cerevisiae. (A) The full primary amino acid sequence of S. cerevisiae Nup49 is aligned with each FG repeat in a line break to align on the left. The FG repeats (green), FXFG (red), and GLFG (blue) are further highlighted. (B) Schematic diagrams for the 11 FG-Nups in S. cerevisiae showing the distribution and type of FG repeats. Single repeats are represented by an oval. FG repeat, green; FXFG repeat, red; GLFG repeat, blue. The diagrams are adapted from Strawn et al. (157) with permission from the publisher. (C) Structural analysis of an FXFG-importin β complex gives a surface view of the FXFG peptide (red) interaction pocket. Hydrophobic residues of importin β are highlighted in yellow. (Reprinted from reference 12 with permission from the publisher.)
TABLE 1.
Amino acid composition of S. cerevisiae FG domains
| Nup protein | Repeat motif(s) | Composition (%) of yeast FG domaina |
|||
|---|---|---|---|---|---|
| Acidic | Basic | Q+N | S+T | ||
| Nup42 | FG | 0.55 | 3.33 | 17.45 | 29.92 |
| Nup159 | FG | 11.14 | 7.74 | 6.29 | 29.54 |
| Nup49 | GLFG | 0 | 2.98 | 21.28 | 24.25 |
| Nup57 | GLFG | 0 | 2.7 | 19.37 | 27.48 |
| Nup116 | FG, GLFG | 0 | 1.98 | 27.6 | 21.49 |
| Nup145 | GLFG | 0 | 2.5 | 21.5 | 30.5 |
| Nup100 | GLFG | 0.18 | 2.28 | 27.76 | 26.89 |
| Nsp1 | FG, FXFG | 10.19 | 11.57 | 12.44 | 24.35 |
| Nup1 | FXFG | 9.9 | 11.88 | 9.11 | 29.11 |
| Nup2 | FXFG | 13.56 | 13.85 | 10.61 | 26.84 |
| Nup60 | FXF | 15.3 | 14.12 | 14.12 | 20 |
FG domain boundaries defined (amino acid residue numbers): Nup42-FG (4 to 364), Nup159-FG (464 to 876), Nup49-GLFG (2 to 236), Nup57-GLFG (2 to 223), Nup145-GLFG (10 to 209), Nup100-GLFG (2 to 570), Nup116-FG,GLFG (2 to 95 and 205 to 715), Nsp1-FG,FXFG (13 to 591), Nup1-FXFG (384 to 888), Nup2-FXFG (189 to 527), and Nup60-FXF (397 to 512).
In metazoans, FG domains are glycosylated. Glycosylation is specifically mediated by O-linked N-acetylglucosamine transferase, which attaches an N-acetylglucosamine (GlcNAc) moiety to Ser or Thr (113). This O-linked glycosylation of Nups is not essential for proper Nup localization at the NPC (87); however, O-linked GlcNAc residues might play a role in translocation through the NPC (36, 57, 70, 113) or regulate the phosphorylation state of specific O-glycosylated Nups (113). These sugar moieties have also been proposed to serve as binding sites for transport of lectins (169). The biological importance of glycosylation of FG-Nups in metazoan cells and the impact of these posttranslational modifications on nucleocytoplasmic transport has not been fully resolved.
FG-Nup ASSEMBLY INTO THE NPC
In the intact NPC, FG-Nups occupy peripheral, surface accessible positions in the NPC; they are predicted to line the innermost layer of the NPC central channel (3, 139). In S. cerevisiae, three FXFG repeat-containing Nups are found exclusively on the nuclear basket face of the NPC; these are Nup1, Nup2, and the FXF-containing Nup60 (Fig. 1B and Table 2) (139). The FG-Nups Nup42 and Nup159 are components of the cytoplasmic fibrils. The FXFG repeat-containing Nsp1 and the GLFG-containing Nup49 and Nup57 are distributed centrally or symmetrically in the NPC. The GLFG-containing Nup100 and Nup116 are biased toward the cytoplasmic face of the pore, whereas Nup145N, also a GLFG repeat Nup, localization is biased toward the nuclear face of the pore (139). Given the homology of both sequence and function for Nup100, Nup116, and Nup145N and their apparent evolutionary relationships (110, 174) (see below), the net distribution of these can be considered effectively symmetrical (157). The FG-Nups of higher eukaryotes also arrange in distinct substructural locations within the pore (159).
TABLE 2.
Properties and homologues of FG-Nupsa
| S. cerevisiae Nup | Essential in S. cerevisiae | Localization in S. cerevisiae | Repeat motif(s) | Abundance/ pore in S. cerevisiaeb | No. of FG repeats in S. cerevisiaec | Homologue in vertebrates | Abundance/pore | Homologue in: |
|
|---|---|---|---|---|---|---|---|---|---|
| C. elegans | D. melanogaster | ||||||||
| Nup42 | No | Cytoplasmic | FG | 8 | 28 | hCG1/NLP1 | 16 | ||
| Nup159 | Yes | Cytoplasmic | FG | 8 | 25 | Nup214 | 8 | npp-14 | Nup214 |
| Nup49 | Yes | Symmetric | GLFG | 16 | 17 | Nup58, Nup45 | 48 | Nup58 | |
| Nup57 | Yes | Symmetric | GLFG | 16 | 15 | Nup54 | 32-48 | npp-1 | Nup54 |
| Nsp1 | Yes | Symmetric | FG, FXFG | 32 | 12, 22 | Nup62 | 16 | npp-11 | Nup62 |
| Nup100 | No | Cytoplasmic bias | GLFG | 8 | 44 | Nup98 | 8 | npp-10 | Nup98 |
| Nup116 | No | Cytoplasmic bias | GLFG | 8 | 9, 40 | 8 | npp-10 | Nup98 | |
| Nup145N | No | Nuclear bias | GLFG | 16* | 13 | 8 | npp-10 | Nup98 | |
| Nup1 | Nod | Nuclear | FXFG | 8 | 22 | Nup153 | 8 | npp-7 | Nup153 |
| Nup2 | No | Nuclear | FXFG | 8* | 14 | Nup50 | 32 | ||
| Nup60 | No | Nuclear | FXF | 8 | 4 | ||||
| Symmetric, integral membranee | FG | 23† | Pom121 | 8 | |||||
| Cytoplasmice | FXFG | 21† | Nup358/RanBP2 | 8 | npp-9 | Nup358 | |||
Estimates of localization and abundance were as published previously (32, 139). Homologues are based on summaries published elsewhere (78, 159).
*, estimate.
†, values that represent the number of repeats in the Homo sapiens protein.
Nup1 is essential in certain S. cerevisiae genetic backgrounds (37).
Localization of Homo sapiens protein.
FG-Nups are anchored into these specific NPC locations and Nup subcomplexes by their non-FG domains, and deletion of these non-FG domains results in mistargeting, (8, 39, 45, 52, 80, 86, 171, 181). Coiled-coil motifs in the non-FG domains of FG-Nups are predicted to provide this NPC anchoring function (44). FG-Nups appear to be among the last proteins recruited to a nascently forming NPC (48). Consistent with their occupying peripheral positions in the NPC, FG-Nups also generally have shorter residence times than Nups predicted to have more structural roles, and some are considered to be transient or shuttling components of the NPC (45, 48, 71, 105, 131). The most prominent exception to this order is that metazoan (m)Pom121 is both an integral membrane protein, intimately connected to anchoring the NPC in the NE lipid bilayer, and also an FG-Nup (154).
STRUCTURAL FEATURES OF FG-Nup DOMAINS
Although FG-Nups are anchored in discrete subcomplex structures throughout the NPC (139, 159), biochemical, biophysical, and cell biological studies reveal shared, unusual properties of these proteins. Single-molecule atomic force spectroscopy studies demonstrate that isolated FG domains are natively unfolded (103), in agreement with biochemical studies (40, 41), and their flexible filaments can occupy a dynamic range of topological positions (53, 103). The FG domains are characterized by a large hydrodynamic (Stokes) radius, are enriched in amino acid residues associated with structural disorder and flexibility, and exhibit high in vitro proteolytic sensitivity (40, 41, 46). Although unfolded regions are predicted in a substantial portion (∼30%) of the S. cerevisiae proteome (49), the FG domains are particularly large spans of unfolded regions. The cellular mechanisms that protect the unfolded FG domains from proteolysis or aggregation in vivo are not fully understood. NPC assembly of S. cerevisiae Nup53 is mediated in part by Kap121 (107), thus suggesting that Kaps might serve as chaperones for FG-Nup assembly. However, Kap95/Kap60 failed to protect FG domains from proteolysis in vitro (40). Thus, it remains unclear whether FG domains are protected by a chaperoning factor prior to being assembled into the NPC.
Unfolded protein domains favor binding to multiple partners and can facilitate rapid association and dissociation rates (164). The flexibility of these domains likely favors repeated collisions with binding partners and means that an FG domain is accessible from various directions. In support of the flexibility of FG domains, immunoelectron microscopy with an antibody specific to the FG domain of Xenopus Nup153 finds that this domain occupies multiple topological positions (53). In contrast, the non-FG domains of Nup153 are anchored at specific points in the NPC. Although some have suggested that FG domains alter their topology (123) or collapse (104) upon transport receptor binding (see below), how this contributes to the transport mechanism remains unknown.
EVOLUTIONARY CONSERVATION OF FG-Nups
The conservation of FG-repeat motifs between Nups and across multiple species directly facilitated the early cloning and characterization of this protein family. For example, FG repeat motifs in FG-Nups from both yeast and metazoans are recognized specifically by the same monoclonal antibodies (7), reflecting that the motifs have shared epitopes. Evolutionary modeling studies have identified repetitive folds and motifs among non-FG-Nup domains and suggest that Nups arose from gene duplication and diversification events over evolutionary time (43). Protein structure prediction analysis of Nups finds very few total structural folds are represented, and the NPC is predominantly built of alpha-helices and beta-sheets (44). These studies also hypothesize that non-FG-Nups are related to coated vesicle components and potentially have the capacity to stabilize highly curved membrane surfaces (23, 43). A high level of redundancy and structural duplication also suggests that the evolution of the NPC and diversification of Nups have been quite rapid. However, there is no clear prokaryotic structural ancestor for FG domains, which makes understanding their evolutionary appearance challenging (110).
Within the Saccharomyces genus, sequence analysis indicates that there is overall rapid evolution and substitution of amino acids (40a). Remarkably, discrete clusters of polar or charged residues adjacent to FG motifs appear to be conserved. From yeast to metazoans, some Nups are fairly highly conserved in both sequence and structure, whereas others have divergent sequences and yet retain similar tertiary structures and functions (110). Taken together, structural elements and subcomplex shapes are maintained in such a way that the ultrastructure of NPCs is highly similar between divergent species (2, 3, 14, 79, 96, 110). An interesting example of gene duplication and divergence is illustrated by the S. cerevisiae FG-Nups Nup100, Nup116, and Nup145 versus their vertebrate counterparts Nup96 and Nup98. Phylogenetic analysis suggests that S. cerevisiae Nup100, Nup116, and Nup145 are lineage-specific derivatives of an ancestral Nup98 (38, 110). Evidence for evolutionary gene duplication events among these three Nups within S. cerevisiae comes from genomic sequences; the same tRNA and transposon sequence elements are adjacent to both NUP100 and NUP116 loci (174), and the N-terminal GLFG repeats of Nup145 are similar to the sequence of repeats in Nup100 and Nup116 GLFG domains (173). A second line of evidence for evolutionary gene duplication and divergence among these three Nups comes from examining the protein domain organization (142). The S. cerevisiae Nup145 polypeptide is a precursor to two proteins found in the NPC; the peptide is autocatalytically cleaved posttranslationally to Nup145N (∼65 kDa) and Nup145C (∼80 kDa) (132, 136, 161, 173), which each assemble into different substructural positions in the NPC (81). Remarkably, this unusual event is conserved; the cleavage motif and event also occurs with the vertebrate homologs Nup96/Nup98, which are transcribed and translated as an ∼195-kDa fusion polypeptide (58, 132). The uncleaved Nup96/Nup98 fusion protein is impaired for assembly into the NPC (81), thus raising interesting questions about whether this proteolytic processing event is involved in a regulatory step of NPC biogenesis or in preventing premature activity linked to either of these polypeptides.
NPC FUNCTIONS MEDIATED BY FG-Nups
FG-Nups have been implicated in a number of NPC functions, including receptor-mediated transport, permeability barrier integrity, gene gating, and directionality of transport. FG domains have been studied extensively for their role in interacting with transport receptors during nucleocytoplasmic transport (1, 4, 5, 20, 34, 91, 111, 140, 142, 145, 156-158, 162) and are required in specific combinations for efficient transport (157, 162) (discussed further below). At least some aspects of transport directionality might be facilitated by FG-Nups (166), although the prevailing model is that the primary determinant for directionality is, instead, the Ran GTP/GDP gradient (115). For both the Kap95/Kap60 import and the mRNA export pathways, motifs adjacent to FG repeats coordinate termination of transport and release of transporting complexes from the NPC (155, 165).
In addition to their role in mediating transport for soluble macromolecules through the central NPC channel, FG-Nups are necessary for the targeting of inner nuclear membrane proteins from the outer nuclear membrane/endoplasmic reticulum (95). In addition, FG-Nups are critical components of the permeability barrier, and NPCs lacking specific FG domains are “leaky,” permitting diffusion of inappropriate molecules (121) (see additional discussion below). FG-Nups are also linked to gene gating, the process of chromatin association with NPCs (24); however, it is not clear whether this association is through their FG-repeat domain or through functions of non-FG domains of these Nups. As a whole, this diversity of functional roles underscores the importance of FG-Nups to the NPC but also increases the complexity of studying the FG-Nups.
TRANSPORT RECEPTORS INVOLVED IN NUCLEOCYTOPLASMIC TRAFFICKING
Nuclear import and export of signal-containing cargoes larger than the permeability barrier limit are generally facilitated by a transport receptor (65). Interestingly, some nucleus-localized macromolecules have an intrinsic capacity to bind FG-Nups and can therefore pass through the permeability barrier without a transport receptor (51, 169). The key molecular determinant of a transport receptor is the ability to interact with both FG-Nups and with cargo(es). The major family of transport receptors is the karyopherins (Kaps), also termed importins, exportins, and transportins. There are 14 known members of the Kap family in S. cerevisiae and more than 21 identified in humans (65, 75, 108, 114). High resolution structural analysis of Kaps reveals an arch built of typically 20 HEAT repeats (30, 124). Structurally, a HEAT repeat forms paired antiparallel alpha helices connected by a short loop. HEAT repeats are found in other cellular proteins, including those from which they derive their name: huntingtin, elongation factor 3, “A” subunit of protein phosphatase A (PR65/A), and TOR1 lipid kinase (6). The arch structure formed by the array of tandem HEAT repeats in Kaps is highly flexible, and this flexibility potentially allows Kaps to adapt to carry a variety of NLS- or NES-containing cargoes and/or to interact with differently spaced FG repeats (see below) (25-27, 29, 99-101, 124). Kaps also interact with the small GTPase Ran (155).
Ran is a member of the Ras superfamily of proteins and, as such, functions as a binary molecular switch between GDP- and GTP-bound forms (172). Ran is essential for assembly and disassembly of transport complexes and provides directionality to Kap-mediated nucleocytoplasmic transport (165). The nucleotide-bound state of Ran is spatially regulated by the Ran GTPase activating protein (RanGAP) and the Ran guanine nucleotide exchange factor (RanGEF) proteins (159). RanGAP is localized to the cytoplasm, and thus the cytoplasm is a RanGDP-rich environment. The RanGEF is nucleus localized, and thus the predominant nuclear form of Ran is in the GTP-bound state. Nuclear RanGTP binds import Kap-cargo complexes to trigger their disassembly in the nucleus. The now-empty import Kap is recycled to the cytoplasm bound to RanGTP. Export Kaps assemble a trimeric complex with a NES-containing cargo and RanGTP prior to nuclear export. Since both the export/recycling step of an import Kap and the export of an export Kap-cargo complex carry RanGTP out of the nucleus—at a rate of efflux estimated at more than 105 molecules per s per nucleus (67, 152)—there must be a countermeasure to import and supply Ran to the nucleus. Indeed, RanGDP is imported to the nucleus by a non-Kap transport receptor, Ntf2. Ntf2 is structurally unrelated to Kaps and functions as a homodimer (124). Importantly, Ntf2 has at least two FG-binding sites (10).
Whereas Kaps are the transport receptors for most proteins and RNAs (including rRNA, tRNA, miRNA, and snRNA) (97, 135), bulk mRNA export employs a nonkaryopherin transport receptor. The mRNA export receptor is the heterodimer Mex67-Mtr2 (S. cerevisiae; in metazoans, TAP/NXF1-p15/NXT1) (92, 143, 146). Mex67-Mtr2 in yeast and NXF1-NXT1 in metazoans are each essential for bulk mRNA export (76, 92, 143, 146, 160, 175). Mex67-Mtr2 and NXF1-NXT1 are structurally distinct from the Kap family of transport receptors and function independently of the RanGTP system (28, 68, 69, 77, 147). However, as with Kap-dependent transport, Mex67-Mtr2 (NXF1-NXT1) interacts FG-Nups, and Mex67-Mtr2 has been demonstrated to bind to at least nine different FG-Nups (4, 156, 158, 162).
STRUCTURAL FEATURES OF TRANSPORT RECEPTOR INTERACTION WITH FG-Nups
Multiple crystallographic studies of the interaction between an FG repeat and transport receptor show that the Phe residue of the FG repeat is buried in a hydrophobic pocket on the outer face of the transport receptor (Fig. 2C) (9-13, 61, 68, 69, 147). This paradigm applies to Kaps, Ntf2, Mex67-Mtr2, and NXF1-NXT1. Extensive domain mapping and structural studies have characterized the outer backbone of each Kap as the platform for interaction with FG-Nups during transport. Specifically, crystallographic and modeling studies show that the Phe side chain of an FG repeat fits into hydrophobic pockets formed by the HEAT repeats of each Kap (11, 12). Multiple FG-binding sites have been identified on the outer face of Kaps (11, 12, 16, 83, 85). Thus, these studies provide direct evidence for the role of the individual FG-repeat unit in interacting with a transport receptor.
In vitro assays demonstrate that hydrophobic residues can be substituted in FG repeats (e.g., Phe to Trp or Phe to Tyr) with only modest effects on Kap95 binding; however, replacing the Phe with Ala in repeats abolishes binding (122). Additional factors may also contribute to binding site specificity, however, including adjacent non-FG binding sites, the substructural location of the FG domain within the NPC, contributions from spacer regions, and the occupancy of neighboring FG-binding sites. Analyzing the potential contributions of each of these has been difficult. Due to the flexibility from the inherently unfolded FG peptides used in crystallization studies to date, interactions between the spacer regions and transport receptors have not been visualized at the atomic or structural level. Thus, it is unclear what role spacer sequences might play.
AFFINITY AND AVIDITY OF FG-TRANSPORT RECEPTOR INTERACTIONS
Transport receptor interaction with FG-Nups is a critical determinant to nucleocytoplasmic translocation, and current evidence supports a model of multiple low-affinity binding events between a transport receptor and FG-Nups during translocation. In accordance with the eightfold radial symmetry of the NPC, FG-Nups are present in multiples of 8 (139). This property simultaneously presents multiple copies of the same FG-binding sites to transport receptors, potentially increasing the number of similar receptors that can occupy the NPC at any given time. Imaging of single-molecule transport in a permeabilized cell system (180) demonstrates that a receptor-cargo complex does not move through the NPC in a directed or linear fashion but instead it proceeds in a Brownian manner, potentially engaging in multiple NPC-receptor interactions during its ∼10-ms transport time. Given this time scale and the motion of the complexes visualized in the NPC, these studies are consistent with multiple, low-affinity interactions occurring between FG repeats and the transport complex. Indeed, interactions between transport receptors and FG domains have typically nanomolar to micromolar binding affinities and are likely transient (129, 163). In fact, transport receptor mutants with increased affinity for binding FG repeats, such as the ntf2-N77Y mutant, impair nucleocytoplasmic transport (98, 130). Overall, rapid, low-affinity interactions between transport receptors and FG repeats are necessary for proper and efficient transport.
Despite the apparent low affinity of FG-receptor interactions, there remains preference for binding specific FG domains. Biochemical approaches have demonstrated that every FG-Nup in S. cerevisiae is capable of binding at least one transport receptor, and each transport receptor can bind at least one FG domain. Overall, each transport receptor appears to have a preference for binding specific FG-Nups or repeat-types (Table 3 and see Table S1 in the supplemental material) (1, 4, 5, 34, 111, 140, 142, 145, 157, 158, 162). However, the mechanistic determinants of these preferences remain elusive. For example, in S. cerevisiae, Mex67 and Kap95 interact preferentially with different domains of Nup116 (158), indicating that there are subtle differences between domains in vivo and also suggesting that a single FG domain could provide binding sites for multiple transport factors. It is not known whether these binding events could be simultaneous. In striking parallel, NXF1 appears to preferentially bind a subset of the GLFG repeats of Nup98, the vertebrate homolog of Nup116 (19).
TABLE 3.
Documented interactions between transport receptors and FG-Nups in S. cerevisiae
| Category | Receptor | Method(s) used to identify interactionsa |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nup42 | Nup159 | Nup49 | Nup57 | Nsp1 | Nup100 | Nup116 | Nup145 | Nup1 | Nup2 | Nup60 | ||
| Import karyopherins | Kap95-Kap60 | A, D, E, G | D | D, E | D, E | D | D, E, G | A, C, D, E, H | D | A, D, E, G, H | A, D, E, G | E, G |
| Pse1 (Kap121) | D | E | A, E | D | E | B, D, E | B, D | A, B | B | |||
| Kap122 | C, E | D | D | |||||||||
| Kap119 (Nmd5) | D | D | D | |||||||||
| Kap104 | A, E | A | A, D | E | D, E | A, D | A | |||||
| Kap123 | A, E | E | E | A, E | D | E | C, E | A, D | E | |||
| Kap114 | G | |||||||||||
| Kap108 (Sxm1) | D, E | E | D | E | E | D | ||||||
| Mtr10 | A | D | A | A | E | A | A, D | |||||
| Import and export | Msn5 | E | E | E | E | B, E | B | B | B | |||
| Export | Xpo1 (Crm1) | A, C, E | A, C, D, E, F | A, E | A, E | E | E | E | A | A | ||
| Los1 | D, F | A, D | ||||||||||
| Cse1 | F | |||||||||||
| Unknown | Kap120 | E | E | C, E | ||||||||
| Other transport receptors | Ntf2 | D | A, D | D | A, D | A | ||||||
| Arx1 | E, G | G | G | A | G | A, E, G | A, G | G | G | G | ||
| Mex67-Mtr2 | D, E | E, G | D, E | E, G | C, D, E, F, G | G | E, G | E, G | ||||
A, yeast two-hybrid assay; B, fluorescence resonance energy transfer; C, copurification/biochemical pulldown; D, affinity copurification-immunoblot detection; E, affinity copurification-mass spectrometry detection; F, genetic interaction; G, reconstituted complex analysis; H, cocrystal structure analysis.
Because there are likely multiple FG-binding sites on a single Kap, it is possible that transport through the NPC is accomplished by pivoting through FG-Nups by binding with different hydrophobic pockets on the Kap. In addition, although FG-Kap binding is measured to be low affinity (e.g., ∼100 nM to 1 μM) (17, 129), there are multiple FG repeats on each FG-Nup and multiple FG-binding sites on each Kap; therefore, the avidity of binding sites may also contribute to transport. These paradigms are predicted to be true for both import and export Kaps (124).
There are at least two FG binding sites on Mex67-Mtr2 and also on NXF1-NXT1 (68, 69, 147). One of these FG binding sites has structural similarity to Ntf2, whereas the other is similar to a ubiquitin-associated motif (21, 22, 61, 62, 68, 69). Within NXF1, the two FG binding sites are structurally different motifs. Interestingly, NXF1 mutants with two Ntf2-like motifs or two UBA-like motifs are competent for mRNA export; NXF1 truncations with just one FG binding motif are nonfunctional (22, 32). Likewise, a mutant form of Mex67 that uncouples the Mex67-Mtr2 heterodimer causes mRNA export defects (143, 146). This supports a model wherein successful NXF1-NXT1 or Mex67-Mtr2 translocation through the NPC requires multiple FG binding sites on the transport receptor and reinforces the notion that avidity is a driving mechanism for FG-transport receptor interactions in the nucleocytoplasmic transport mechanism.
Avidity of FG repeats does impact Kap95 binding to purified FG domains in vitro (122). Furthermore, recent mathematical and computational modeling predicts that transport receptors have more FG binding sites than previously detected (83-85).
These observations were made via molecular dynamics simulations and must be verified biochemically. In addition, the functional importance of avidity of FG repeats within a given domain has not been examined in vivo. Since all known transport receptors have more than one binding site for FG repeats on their surface, it is likely that the avidity of FG repeats within the NPC and in interacting with these receptors is an important factor influencing the transport mechanism.
NPC PERMEABILITY BARRIER
Although the NPC faithfully impedes the transport of molecules larger than the ∼40-kDa permeability limit, it is an effective selective barrier (63). Molecules smaller than this permeability limit diffuse through at a rate that is inversely proportional to their size (119). Receptor-bound molecules greater than the barrier limit size move through the pore at a rate that approaches the rate of diffusion (64). Thus, the NPC does not significantly slow the passage of appropriate, transport-competent large molecules. Paraxodically, binding Nups actually accelerates transport efficiency. The rate of transport through the NPC for similarly sized molecules is significantly different if one of them binds Nups. Specifically, the transport receptor Ntf2 enters the nucleus ∼30-fold faster than green fluorescent protein, even though these two molecules are of similar size (150). Therefore, interactions between transport complexes and the NPC must be transient and in a manner that does not slow the movement of the transport complex through the NPC. Although molecules under the diffusive permeability barrier size limit can move across the NPC independent of a receptor, it is interesting that there are no known essential factors that rely solely on diffusion for nuclear entry during interphase. This underscores the functional efficiency and importance of receptor-facilitated nucleocytoplasmic transport.
The integrity of the NPC is necessary to maintain the permeability barrier. For example, in S. cerevisiae, deletion of the structural proteins Nup170 or Nup188 results in NPCs that are “leaky” to diffusion of molecules larger than wild-type pores permit (149). Both Nup170 and Nup188 are linked to structural roles in NPC assembly, and at least in the case of the nup170Δ mutant, NPC assembly is impaired, preventing incorporation of a subset of structural Nups and FG-Nups (94, 116). Thus, Nup deletions from the NPC can alter permeability barrier integrity. Further, the barrier that remains in nup170Δ cells has increased sensitivity to aliphatic alcohols (148). This suggests that the barrier is likely maintained by hydrophobic interactions between FG-Nups (121).
The precise role of FG domains in forming the permeability barrier in vivo is unclear and remains a subject of much debate. Patel et al. detect an impaired permeability barrier upon removal of a single FG domain from budding yeast (121). However, Strawn et al. reported that the barrier was intact in a mutant with more than half of its FG domains deleted (157). The discrepancy between these results might be due to differences in the assay system used or precise FG domain boundaries deleted. It is, however, clear that FG domains are sufficient to form a rudimentary permeability barrier in an in vitro nanopore system (89). This artificial system consists of a polycarbonate filter separating two fluid chambers, with the filter perforated by cylindrical holes of a diameter comparable to that of the NPC. When these filters are coated with the FXFG domain of Nsp1, transport receptor-bound macromolecules move through the nanopore, whereas macromolecules that do not interact with the FXFG domains are not rapidly transported. This demonstrates that FG domains are minimally sufficient to form a selective permeability barrier.
At a physiological level, alterations to the nuclear permeability barrier can regulate transport. The filamentous fungus Aspergillus nidulans partially disassembles its NPC, removing both structural and FG-Nups, in a cell-cycle-dependent manner (42, 118). The direct consequence is that these nuclei have a relaxed permeability barrier that correlates with nuclear influx of cell cycle machinery. The nuclear entry of these cell cycle regulators is thus potentially controlled at the level of the NPC permeability barrier.
FUNCTIONAL REDUNDANCY AND DIVERGENCE OF FG DOMAINS
Although there are some striking examples of FG-Nups conserved across species, there are also many examples of functional redundancy within the context of a single NPC. The complexity and redundancy of FG-Nups within the NPC has made it difficult to study their roles in vivo in metazoans. The genetically tractable budding yeast has allowed the most comprehensive analyses of redundancy between FG domains and FG-Nups. This system demonstrated that transport defects are only detectible when multiple specific FG domains are deleted (157, 162). Redundancy is also exemplified by the fact that many FG-Nup genes are not essential in S. cerevisiae when deleted singly, only displaying lethality when combined as double or higher-order nulls (47). Even within a single FG domain there is evidence for redundancy among FG repeats. For example, the Tf1 retrotransposon of fission yeast requires FXFG repeats within Nup124 for nuclear import but shows no discernible preference for any single repeat within the FXFG domain (151).
Despite their potential roles in terminal events of nuclear export or in initial events in nuclear import, the cytoplasmic filament Nups and their FG domains are dispensable (157, 170). In addition, direct swapping of the FG domains between S. cerevisiae Nup1 (FXFG domain; nuclear basket localized) and Nup159 (FG domain; cytoplasmic filament localized) does not cause any detectable perturbations of transport (181). Indeed, cells with deletions of all asymmetric FG domains (i.e., those of Nup1, Nup2, Nup60, Nup42, and Nup159) in S. cerevisiae are viable and have no significant transport defects (157). Given that none of the five asymmetric FG domains of S. cerevisiae are essential for transport, it is perhaps not surprising that a direct swap between the FG domain of Nup159 and the FXFG domain of Nup1 did not have any detectable effects (181).
In spite of this evidence for global functional redundancy of FG domains, there is a growing body of literature identifying specific FG domains as critical determinants for single transport receptors. Many laboratories have demonstrated that transport receptors bind different FG domains preferentially (1, 4, 5, 34, 111, 140, 145, 157, 158). Even within a single FG domain, there are specific binding sites for different receptors (158) and perhaps subtle differences between spacer sequences contribute to this. In support of this, the sequence composition and length of a linker sequence in Nup1 affects Kap binding (33, 106). Importantly, though, these studies used in vitro binding and did not consider the transport event in the context of an intact NPC. In vivo evidence for preferred binding sites for each transport receptor comes from other studies. Antibodies to mNup98 or mNup153 block only a subset of transport events (128, 166); however, these antibodies are not directed against FG domains. A fluorescence resonance energy transfer-based assay for Kap-Nup interactions in vivo suggested that Kap121 and Msn5 have both overlapping and specific Nup interactions during transport (34).
In budding yeast, a combinatorial deletion strategy has identified the FG domains required for nucleocytoplasmic shuttling of transport receptors (157, 162). In vivo, the central or symmetrically distributed FG domains are required in specific combinations (157, 162). There are functional differences among the GLFG domains of Nup100, Nup116, Nup145N, Nup49, and Nup57, and the FG-FXFG domain of Nsp1 with respect to their requirements for transport. NPCs with only GLFG domains are viable; however, deletion of multiple GLFG domains generally results in multiple transport defects (157). In the absence of the asymmetric FG domains, there are also functional differences between single, central GLFG domains (162): the GLFG domain of Nup57 is required for mRNA export, while the GLFG domains of Nup100 and Nup145N are required for Kap121-mediated import. In murine cells, Nup98-deficient cells fail to assemble at least three FG-Nups properly (Nup358, Nup214, and Nup62) and yet have defects in transport of only a subset of transport receptors (177). Taken together, there are in vivo preferences for specific transport receptors for distinct FG domains.
A dramatic example of how FG-domain composition can regulate transport is observed in the binucleate Tetrahymena. There are multiple Nup96-Nup98 homologues in Tetrahymena, including two GLFG-repeat-containing and two FG-domain variants that harbor repeats of NIFN. The former assembles into the macronucleus, while the NIFN-variant assembles specifically into the NPCs of the Tetrahymena micronucleus. Direct swapping between the GLFG and NIFN domains resulted in a macronucleus with NIFN repeats and a micronucleus with GLFG repeats. This was sufficient to drive import of micronucleus-specific factors into the macronucleus and vice versa. Thus, the FG-domain composition of NPCs differentially directs macronuclear versus micronuclear import in Tetrahymena (86, 109). These in vivo studies indicate that there are complex requirements for combinations of FG domains for transport via different transport receptors and that the FG repeat composition of an NPC dictates its transport permissibility.
ALTERATION OF FG-Nup COMPOSITION IN DISEASE AND DEVELOPMENT
Changing the FG-Nup composition of the NPC is a potentially rapid and dramatic strategy for modulating the flux of all traffic through the NPC. Evidence for the importance of individual FG-Nups in affecting nucleocytoplasmic transport comes from studies demonstrating changes in FG-Nup composition during disease and development. Disassociation of FG-Nups from the NPC in A. nidulans is used to alter the transport capacity of the pore (42, 118). Likewise, the degradation of Nups by many viruses highlights the modularity of the system to favor specific trafficking events (54, 72-74, 144). Classic electron microscopy experiments have detected an increased number of NPCs in the NE of a stimulated lymphocyte (112), suggesting that there are global mechanisms to regulate the total number of NPCs and to make rapid changes in NPC abundance.
Are there more subtle differences in Nup expression and NPC structure or pathways during organism development? Tissue-specific expression of two Nups has been detected during mouse development (117, 153), although the molecular consequences of this altered NPC composition on signaling and trafficking is not fully understood. In Drosophila, expression of the structural Nup mbo is spatially restricted, and mbo has an inhibitory effect on Crm1-mediated export (167). Further evidence for Nup roles in disease come from studies of a NUP98 knockout mouse (177). NUP98−/− murine cells have defects in a subset of transport pathways (177), and the NUP98+/− mice have defects in interferon responsiveness (50). This thwarted interferon response increases the susceptibility of the mice to lethal viral infection (144), thus demonstrating the importance of functional nucleocytoplasmic transport in immune response. In addition, chromosomal translocations fusing NUP98 to the homeobox transcription factor HOXA9 or NUP214 to the DNA-binding protein DEK have been identified in cases of acute myeloid leukemia (reviewed in references 93, 127, and 179). Fusions of these FG-Nups to other cellular factors have also been reported in other cancers (179). Thus, the expression and localization of FG-Nups can dramatically impact cellular physiology. We predict that future analysis of gene expression patterns in varied tissues and developmental states will detect altered expression of FG-Nups, and the transport components that bind them, with resulting regulatory impacts on cellular processes.
PROPOSED MODELS OF THE TRANSPORT MECHANISM
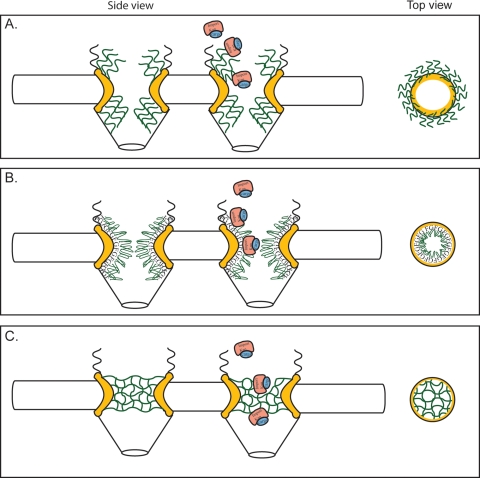
The complexity of the NPC and the dynamic nature of transporting molecules has made it difficult to define the mechanism of nucleocytoplasmic transport. In addition, it has been difficult to develop or design experimental systems to validate proposed models of the transport mechanism. Multiple NPC translocation models have been proposed; however, none completely account for all known NPC properties. Overall, the key differences between proposed models are in the nature of interactions between FG repeats and in the biophysical consequences of FG-receptor interaction (Fig. 3).
FIG. 3.
Models for the mechanism of NPC selectivity and transport. Based on the different features of the respective models, the distribution and physical features of the FG domains are distinct. This is represented in structural models of both a side view (perpendicular to the NE) and top view (cross-section through center of NPC; e.g., from cytoplasm onto plane of NE). NE, black; FG domains, green; structural NPC elements, yellow; importing karyopherin transport receptor, pink; NLS-bearing cargo, blue. (A) Brownian virtual gating model (138). The center of the NPC is a narrow channel, from which FG domains extend to form an entropic barrier to transport. Transport receptors bind these FG domains, overcoming the entropic barrier. By collecting on the NPC periphery, transport complexes increase the probability that they will spontaneously move across the barrier. (B) Reduction of Dimensionality model (125, 126). FG repeats form a continuous surface along the inner face of the NPC, and transport complexes pivot along this surface. The spacer sequence between FG repeats loop outward, forming a physical barrier to diffusion of large molecules; transport complexes might transiently displace these as they move along the FG surface. (C) Selective phase-partitioning model (133, 134). Hydrophobic interactions between FG repeats form a physical meshwork with gel-like properties. Transport receptors bind and transiently dissolve the meshwork in order to translocate through the NPC.
The Brownian/virtual gate model suggests that the NPC is an energy/entropy barrier (138, 139). As such, FG domains form an entropic barrier at each face of the NPC in a way that makes barrier passage energetically unfavorable for molecules in a size-dependent manner (i.e., the larger the molecule, the more entropically unfavorable barrier passage is). These FG domains are presumably mobile and unstructured. Transport receptors overcome this barrier by stochastically interacting with FG-Nups, directly increasing the local concentration of receptor-cargo complexes on FG-Nups and therefore also increasing the probability that a given receptor-cargo complex will randomly diffuse through the NPC (Fig. 3A). In support of this model, a layer of mNup153 FXFG domains is entropically repulsive (103, 104). Furthermore, in follow-up studies, the addition of mImportin β to this system collapsed this entropic layer, as is predicted for the virtual gating model (102). The topological flexibility of FG domains viewed by electron microscopy is consistent with a model in which FG domains do not stably interact, but these data are inconsistent with a recent study that suggests that FG domains form a physically rigorous gel in vitro (60). The Brownian/virtual gate mechanism requires that an adequately high concentration of FG domains be present to form a strong energetic barrier. Surprisingly, deletion of up to half of the FG mass from the NPC does not cause the permeability barrier to collapse (157). Thus, either the NPC permeability barrier is highly resilient to substantial losses of FG domains, or other factors can compensate for these losses.
In contrast to the energetic barrier proposed in the Brownian/virtual gate mechanism, two other models propose a physical barrier to transport. The “reduction of dimensionality” model (125, 126) proposes that FG domains form a continuous surface of potential transport binding sites with the Phe residues aligning along the inner surface of the NPC (Fig. 3B). The spacer sequences between FG repeats and other Nups are proposed to form a selectivity filter that occludes the free diffusion of large molecules (125). Asymmetric FG repeats collect transport complexes, which then move along this Phe surface via a two-dimensional walk, pivoting from one binding site to the next. Mathematical modeling has previously suggested that reduction of dimensionality expedites the rate at which a ligand finds its receptor (see reference 125 and references therein). Thus, this model predicts that removal of FG repeats might cause gaps and disrupt the continuity of the FG surface; such gaps would reintroduce a third dimension for molecular movement through the NPC. This could be the cause of transport defects in certain FG domain deletion strains (157). In addition, the reduction-of-dimensionality model predicts that removal of asymmetric FG repeats would diminish the efficiency of NPCs to collect transport complexes. Curiously, this is not observed in cases where the cytoplasmic filaments are absent (170) or all asymmetric FG domains are removed (157). For the model to hold, functional compensation by the remaining FG repeats would be required.
The selective phase-partitioning model proposes that FG repeats form a physical meshwork (Fig. 3C) (133). This mesh would be assembled by weak hydrophobic interactions between the Phe side chains of FG repeats, and the entirety of the mesh throughout the NPC would resemble a hydrophobic phase or gel. The spacing between Phe-Phe contacts in the mesh is proposed to be such that small molecules can diffuse through without disturbing these contacts. Transport complexes are suggested to traverse the mesh phase by transiently binding to FG repeats and locally disrupting the meshwork. Therefore, this model predicts that FG repeats directly interact and that transport receptors can compete and temporarily disrupt the Phe-Phe hydrophobic interactions. Recent experiments have demonstrated that high concentrations of FXFG domains from Nsp1 can form a gel substance in vitro (60). Indeed, a fluorescently tagged transport receptor can partition into an FXFG gel substance in vitro (59), whereas a protein that cannot interact with FG repeats does not enter this gel efficiently. Although it is impressive that an FXFG gel can discriminate between an inert and an FG-interacting protein, it is not clear whether such a gel barrier could form under physiological conditions or in vivo since formation of the in vitro hydrogel was initiated using harsh chemical conditions. Further, mathematical modeling predicts that binding to and moving through a hydrogel will retard the mobility of transport receptor complexes and will decrease transport efficiency of cargo-bound receptors (i.e., larger complexes) more than free transport receptors (18). Thus, the ability of this proposed FG hydrogel to form in vivo and support known transport rates remains controversial.
RECONCILING DIFFERENCES BETWEEN MODELS
Recently, a novel in vitro assay for detecting low-affinity interactions has shown that certain FG domains are cohesive (121). These assays found that the FG domain of Nup42 and the GLFG domains of Nup116, Nup100, Nup57, Nup145N, and Nup49 can all interact with each other in pair-wise tests. Curiously, however, these experiments did not detect interaction between the Nsp1 FXFG domains (121), in direct contradiction with the proposed self-interaction of these domains in the above FXFG hydrogel (59, 60). Reconciling these discrepancies will require further refining of assays for detecting interactions and developing techniques that can test these properties in vivo at the NPC. It is possible that a hybrid mechanism exists, such as a dually gated system with entropic barriers on either side of the NPC and a physical meshwork barrier in the center of the pore (121). The recently reported FXFG nanopore system (89) may prove a useful tool for probing the conformation and biophysical state of FG domains in the presence of transport receptors. High-resolution microscopic analysis of transport receptor binding to NPCs has recently demonstrated that transport receptors are distributed relatively uniformly along the NPC channel (90). This observation suggests that FG domains are distributed and accessible throughout the NPC, rather than predominantly directed toward the ends of the pore. Continued experiments using these advances in microscopy (90) and artificial selective nanopores (89) have the potential to provide further insights into the NPC gating and translocation mechanism.
FUTURE CONSIDERATIONS
As a whole, the two key differences between the NPC translocation models are in the nature of interactions among FG repeats and how these interactions are altered by transport receptors. Recent work has made progress in understanding the nature of FG-FG interactions, and current evidence supports elements of each of these proposed models in forming the selective yet efficient transport channel of the NPC. Future goals will likely include answering several key questions raised by the FG interaction experiments, such as what dictates each individual FG domain forming an intramolecular or intermolecular network? How would a gelatinous meshwork form in a newly assembling NPC? How does the heterogeneity of FG repeat types in the NPC or the glycosylation of vertebrate FG domains affect the stability of the FG environment? How does the local and native environment of structural Nups and transport factors affect the FG domains? What are the full in vivo consequences of regarding the apparent preferential FG domains for specific transport receptors? Answering these questions will help to resolve the biophysical nature of the center of the NPC translocation channel in the context of the physiological environment.
Supplementary Material
Acknowledgments
This study was supported by grant R01 GM051219 from the National Institutes of Health (S.R.W.).
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Aitchison, J. D., G. Blobel, and M. P. Rout. 1996. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science 274:624-627. [DOI] [PubMed] [Google Scholar]
- 2.Akey, C. W., and M. Radermacher. 1993. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122:1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alber, F., S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2007. The molecular architecture of the nuclear pore complex. Nature 450:695-701. [DOI] [PubMed] [Google Scholar]
- 4.Allen, N. P., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268-29274. [DOI] [PubMed] [Google Scholar]
- 5.Allen, N. P., S. S. Patel, L. Huang, R. J. Chalkley, A. Burlingame, M. Lutzmann, E. C. Hurt, and M. Rexach. 2002. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell. Proteomics 1:930-946. [DOI] [PubMed] [Google Scholar]
- 6.Andrade, M. A., C. Petosa, S. I. O'Donoghue, C. W. Muller, and P. Bork. 2001. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309:1-18. [DOI] [PubMed] [Google Scholar]
- 7.Aris, J. P., and G. Blobel. 1989. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J. Cell Biol. 108:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailer, S. M., S. Siniossoglou, A. Podtelejnikov, A. Hellwig, M. Mann, and E. Hurt. 1998. Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J. 17:1107-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayliss, R., H. M. Kent, A. H. Corbett, and M. Stewart. 2000. Crystallization and initial X-ray diffraction characterization of complexes of FxFG nucleoporin repeats with nuclear transport factors. J. Struct. Biol. 131:240-247. [DOI] [PubMed] [Google Scholar]
- 10.Bayliss, R., S. W. Leung, R. P. Baker, B. B. Quimby, A. H. Corbett, and M. Stewart. 2002. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21:2843-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell 102:99-108. [DOI] [PubMed] [Google Scholar]
- 12.Bayliss, R., T. Littlewood, L. A. Strawn, S. R. Wente, and M. Stewart. 2002. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 277:50597-50606. [DOI] [PubMed] [Google Scholar]
- 13.Bayliss, R., K. Ribbeck, D. Akin, H. M. Kent, C. M. Feldherr, D. Gorlich, and M. Stewart. 1999. Interaction between NTF2 and xFxFG-containing nucleoporins is required to mediate nuclear import of RanGDP. J. Mol. Biol. 293:579-593. [DOI] [PubMed] [Google Scholar]
- 14.Beck, M., F. Forster, M. Ecke, J. M. Plitzko, F. Melchior, G. Gerisch, W. Baumeister, and O. Medalia. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306:1387-1390. [DOI] [PubMed] [Google Scholar]
- 15.Beck, M., V. Lucic, F. Forster, W. Baumeister, and O. Medalia. 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 449:611-615. [DOI] [PubMed] [Google Scholar]
- 16.Bednenko, J., G. Cingolani, and L. Gerace. 2003. Importin beta contains a COOH-terminal nucleoporin binding region important for nuclear transport. J. Cell Biol. 162:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bickel, T., and R. Bruinsma. 2002. The nuclear pore complex mystery and anomalous diffusion in reversible gels. Biophys. J. 83:3079-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blevins, M. B., A. M. Smith, E. M. Phillips, and M. A. Powers. 2003. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J. Biol. Chem. 278:20979-20988. [DOI] [PubMed] [Google Scholar]
- 20.Bradatsch, B., J. Katahira, E. Kowalinski, G. Bange, W. Yao, T. Sekimoto, V. Baumgartel, G. Boese, J. Bassler, K. Wild, R. Peters, Y. Yoneda, I. Sinning, and E. Hurt. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell 27:767-779. [DOI] [PubMed] [Google Scholar]
- 21.Braun, I. C., A. Herold, M. Rode, E. Conti, and E. Izaurralde. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536-20543. [DOI] [PubMed] [Google Scholar]
- 22.Braun, I. C., A. Herold, M. Rode, and E. Izaurralde. 2002. Nuclear export of mRNA by TAP/NXF1 requires two nucleoporin-binding sites but not p15. Mol. Cell. Biol. 22:5405-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brohawn, S. G., N. C. Leksa, E. D. Spear, K. R. Rajashankar, and T. U. Schwartz. 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 25.Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703-715. [DOI] [PubMed] [Google Scholar]
- 26.Cingolani, G., J. Bednenko, M. T. Gillespie, and L. Gerace. 2002. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell 10:1345-1353. [DOI] [PubMed] [Google Scholar]
- 27.Cingolani, G., C. Petosa, K. Weis, and C. W. Muller. 1999. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature 399:221-229. [DOI] [PubMed] [Google Scholar]
- 28.Clouse, K. N., M. J. Luo, Z. Zhou, and R. Reed. 2001. A Ran-independent pathway for export of spliced mRNA. Nat. Cell Biol. 3:97-99. [DOI] [PubMed] [Google Scholar]
- 29.Conti, E., C. W. Muller, and M. Stewart. 2006. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 16:237-244. [DOI] [PubMed] [Google Scholar]
- 30.Cook, A., F. Bono, M. Jinek, and E. Conti. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76:647-671. [DOI] [PubMed] [Google Scholar]
- 31.Cronshaw, J. M., A. N. Krutchinsky, W. Zhang, B. T. Chait, and M. J. Matunis. 2002. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cullen, B. R. 2003. Nuclear RNA export. J. Cell Sci. 116:587-597. [DOI] [PubMed] [Google Scholar]
- 33.Cushman, I., T. Palzkill, and M. S. Moore. 2006. Using peptide arrays to define nuclear carrier binding sites on nucleoporins. Methods 39:329-341. [DOI] [PubMed] [Google Scholar]
- 34.Damelin, M., and P. A. Silver. 2000. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell 5:133-140. [DOI] [PubMed] [Google Scholar]
- 35.Daneholt, B. 2001. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. USA 98:7012-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis, L. I., and G. Blobel. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84:7552-7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis, L. I., and G. R. Fink. 1990. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell 61:965-978. [DOI] [PubMed] [Google Scholar]
- 38.Degrasse, J. A., K. N. Dubois, D. Devos, T. N. Siegel, A. Sali, M. C. Field, M. P. Rout, and B. T. Chait. 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8:2119-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denning, D., B. Mykytka, N. P. Allen, L. Huang, B. Al, and M. Rexach. 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154:937-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denning, D. P., S. S. Patel, V. Uversky, A. L. Fink, and M. Rexach. 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 100:2450-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Denning, D. P., and M. F. Rexach. 2007. Rapid evolution exposes the boundaries of domain structure and function in natively unfolded FG nucleoporins. Mol. Cell. Proteomics 6:272-282. [DOI] [PubMed] [Google Scholar]
- 41.Denning, D. P., V. Uversky, S. S. Patel, A. L. Fink, and M. Rexach. 2002. The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J. Biol. Chem. 277:33447-33455. [DOI] [PubMed] [Google Scholar]
- 42.De Souza, C. P., A. H. Osmani, S. B. Hashmi, and S. A. Osmani. 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14:1973-1984. [DOI] [PubMed] [Google Scholar]
- 43.Devos, D., S. Dokudovskaya, F. Alber, R. Williams, B. T. Chait, A. Sali, and M. P. Rout. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devos, D., S. Dokudovskaya, R. Williams, F. Alber, N. Eswar, B. T. Chait, M. P. Rout, and A. Sali. 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. USA 103:2172-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dilworth, D. J., A. Suprapto, J. C. Padovan, B. T. Chait, R. W. Wozniak, M. P. Rout, and J. D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dokudovskaya, S., R. Williams, D. Devos, A. Sali, B. T. Chait, and M. P. Rout. 2006. Protease accessibility laddering: a proteomic tool for probing protein structure. Structure 14:653-660. [DOI] [PubMed] [Google Scholar]
- 47.Doye, V., and E. Hurt. 1997. From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol. 9:401-411. [DOI] [PubMed] [Google Scholar]
- 48.Dultz, E., E. Zanin, C. Wurzenberger, M. Braun, G. Rabut, L. Sironi, and J. Ellenberg. 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180:857-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunker, A. K., J. D. Lawson, C. J. Brown, R. M. Williams, P. Romero, J. S. Oh, C. J. Oldfield, A. M. Campen, C. M. Ratliff, K. W. Hipps, J. Ausio, M. S. Nissen, R. Reeves, C. Kang, C. R. Kissinger, R. W. Bailey, M. D. Griswold, W. Chiu, E. C. Garner, and Z. Obradovic. 2001. Intrinsically disordered protein. J. Mol. Graph Model. 19:26-59. [DOI] [PubMed] [Google Scholar]
- 50.Enninga, J., D. E. Levy, G. Blobel, and B. M. Fontoura. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295:1523-1525. [DOI] [PubMed] [Google Scholar]
- 51.Fagotto, F., U. Gluck, and B. M. Gumbiner. 1998. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr. Biol. 8:181-190. [DOI] [PubMed] [Google Scholar]
- 52.Fahrenkrog, B., E. C. Hurt, U. Aebi, and N. Pante. 1998. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol. 143:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fahrenkrog, B., B. Maco, A. M. Fager, J. Koser, U. Sauder, K. S. Ullman, and U. Aebi. 2002. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 140:254-267. [DOI] [PubMed] [Google Scholar]
- 54.Faria, P. A., P. Chakraborty, A. Levay, G. N. Barber, H. J. Ezelle, J. Enninga, C. Arana, J. van Deursen, and B. M. Fontoura. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17:93-102. [DOI] [PubMed] [Google Scholar]
- 55.Feldherr, C. M., and D. Akin. 1997. The location of the transport gate in the nuclear pore complex. J. Cell Sci. 110:3065-3070. [DOI] [PubMed] [Google Scholar]
- 56.Feldherr, C. M., E. Kallenbach, and N. Schultz. 1984. Movement of a karyophilic protein through the nuclear pores of oocytes. J. Cell Biol. 99:2216-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finlay, D. R., and D. J. Forbes. 1990. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell 60:17-29. [DOI] [PubMed] [Google Scholar]
- 58.Fontoura, B. M., G. Blobel, and M. J. Matunis. 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144:1097-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frey, S., and D. Gorlich. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130:512-523. [DOI] [PubMed] [Google Scholar]
- 60.Frey, S., R. P. Richter, and D. Gorlich. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314:815-817. [DOI] [PubMed] [Google Scholar]
- 61.Fribourg, S., I. C. Braun, E. Izaurralde, and E. Conti. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 8:645-656. [DOI] [PubMed] [Google Scholar]
- 62.Fribourg, S., and E. Conti. 2003. Structural similarity in the absence of sequence homology of the messenger RNA export factors Mtr2 and p15. EMBO Rep. 4:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fried, H., and U. Kutay. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilchrist, D., B. Mykytka, and M. Rexach. 2002. Accelerating the rate of disassembly of karyopherin-cargo complexes. J. Biol. Chem. 277:18161-18172. [DOI] [PubMed] [Google Scholar]
- 65.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 66.Gorlich, D., and I. W. Mattaj. 1996. Nucleocytoplasmic transport. Science 271:1513-1518. [DOI] [PubMed] [Google Scholar]
- 67.Gorlich, D., M. J. Seewald, and K. Ribbeck. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22:1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grant, R. P., E. Hurt, D. Neuhaus, and M. Stewart. 2002. Structure of the C-terminal FG-nucleoporin binding domain of Tap/NXF1. Nat. Struct. Biol. 9:247-251. [DOI] [PubMed] [Google Scholar]
- 69.Grant, R. P., D. Neuhaus, and M. Stewart. 2003. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J. Mol. Biol. 326:849-858. [DOI] [PubMed] [Google Scholar]
- 70.Greber, U. F., and L. Gerace. 1992. Nuclear protein import is inhibited by an antibody to a lumenal epitope of a nuclear pore complex glycoprotein. J. Cell Biol. 116:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Griffis, E. R., N. Altan, J. Lippincott-Schwartz, and M. A. Powers. 2002. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13:1282-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gustin, K. E. 2003. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 95:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gustin, K. E., and P. Sarnow. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 76:8787-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harel, A., and D. J. Forbes. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319-330. [DOI] [PubMed] [Google Scholar]
- 76.Herold, A., T. Klymenko, and E. Izaurralde. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7:1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 77.Herold, A., M. Suyama, J. P. Rodrigues, I. C. Braun, U. Kutay, M. Carmo-Fonseca, P. Bork, and E. Izaurralde. 2000. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell. Biol. 20:8996-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hetzer, M. W., T. C. Walther, and I. W. Mattaj. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347-380. [DOI] [PubMed] [Google Scholar]
- 79.Hinshaw, J. E., B. O. Carragher, and R. A. Milligan. 1992. Architecture and design of the nuclear pore complex. Cell 69:1133-1141. [DOI] [PubMed] [Google Scholar]
- 80.Ho, A. K., T. X. Shen, K. J. Ryan, E. Kiseleva, M. A. Levy, T. D. Allen, and S. R. Wente. 2000. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol. 20:5736-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hodel, A. E., M. R. Hodel, E. R. Griffis, K. A. Hennig, G. A. Ratner, S. Xu, and M. A. Powers. 2002. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell 10:347-358. [DOI] [PubMed] [Google Scholar]
- 82.Hurt, E., K. Strasser, A. Segref, S. M. Bailer, N. Schlaich, C. Presutti, D. Tollervey, and R. Jansen. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 275:8361-8368. [DOI] [PubMed] [Google Scholar]
- 83.Isgro, T. A., and K. Schulten. 2005. Binding dynamics of isolated nucleoporin repeat regions to importin-beta. Structure 13:1869-1879. [DOI] [PubMed] [Google Scholar]
- 84.Isgro, T. A., and K. Schulten. 2007. Association of nuclear pore FG-repeat domains to NTF2 import and export complexes. J. Mol. Biol. 366:330-345. [DOI] [PubMed] [Google Scholar]
- 85.Isgro, T. A., and K. Schulten. 2007. Cse1p-binding dynamics reveal a binding pattern for FG-repeat nucleoporins on transport receptors. Structure 15:977-991. [DOI] [PubMed] [Google Scholar]
- 86.Iwamoto, M., C. Mori, T. Kojidani, F. Bunai, T. Hori, T. Fukagawa, Y. Hiraoka, and T. Haraguchi. 2009. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr. Biol. 19:843-847. [DOI] [PubMed] [Google Scholar]
- 87.Jinek, M., J. Rehwinkel, B. D. Lazarus, E. Izaurralde, J. A. Hanover, and E. Conti. 2004. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11:1001-1007. [DOI] [PubMed] [Google Scholar]
- 88.Johnson, A. W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27:580-585. [DOI] [PubMed] [Google Scholar]
- 89.Jovanovic-Talisman, T., J. Tetenbaum-Novatt, A. S. McKenney, A. Zilman, R. Peters, M. P. Rout, and B. T. Chait. 2009. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457:1023-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kahms, M., P. Lehrich, J. Huve, N. Sanetra, and R. Peters. 2009. Binding site distribution of nuclear transport receptors and transport complexes in single nuclear pore complexes. Traffic 10:1228-1242. [DOI] [PubMed] [Google Scholar]
- 91.Katahira, J., K. Straesser, T. Saiwaki, Y. Yoneda, and E. Hurt. 2002. Complex formation between Tap and p15 affects binding to FG-repeat nucleoporins and nucleocytoplasmic shuttling. J. Biol. Chem. 277:9242-9246. [DOI] [PubMed] [Google Scholar]
- 92.Katahira, J., K. Strasser, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kau, T. R., J. C. Way, and P. A. Silver. 2004. Nuclear transport and cancer: from mechanism to intervention. Nat. Rev. Cancer 4:106-117. [DOI] [PubMed] [Google Scholar]
- 94.Kenna, M. A., J. G. Petranka, J. L. Reilly, and L. I. Davis. 1996. Yeast Nle3p/Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex. Mol. Cell. Biol. 16:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.King, M. C., C. P. Lusk, and G. Blobel. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442:1003-1007. [DOI] [PubMed] [Google Scholar]
- 96.Kiseleva, E., T. D. Allen, S. Rutherford, M. Bucci, S. R. Wente, and M. W. Goldberg. 2004. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 145:272-288. [DOI] [PubMed] [Google Scholar]
- 97.Kohler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell. Biol. 8:761-773. [DOI] [PubMed] [Google Scholar]
- 98.Lane, C. M., I. Cushman, and M. S. Moore. 2000. Selective disruption of nuclear import by a functional mutant nuclear transport carrier. J. Cell Biol. 151:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee, S. J., N. Imamoto, H. Sakai, A. Nakagawa, S. Kose, M. Koike, M. Yamamoto, T. Kumasaka, Y. Yoneda, and T. Tsukihara. 2000. The adoption of a twisted structure of importin-beta is essential for the protein-protein interaction required for nuclear transport. J. Mol. Biol. 302:251-264. [DOI] [PubMed] [Google Scholar]
- 100.Lee, S. J., T. Sekimoto, E. Yamashita, E. Nagoshi, A. Nakagawa, N. Imamoto, M. Yoshimura, H. Sakai, K. T. Chong, T. Tsukihara, and Y. Yoneda. 2003. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science 302:1571-1575. [DOI] [PubMed] [Google Scholar]
- 101.Lee, S. J., T. Sekimoto, E. Yamashita, E. Nagoshi, A. Nakagawa, H. Tanaka, Y. Yoneda, and T. Tsukihara. 2003. Crystallization and preliminary crystallographic analysis of the importin-beta-SREBP-2 complex. Acta Crystallogr. D Biol. Crystallogr 59:1866-1868. [DOI] [PubMed] [Google Scholar]
- 102.Lim, R. Y., B. Fahrenkrog, J. Koser, K. Schwarz-Herion, J. Deng, and U. Aebi. 2007. Nanomechanical basis of selective gating by the nuclear pore complex. Science 318:640-643. [DOI] [PubMed] [Google Scholar]
- 103.Lim, R. Y., N. P. Huang, J. Koser, J. Deng, K. H. Lau, K. Schwarz-Herion, B. Fahrenkrog, and U. Aebi. 2006. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc. Natl. Acad. Sci. USA 103:9512-9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lim, R. Y., J. Koser, N. P. Huang, K. Schwarz-Herion, and U. Aebi. 2007. Nanomechanical interactions of phenylalanine-glycine nucleoporins studied by single molecule force-volume spectroscopy. J. Struct. Biol. 159:277-289. [DOI] [PubMed] [Google Scholar]
- 105.Lindsay, M. E., K. Plafker, A. E. Smith, B. E. Clurman, and I. G. Macara. 2002. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell 110:349-360. [DOI] [PubMed] [Google Scholar]
- 106.Liu, S. M., and M. Stewart. 2005. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J. Mol. Biol. 349:515-525. [DOI] [PubMed] [Google Scholar]
- 107.Lusk, C. P., T. Makhnevych, M. Marelli, J. D. Aitchison, and R. W. Wozniak. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Malone, C. D., K. A. Falkowska, A. Y. Li, S. E. Galanti, R. C. Kanuru, E. G. LaMont, K. C. Mazzarella, A. J. Micev, M. M. Osman, N. K. Piotrowski, J. W. Suszko, A. C. Timm, M. M. Xu, L. Liu, and D. L. Chalker. 2008. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot. Cell 7:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mans, B. J., V. Anantharaman, L. Aravind, and E. V. Koonin. 2004. Comparative genomics, evolution, and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3:1612-1637. [DOI] [PubMed] [Google Scholar]
- 111.Marelli, M., J. D. Aitchison, and R. W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Maul, G. G., H. M. Maul, J. E. Scogna, M. W. Lieberman, G. S. Stein, B. Y. Hsu, and T. W. Borun. 1972. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 55:433-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miller, M. W., M. R. Caracciolo, W. K. Berlin, and J. A. Hanover. 1999. Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 367:51-60. [DOI] [PubMed] [Google Scholar]
- 114.Mosammaparast, N., and L. F. Pemberton. 2004. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 14:547-556. [DOI] [PubMed] [Google Scholar]
- 115.Nachury, M. V., and K. Weis. 1999. The direction of transport through the nuclear pore can be inverted. Proc. Natl. Acad. Sci. USA 96:9622-9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nehrbass, U., M. P. Rout, S. Maguire, G. Blobel, and R. W. Wozniak. 1996. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J. Cell Biol. 133:1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Olsson, M., S. Scheele, and P. Ekblom. 2004. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 292:359-370. [DOI] [PubMed] [Google Scholar]
- 118.Osmani, A. H., J. Davies, H. L. Liu, A. Nile, and S. A. Osmani. 2006. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17:4946-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Paine, P. L., L. C. Moore, and S. B. Horowitz. 1975. Nuclear envelope permeability. Nature 254:109-114. [DOI] [PubMed] [Google Scholar]
- 120.Pante, N., and M. Kann. 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell 13:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patel, S. S., B. J. Belmont, J. M. Sante, and M. F. Rexach. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129:83-96. [DOI] [PubMed] [Google Scholar]
- 122.Patel, S. S., and M. F. Rexach. 2008. Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol. Cell. Proteomics 7:121-131. [DOI] [PubMed] [Google Scholar]
- 123.Paulillo, S. M., E. M. Phillips, J. Koser, U. Sauder, K. S. Ullman, M. A. Powers, and B. Fahrenkrog. 2005. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J. Mol. Biol. 351:784-798. [DOI] [PubMed] [Google Scholar]
- 124.Pemberton, L. F., and B. M. Paschal. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187-198. [DOI] [PubMed] [Google Scholar]
- 125.Peters, R. 2005. Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic 6:421-427. [DOI] [PubMed] [Google Scholar]
- 126.Peters, R. 2009. Translocation through the nuclear pore: Kaps pave the way. Bioessays 31:466-477. [DOI] [PubMed] [Google Scholar]
- 127.Poon, I. K., and D. A. Jans. 2005. Regulation of nuclear transport: central role in development and transformation? Traffic 6:173-186. [DOI] [PubMed] [Google Scholar]
- 128.Powers, M. A., D. J. Forbes, J. E. Dahlberg, and E. Lund. 1997. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 136:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pyhtila, B., and M. Rexach. 2003. A gradient of affinity for the karyopherin Kap95p along the yeast nuclear pore complex. J. Biol. Chem. 278:42699-42709. [DOI] [PubMed] [Google Scholar]
- 130.Quimby, B. B., S. W. Leung, R. Bayliss, M. T. Harreman, G. Thirumala, M. Stewart, and A. H. Corbett. 2001. Functional analysis of the hydrophobic patch on nuclear transport factor 2 involved in interactions with the nuclear pore in vivo. J. Biol. Chem. 276:38820-38829. [DOI] [PubMed] [Google Scholar]
- 131.Rabut, G., P. Lenart, and J. Ellenberg. 2004. Dynamics of nuclear pore complex organization through the cell cycle. Curr. Opin. Cell Biol. 16:314-321. [DOI] [PubMed] [Google Scholar]
- 132.Ratner, G. A., A. E. Hodel, and M. A. Powers. 2007. Molecular determinants of binding between Gly-Leu-Phe-Gly nucleoporins and the nuclear pore complex. J. Biol. Chem. 282:33968-33976. [DOI] [PubMed] [Google Scholar]
- 133.Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ribbeck, K., and D. Gorlich. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rodriguez, M. S., C. Dargemont, and F. Stutz. 2004. Nuclear export of RNA. Biol. Cell 96:639-655. [DOI] [PubMed] [Google Scholar]
- 136.Rosenblum, J. S., and G. Blobel. 1999. Autoproteolysis in nucleoporin biogenesis. Proc. Natl. Acad. Sci. USA 96:11370-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 138.Rout, M. P., J. D. Aitchison, M. O. Magnasco, and B. T. Chait. 2003. Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 13:622-628. [DOI] [PubMed] [Google Scholar]
- 139.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rout, M. P., G. Blobel, and J. D. Aitchison. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell 89:715-725. [DOI] [PubMed] [Google Scholar]
- 141.Rout, M. P., and S. R. Wente. 1994. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 4:357-365. [DOI] [PubMed] [Google Scholar]
- 142.Ryan, K. J., and S. R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361-371. [DOI] [PubMed] [Google Scholar]
- 143.Santos-Rosa, H., H. Moreno, G. Simos, A. Segref, B. Fahrenkrog, N. Pante, and E. Hurt. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell. Biol. 18:6826-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzveig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. USA 104:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Seedorf, M., M. Damelin, J. Kahana, T. Taura, and P. A. Silver. 1999. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol. Cell. Biol. 19:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Segref, A., K. Sharma, V. Doye, A. Hellwig, J. Huber, R. Luhrmann, and E. Hurt. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16:3256-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Senay, C., P. Ferrari, C. Rocher, K. J. Rieger, J. Winter, D. Platel, and Y. Bourne. 2003. The Mtr2-Mex67 NTF2-like domain complex: structural insights into a dual role of Mtr2 for yeast nuclear export. J. Biol. Chem. 278:48395-48403. [DOI] [PubMed] [Google Scholar]
- 148.Shulga, N., and D. S. Goldfarb. 2003. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol. Cell. Biol. 23:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shulga, N., N. Mosammaparast, R. Wozniak, and D. S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Siebrasse, J. P., and R. Peters. 2002. Rapid translocation of NTF2 through the nuclear pore of isolated nuclei and nuclear envelopes. EMBO Rep. 3:887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sistla, S., J. V. Pang, C. X. Wang, and D. Balasundaram. 2007. Multiple conserved domains of the nucleoporin Nup124p and its orthologs Nup1p and Nup153 are critical for nuclear import and activity of the fission yeast Tf1 retrotransposon. Mol. Biol. Cell 18:3692-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Smith, A. E., B. M. Slepchenko, J. C. Schaff, L. M. Loew, and I. G. Macara. 2002. Systems analysis of Ran transport. Science 295:488-491. [DOI] [PubMed] [Google Scholar]
- 153.Smitherman, M., K. Lee, J. Swanger, R. Kapur, and B. E. Clurman. 2000. Characterization and targeted disruption of murine Nup50, a p27Kip1-interacting component of the nuclear pore complex. Mol. Cell. Biol. 20:5631-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Soderqvist, H., and E. Hallberg. 1994. The large C-terminal region of the integral pore membrane protein, POM121, is facing the nuclear pore complex. Eur. J. Cell Biol. 64:186-191. [PubMed] [Google Scholar]
- 155.Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8:195-208. [DOI] [PubMed] [Google Scholar]
- 156.Strasser, K., J. Bassler, and E. Hurt. 2000. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J. Cell Biol. 150:695-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Strawn, L. A., T. Shen, N. Shulga, D. S. Goldfarb, and S. R. Wente. 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6:197-206. [DOI] [PubMed] [Google Scholar]
- 158.Strawn, L. A., T. Shen, and S. R. Wente. 2001. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J. Biol. Chem. 276:6445-6452. [DOI] [PubMed] [Google Scholar]
- 159.Suntharalingam, M., and S. R. Wente. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell 4:775-789. [DOI] [PubMed] [Google Scholar]
- 160.Tan, W., A. S. Zolotukhin, J. Bear, D. J. Patenaude, and B. K. Felber. 2000. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA 6:1762-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Teixeira, M. T., S. Siniossoglou, S. Podtelejnikov, J. C. Benichou, M. Mann, B. Dujon, E. Hurt, and E. Fabre. 1997. Two functionally distinct domains generated by in vivo cleavage of Nup145p: a novel biogenesis pathway for nucleoporins. EMBO J. 16:5086-5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Terry, L. J., and S. R. Wente. 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178:1121-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Timney, B. L., J. Tetenbaum-Novatt, D. S. Agate, R. Williams, W. Zhang, B. T. Chait, and M. P. Rout. 2006. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 175:579-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tompa, P. 2005. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 579:3346-3354. [DOI] [PubMed] [Google Scholar]
- 165.Tran, E. J., and S. R. Wente. 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125:1041-1053. [DOI] [PubMed] [Google Scholar]
- 166.Ullman, K. S., S. Shah, M. A. Powers, and D. J. Forbes. 1999. The nucleoporin Nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell 10:649-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Uv, A. E., P. Roth, N. Xylourgidis, A. Wickberg, R. Cantera, and C. Samakovlis. 2000. members only encodes a Drosophila nucleoporin required for rel protein import and immune response activation. Genes Dev. 14:1945-1957. [PMC free article] [PubMed] [Google Scholar]
- 168.Vasu, S., S. Shah, A. Orjalo, M. Park, W. H. Fischer, and D. J. Forbes. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wagstaff, K. M., and D. A. Jans. 2009. Importins and beyond: non-conventional nuclear transport mechanisms. Traffic 10:1188-1198. [DOI] [PubMed] [Google Scholar]
- 170.Walther, T. C., H. S. Pickersgill, V. C. Cordes, M. W. Goldberg, T. D. Allen, I. W. Mattaj, and M. Fornerod. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158:63-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Weirich, C. S., J. P. Erzberger, J. M. Berger, and K. Weis. 2004. The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16:749-760. [DOI] [PubMed] [Google Scholar]
- 172.Wennerberg, K., K. L. Rossman, and C. J. Der. 2005. The Ras superfamily at a glance. J. Cell Sci. 118:843-846. [DOI] [PubMed] [Google Scholar]
- 173.Wente, S. R., and G. Blobel. 1994. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J. Cell Biol. 125:955-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wente, S. R., M. P. Rout, and G. Blobel. 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119:705-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wiegand, H. L., G. A. Coburn, Y. Zeng, Y. Kang, H. P. Bogerd, and B. R. Cullen. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Winey, M., D. Yarar, T. H. Giddings, Jr., and D. N. Mastronarde. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell 8:2119- 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wu, X., L. H. Kasper, R. T. Mantcheva, G. T. Mantchev, M. J. Springett, and J. M. van Deursen. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl. Acad. Sci. USA 98:3191-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wurtz, T., A. Lonnroth, L. Ovchinnikov, U. Skoglund, and B. Daneholt. 1990. Isolation and initial characterization of a specific premessenger ribonucleoprotein particle. Proc. Natl. Acad. Sci. USA 87:831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xu, S., and M. A. Powers. 2009. Nuclear pore proteins and cancer. Semin. Cell Dev. Biol. 20:620-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang, W., J. Gelles, and S. M. Musser. 2004. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA 101:12887-12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zeitler, B., and K. Weis. 2004. The FG-repeat asymmetry of the nuclear pore complex is dispensible for bulk nucleocytoplasmic transport in vivo. J. Cell Biol. 167:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.