Abstract
Candida albicans Hwp1, Hwp2, and Rbt1 are related cell wall proteins expressed during the programs of sexual differentiation and filamentous growth. In this study, we compare strains lacking either single factors or a combination of these genes, and we demonstrate distinct but overlapping roles in mating and biofilm formation.
The fungus Candida albicans is often a harmless commensal in humans yet has the capacity to cause life-threatening infections, particularly in the immunocompromised host (27). The transition between commensal and pathogenic states is associated with morphological changes, among which the yeast-hypha switch is paramount. During this transition, cells switch from growing as budding yeast cells to growing as filamentous hyphae. Importantly, many genes associated with this transition are essential for virulence, including both cell wall proteins and secreted enzymes (7, 18, 33, 35).
Originally classified as an obligately asexual organism, a sexual (or parasexual) program has recently been uncovered in C. albicans (2, 11, 20, 31). The mating cycle is unique in that it is regulated by phenotypic switching; a and α cells only undergo efficient mating if they switch from the common “white” phase to the alternative “opaque” phase (19, 21). Furthermore, transcriptional profiling analyses have revealed an unexpected overlap between genes induced during mating (of opaque cells) and those induced during filamentation (in white cells) (4, 36). One hypothesis for this overlap is that genes originally involved in mating were co-opted during evolution for adherence and invasion of the host (4). It is therefore likely that studying the role of these genes in mating will also provide insight into their functions during pathogenesis.
Hwp1, Hwp2, and Rbt1 are three hypha-specific cell wall proteins that are also upregulated during mating of opaque cells (4, 5, 14, 32). Hwp1 is a well-characterized adhesin required for covalent attachment to host epithelial cells and virulence (32), as well as biofilm formation (5). Interestingly, expression and localization of Hwp1 during mating has been reported as being mating type specific; opaque a cells, but not α cells, were shown to express Hwp1 on their cell surface (8). This is reminiscent of agglutinin-type activity in Saccharomyces cerevisiae, where mating-type-specific proteins promote cell-cell adhesion between a and α cells (16). In this study, we examined the role of Hwp1 and the related proteins Hwp2 and Rbt1 in the mating program of C. albicans and extended this analysis to in vitro models of biofilm formation. Our results indicate the importance of these proteins in both mating and biofilm formation and, in particular, their nonredundant roles in these processes.
Hwp1 is expressed in both a and α cells of SC5314 during mating.
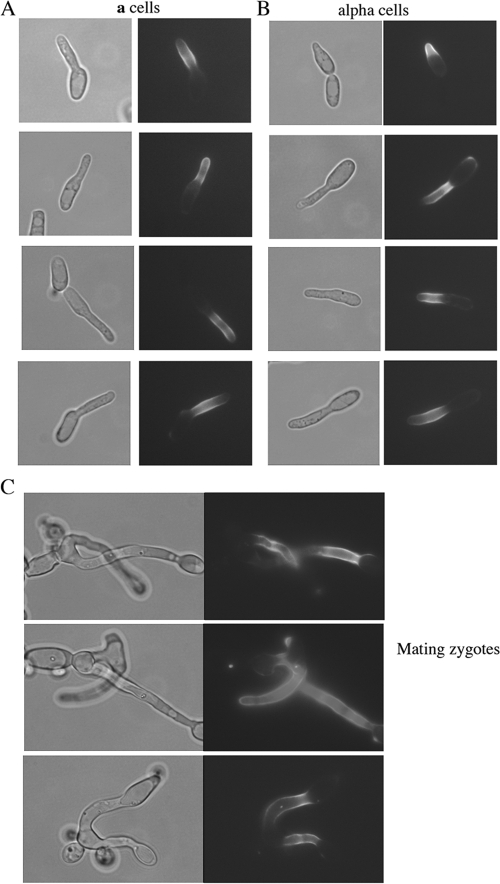
Previous studies indicated that Hwp1 is expressed on the surface of conjugation tubes produced by opaque a cells, but not α cells, during mating (8). However, these experiments were performed using nonisogenic a and α clinical isolates of C. albicans. Furthermore, transcriptional profiling studies using derivatives of SC5314, the standard laboratory strain of C. albicans, suggested that the HWP1 gene was expressed in both cell types during mating (34). To establish the pattern of Hwp1 expression and localization in a and α strains of SC5314, Hwp1-green fluorescent protein (GFP)-Hwp1 fusion constructs were introduced into both cell types. In all cases, isogenic a and α strains were obtained by growth of C. albicans strains on sorbose medium, which selects for loss of one copy of chromosome 5 (containing the MTL locus), followed by reduplication of the remaining copy of chromosome 5 during growth on yeast extract-peptone-dextrose (12). The strains used in this study are listed in Table 1, and a list of the oligonucleotides is provided in Table 2. As shown in Fig. 1, Hwp1 protein was observed on the cell surface of both a and α cells undergoing mating and was also detectable in both halves of the conjugation bridge in zygotes (Fig. 1). Thus, in the SC5314 background, isogenic a and α cells express Hwp1 during mating, and the protein localizes to the cell surface of both of these cell types.
TABLE 1.
C. albicans strains used in this studya
| Strain (white) | Strain (opaque) | Genotype | Mating type | Reference |
|---|---|---|---|---|
| RBY 1040 | ura3::imm434/ura3::imm434 hwp1::HWP1-GFP-HWP1/HWP1 | a/a | This study | |
| RBY 1042 | ura3::imm434/ura3::imm434 hwp1::HWP1-GFP-HWP1/HWP1 | a/a | This study | |
| RBY 1045 | ura3::imm434/ura3::imm434 hwp1::HWP1-GFP-HWP1/HWP1 | α/α | This study | |
| RBY 1046 | ura3::imm434/ura3::imm434 hwp1::HWP1-GFP-HWP1/HWP1 | α/α | This study | |
| RBY 1132 | leu2/leu2 his1/his1 arg4/arg4 | a/a | This study | |
| RBY 1134 | leu2/leu2 his1/his1 arg4/arg4 | α/α | This study | |
| RZY 48 | RBY 1118 | leu2/leu2 his1/his1 | a/a | 30 |
| RZY 50 | RBY 1119 | leu2/leu2 his1/his1 | α/α | 30 |
| RBY 1175 | RBY 1179 | arg4/arg4 | a/a | 30 |
| RBY 1176 | RBY 1180 | arg4/arg4 | α/α | 30 |
| CAY 168 | CAY 200 | leu2/leu2 his1/his1 arg4/arg4 hwp1::LEU2/hwp1::HIS1 | a/a | This study |
| CAY 169 | CAY 210 | leu2/leu2 his1/his1 arg4/arg4 hwp2::LEU2/hwp2::HIS4 | a/a | This study |
| CAY 170 | CAY 201 | leu2/leu2 his1/his1 arg4/arg4 hwp2::LEU2/hwp2::HIS1 | a/a | This study |
| CAY 171 | CAY 211 | leu2/leu2 his1/his1 arg4/arg4 rbt1::LEU2/rbt1::HIS4 | a/a | This study |
| CAY 173 | CAY 212 | leu2/leu2 his1/his1 arg4/arg4 hwp1::HIS1/hwp1::ARG4 | α/α | This study |
| CAY 175 | CAY 179 | leu2/leu2 his1/his1 arg4/arg4 hwp2::HIS1/hwp2::ARG4 | α/α | This study |
| CAY 178 | CAY 209 | leu2/leu2 his1/his1 arg4/arg4 rbt1::HIS1/rbt1::ARG4 | α/α | This study |
| CAY 418 | CAY 560 | leu2/leu2 his1/his1 arg4/arg4 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 419 | CAY 561 | leu2/leu2 his1/his1 arg4/arg4 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 425 | CAY 562 | leu2/leu2 his1/his1 arg4/arg4 rbt1,hwp2::LEU2/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 426 | CAY 563 | leu2/leu2 his1/his1 arg4/arg4 rbt1,hwp2::LEU2/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 453 | CAY 476 | leu2/leu2 his1/his1 arg4/arg4 als3::LEU2/als3::HIS1 | a/a | This study |
| CAY 454 | CAY 477 | leu2/leu2 his1/his1 arg4/arg4 als3::LEU2/als3::HIS1 | a/a | This study |
| CAY 482 | CAY 494 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS | a/a | This study |
| CAY 483 | CAY 495 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 484 | CAY 505 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 485 | CAY 546 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 486 | CAY 506 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS | a/a | This study |
| CAY 487 | CAY487 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 497 | CAY 547 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 498 | CAY 548 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 503 | CAY 558 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 504 | CAY 529 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::LEU2/rbt1,hwp2::HIS1 | a/a | This study |
| CAY 488 | CAY 496 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::HIS1/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 489 | CAY 507 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::HIS1/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 499 | CAY 528 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::HIS1/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 500 | CAY 549 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::HIS1/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 501 | CAY 508 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::HIS1/rbt1,hwp2::ARG4 | α/α | This study |
| CAY 502 | CAY 509 | leu2/leu2 his1/his1 arg4/arg4 hwp1/hwp1::SAT1 rbt1,hwp2::HIS1/rbt1,hwp2::ARG4 | α/α | This study |
All strains are derivatives of SC5314 and, except for RBY1040, -1042, -1045, and -1046, are URA3/ura3::imm434 IRO1/iro1::imm434.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequencea |
|---|---|
| Hwp1-GFP oligo 1 | CGGTCAAAATAACCGGCTATTTTCAATTTCC |
| Hwp1-GFP oligo 2 | GTACCTATCACCTTTAATGTAGTAAAC |
| Hwp1 oligo1 | GCTATCAACTATGAACCGAAAACAG |
| Hwp1 oligo 3 | CACGGCGCGCCTAGCAGCGGGAAACCTCACCAATTGCTCCA |
| Hwp1 oligo 4 | GTCAGCGGCCGCATCCCTGCCGAAACTAAAAGCGAGTGAC |
| Hwp1 oligo 6 | CAAGGAATTCGGAAATTCTGACG |
| Rbt1 oligo 1 | GGCACAGCATCTTTGTTTCC |
| Rbt1 oligo 3 | CACGGCGCGCCTAGCAGCGGGGCAGTTGCAAATCTCA |
| Rbt1 oligo 4 | GTCAGCGGCCGCATCCCTGCCATCTGCACCAGGTACTGAAAC |
| Rbt1 oligo 6 | GAATTAGATCAAGAATGCAGC |
| Hwp2 oligo 1 | ATCTTCCGAGTTCCTGGAGA |
| Hwp2 oligo 3 | CACGGCGCGCCTAGCAGCGGAGCGAATGACTATCGGAGAC |
| Hwp2 oligo 4 | GTCAGCGGCCGCATCCCTGCATTGCTGGTGTCGCTGCCTT |
| Hwp2 oligo 6 | CTTAAAGCCGACAAGTGATAC |
| Hwp1 (−500) for | GGCGCCGGGCCCGGAATTCGGAAATTCTGACG |
| Hwp1 (0) rev | GCCGGCCTCGAGCTAAAAGCGAGTGACTATAGG |
| Hwp1 (+1900) for | GCCGGCCCGCGGGGTATTGCTGCATTCTTGATC |
| Hwp1 (+2300) rev | GGCGCCGAGCTCCTTCATGCGTCCAGAATAATG |
Sequence portions shown in bold indicate restriction sites.
FIG. 1.
Hwp1 protein is expressed on the cell surface of both a and α cells during mating. A C. albicans strain carrying an HWP1-GFP-URA3-GFP-HWP1 construct (gift of J. Berman, University of Minnesota) was PCR amplified using Hwp1-GFP oligos 1 and 2 (Table 2), and the resulting PCR product was integrated into both a and α derivatives of CAI4 (10), generating RBY1040 and -1042 and RBY1045 and -1046 strains, respectively. Selection of these strains on 5-fluoroorotic acid promoted recombination between the GFP repeats and loss of URA3, leaving an HWP1-GFP-HWP1 construct in the genome. The figure shows mixtures of opaque a and α cells cocultured in Spider medium for 6 h, in which either a cells (A) or α cells (B) carried the fluorescent construct. In both cases, Hwp1 localized specifically to the cell surface of polarized mating projections. (C) Pictures of mating zygotes in which a and α cells carrying the Hwp1-GFP construct were coincubated on Spider plates for 2 days to induce mating and conjugation. Hwp1 is seen localizing to the conjugation bridges originating from both a and α cells in the zygote.
Hwp1, Hwp2, and Rbt1 affect mating efficiency in C. albicans.
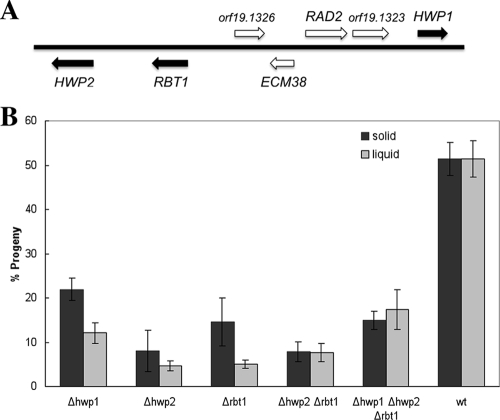
Hwp1 is part of a family of related cell surface factors that includes Hwp2 and Rbt1 (5, 6). The genes encoding these factors are clustered together in the genome, suggestive of a common ancestry (Fig. 2A). All three genes are induced in opaque cells in response to mating pheromones (4, 36), and quantitative PCR revealed a greater-than-100-fold increase in mRNA levels. Deletion strains lacking HWP1, HWP2, or RBT1 were constructed in a and α derivatives of SC5314, along with hwp2/hwp2 rbt1/rbt1 and hwp1/hwp1 hwp2/hwp2 rbt1/rbt1 strains. Quantitative mating assays were performed on the mutants by crossing strains with different auxotrophic markers (1, 3) and revealed that each of the mutant strains exhibited a significant decrease in mating efficiency (Fig. 2B). Since agglutinin function in S. cerevisiae mating is important only for cell-cell adhesion in liquid medium (17, 29), the frequency of C. albicans mating was analyzed in both liquid and solid media. In comparing single gene deletions, loss of HWP2 resulted in the largest decrease in mating efficiency, with only 4% of cells mating in liquid media and 8% on solid media, while 50% of wild-type cells underwent mating under these conditions (Fig. 2B). Loss of HWP1 or RBT1 also reduced the overall mating frequency in both liquid and solid media (Fig. 2B), although double and triple mutants did not show a further reduction in mating. Significantly, the difference between mating efficiencies in liquid and solid media were small in all of the crosses, suggesting that these proteins do not exhibit the classical agglutinin function, with which a much larger reduction in mating efficiency (orders of magnitude) would be expected in liquid culture (16, 17, 29). Alternatively, other cell surface factors may provide redundancy, and only by deletion of these additional, as-yet-unidentified factors will a large mating deficiency in liquid medium be observed.
FIG. 2.
Mating efficiency in strains lacking Hwp1, Hwp2, or Rbt1. (A) Schematic of the region on chromosome 4 containing HWP1, HWP2, and RBT1 genes. The three genes of interest are shown as filled boxes, and intervening genes are open boxes. (B) A quantitative mating assay was used to determine mating efficiency in both liquid and solid media, as previously described (30). Mutant strains were derived from RBY1132 (a/a) or RBY 1134 (α/α) and are listed in Table 1. A PCR fusion technique was used for gene deletion (26) with the oligonucleotides listed in Table 2. For each gene knockout, one flank of the gene was amplified with oligos 1 and 3 and the opposite flank was amplified with oligos 4 and 6. The resulting PCR products were then used to generate a targeting cassette containing the HIS1, LEU2, or ARG4 marker, as described previously (26). Following transformation, correct integration of each construct was confirmed by PCR across the DNA junctions at the site of integration. For the double deletion of RBT1/HWP2, a single construct was used to target both genes, based on their adjacent position in the genome. PCRs of the 5′ flank of RBT1 (oligos 1 and 3) and the 3′ flank of HWP2 (oligos 4 and 6) were combined in the targeting cassette and used to remove both genes simultaneously. For the triple mutant lacking HWP1, HWP2 and RBT1, HWP1 was deleted in the RBT1/HWP2 mutant background by using the SAT1 flipper construct (28). In this case, the 5′ and 3′ flanks of HWP1 were PCR amplified using HWP1 (−500) for/HWP1 (0) rev and HWP1 (+1900) for/HWP1 (+2300) rev, respectively, and cloned into the pSFS2A plasmid (28). The resulting construct was digested with ApaI and SacI and used to target HWP1. The standard practice for analyzing gene disruptions in C. albicans involves either comparing multiple independent disruptions or complementation by reintroducing a wild-type copy of the gene of interest. Due to the analysis of strains with multiple gene deletions, we chose the former approach, and multiple strains were therefore analyzed in this study (Table 2). Each of the mutant strains showed a statistically significant decrease (P < 0.05) in the number of mating products formed in both liquid and solid mating assays compared to the wild-type strain. Results are means ± standard errors of the mean of 2 to 12 experiments with each strain, and statistical analyses were performed using two-sample t tests. All P values are two tailed and are based on comparisons with the wild type.
Hwp1, Hwp2, and Rbt1 are required for efficient biofilm formation.
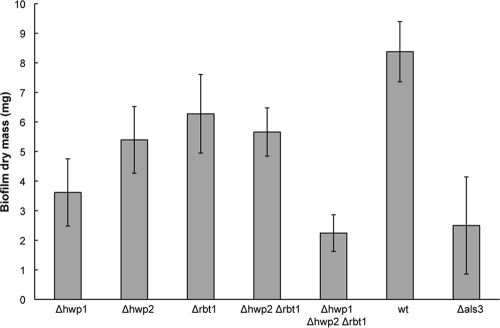
We also examined the role of the three cell surface proteins in biofilm assays. Biofilm formation is dependent on adhesins to mediate both the attachment of cells to the substrate surface as well as the adherence of cells to one another (23, 25). Assays were performed using white-phase cells, as these undergo hypha formation, during which expression of HWP1, HWP2, and RBT1 is upregulated (5, 13), and this mode of growth is necessary for efficient biofilm formation (23). We observed decreased biofilm formation in each of the mutants, with hwp1/hwp1 strains showing a greater decrease than either hwp2/hwp2 or rbt1/rbt1 mutants (Fig. 3). This is also in agreement with recent studies that implicated Hwp1 as having an important role in C. albicans biofilm formation both in vitro and in vivo (24, 25). The triple mutant strain hwp1/hwp1 hwp2/hwp2 rbt1/rbt1 showed the greatest defect in biofilm formation, suggesting that the three surface factors have related, but nonoverlapping, roles in biofilm formation (Fig. 3).
FIG. 3.
Defective biofilm formation in strains lacking Hwp1, Hwp2, or Rbt1. An in vitro biofilm assay was performed on silicone elastomer as previously described (24), except that biofilms were incubated for 60 h instead of 48 h. Biofilm efficacy was evaluated at the end of the experiment as the dry mass of the biofilm. All mutant strains formed statistically significantly smaller biofilms than the wild-type strain (P < 0.05), except for Δrbt1 and Δhwp2. Results are means ± standard errors of the mean of 4 to 16 experiments, and statistical analyses were performed using two-sample t tests. All P values are two tailed and are based on comparisons with the wild type. Mutants lacking als3 were also defective in biofilm formation, as reported elsewhere (22), and were therefore used as an additional control in these experiments.
Concluding remarks.
Our results demonstrate that the three cell surface proteins, Hwp1, Hwp2, and Rbt1, play important roles in both white and opaque phases of C. albicans biology. These factors are induced in the opaque phase by pheromones and enhance mating between a and α cells. However, they do not exhibit classic agglutinin functions, as the mating frequencies of mutants were comparable in both liquid and solid media. Hwp1 was also shown to be expressed on the surface of both a and α cells in isogenic derivatives of SC5314. This result contrasts with that reported in clinical isolates of C. albicans (8), and the fact that both mating types express this gene again distinguishes Hwp1 from the sexual agglutinins of S. cerevisiae (16). Perhaps C. albicans no longer requires classical agglutinins if conjugation occurs on the surface of the skin (15) or in a confined three-dimensional matrix (9). Hwp1, Hwp2, and Rbt1 also play a defined role in C. albicans for promoting biofilm formation by white-phase cells, as biofilm masses were diminished in each of the three mutant backgrounds. Hwp1 was the most important of the three, a result in keeping with recent studies (24, 25), yet strains lacking all three factors showed the greatest defect in biofilm formation. These results indicate that there is partial redundancy between these cell wall proteins and that loss of one factor can be compensated for, at least in part, by another. Our findings also suggest that further analysis of these factors in mating and adhesion will extend their prospective roles in biofilm formation and host pathogenesis.
Acknowledgments
We thank Judith Berman for the gift of the Hwp1-GFP construct used for localization studies, Clarissa Nobile for technical advice with biofilm assays, and Tricia Serio for use of the microscope. We also thank members of the Bennett lab for reading of the manuscript.
R.J.B. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Footnotes
Published ahead of print on 16 October 2009.
REFERENCES
- 1.Bennett, R. J., and A. D. Johnson. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22:2505-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. J., and A. D. Johnson. 2005. Mating in Candida albicans and the search for a sexual cycle. Annu. Rev. Microbiol. 59:233-255. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, R. J., and A. D. Johnson. 2006. The role of nutrient regulation and the Gpa2 protein in the mating pheromone response of C. albicans. Mol. Microbiol. 62:100-119. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, R. J., M. A. Uhl, M. G. Miller, and A. D. Johnson. 2003. Identification and characterization of a Candida albicans mating pheromone. Mol. Cell. Biol. 23:8189-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, G., M. D. Rasmussen, M. F. Lin, M. A. Santos, S. Sakthikumar, C. A. Munro, E. Rheinbay, M. Grabherr, A. Forche, J. L. Reedy, I. Agrafioti, M. B. Arnaud, S. Bates, A. J. Brown, S. Brunke, M. C. Costanzo, D. A. Fitzpatrick, P. W. de Groot, D. Harris, L. L. Hoyer, B. Hube, F. M. Klis, C. Kodira, N. Lennard, M. E. Logue, R. Martin, A. M. Neiman, E. Nikolaou, M. A. Quail, J. Quinn, M. C. Santos, F. F. Schmitzberger, G. Sherlock, P. Shah, K. A. Silverstein, M. S. Skrzypek, D. Soll, R. Staggs, I. Stansfield, M. P. Stumpf, P. E. Sudbery, T. Srikantha, Q. Zeng, J. Berman, M. Berriman, J. Heitman, N. A. Gow, M. C. Lorenz, B. W. Birren, M. Kellis, and C. A. Cuomo. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 8.Daniels, K. J., S. R. Lockhart, J. F. Staab, P. Sundstrom, and D. R. Soll. 2003. The adhesin Hwp1 and the first daughter cell localize to the a/a portion of the conjugation bridge during Candida albicans mating. Mol. Biol. Cell 14:4920-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniels, K. J., T. Srikantha, S. R. Lockhart, C. Pujol, and D. R. Soll. 2006. Opaque cells signal white cells to form biofilms in Candida albicans. EMBO J. 25:2240-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 12.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadosh, D., and A. D. Johnson. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lachke, S. A., S. R. Lockhart, K. J. Daniels, and D. R. Soll. 2003. Skin facilitates Candida albicans mating. Infect. Immun. 71:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipke, P. N., and J. Kurjan. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 56:180-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipke, P. N., D. Wojciechowicz, and J. Kurjan. 1989. AGα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol. Cell. Biol. 9:3155-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H. 2002. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int. J. Med. Microbiol. 292:299-311. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart, S. R., C. Pujol, K. J. Daniels, M. G. Miller, A. D. Johnson, M. A. Pfaller, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 21.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 22.Nobile, C. J., D. R. Andes, J. E. Nett, F. J. Smith, F. Yue, Q. T. Phan, J. E. Edwards, S. G. Filler, and A. P. Mitchell. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobile, C. J., and A. P. Mitchell. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell. Microbiol. 8:1382-1391. [DOI] [PubMed] [Google Scholar]
- 24.Nobile, C. J., J. E. Nett, D. R. Andes, and A. P. Mitchell. 2006. Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot. Cell 5:1604-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobile, C. J., H. A. Schneider, J. E. Nett, D. C. Sheppard, S. G. Filler, D. R. Andes, and A. P. Mitchell. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18:1017-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noble, S. M., and A. D. Johnson. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot. Cell 4:298-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 29.Roy, A., C. F. Lu, D. L. Marykwas, P. N. Lipke, and J. Kurjan. 1991. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein α-agglutinin. Mol. Cell. Biol. 11:4196-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaefer, D., P. Cote, M. Whiteway, and R. J. Bennett. 2007. Barrier activity in Candida albicans mediates pheromone degradation and promotes mating. Eukaryot. Cell 6:907-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soll, D. R. 2004. Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans. Bioessays 26:10-20. [DOI] [PubMed] [Google Scholar]
- 32.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 33.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 34.Tsong, A. E., B. B. Tuch, H. Li, and A. D. Johnson. 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443:415-420. [DOI] [PubMed] [Google Scholar]
- 35.Whiteway, M., and U. Oberholzer. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350-357. [DOI] [PubMed] [Google Scholar]
- 36.Zhao, R., K. J. Daniels, S. R. Lockhart, K. M. Yeater, L. L. Hoyer, and D. R. Soll. 2005. Unique aspects of gene expression during Candida albicans mating and possible G1 dependency. Eukaryot. Cell 4:1175-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]