Abstract
We have identified a new Plasmodium falciparum erythrocyte binding protein that appears to be located in the micronemes of the merozoite stage of the parasite and membrane linked through a glycosylphosphatidylinositol (GPI) anchor. The protein is designated GPI-anchored micronemal antigen (GAMA) and was identified by applying a set of selection criteria to identify previously uncharacterized merozoite proteins that may have a role in cell invasion. The protein is also present in the proteomes of the sporozoite and ookinete micronemes and is conserved throughout the genus. GAMA contains a novel domain that may be constrained by disulfide bonds and a predicted C-terminal hydrophobic sequence that is presumably replaced by the GPI. The protein is synthesized late during schizogony, processed into two fragments that are linked by a disulfide bond, and translocated to an apical location, which is probably the micronemes. In a proportion of free merozoites GAMA can also be detected on the parasite surface. Following erythrocyte invasion the bulk of the protein is shed in a soluble form, although a short C-terminal fragment may be carried into the newly invaded red blood cell. The protein was shown to bind reversibly to erythrocytes and therefore represents a new example of a host cell binding protein.
Malaria is a potentially fatal disease that still devastates poverty-stricken nations more than a century after the protozoan parasite Plasmodium was identified as its causative agent. An accurate estimation of mortality remains elusive, although recent estimates suggest that just over 2.5 billion people live at risk of infection by Plasmodium falciparum (22), the species of the parasite responsible for the vast majority of deaths. It is believed that there are in the range of 500 million clinical episodes of P. falciparum infection each year, with the vast majority of the estimated 1 million fatalities occurring in children under the age of 5 years in sub-Saharan Africa (50). The often-fruitless efforts to develop a licensed malaria vaccine, along with the emergence of drug-resistant parasites and insecticide-resistant mosquitoes, highlight the urgency with which new points of attack to combat malaria need to be identified. The arrival of the P. falciparum genome sequence (15), along with its transcription (8, 34) and proteomic (14, 33) profiles, has provided great opportunities to identify novel drug and vaccine candidates.
The asexual blood stage of the parasite is exclusively responsible for the clinical symptoms of malaria, and so, understandably, great efforts have gone into elucidating the molecular mechanism of erythrocyte invasion that is driven by the parasite's actomyosin motor. Although this process remains largely undefined, it is known that the secretory organelles located at the apical end of the invasive merozoite are pivotal. These organelles consist minimally of micronemes, rhoptries, and dense granules. Proteins belonging to the Duffy binding-like erythrocyte binding protein (18, 37, 49) and P. falciparum reticulocyte binding-like homologue (PfRH) protein families (11, 44, 53, 54, 56), located in the apical organelles, are likely to function as adhesins. These adhesins are believed to be externalized and then bind to erythrocyte receptors. Surface adhesins must also be involved in the formation of the tight junction and connect to the internal molecular motor of the parasite, which generates the force necessary for productive invasion. Apical organelle-resident proteins are also crucial in the later stages of invasion, in particular during the removal of proteins from the parasite surface, formation of a parasitophorous vacuole, and subsequent modifications of the host cell. For example, at least three proteins, merozoite surface protein 1 (MSP1), apical merozoite antigen 1 (AMA1), and Plasmodium thrombospondin-related apical merozoite protein (PTRAMP), are shed from the surface of merozoites at the time of invasion by the action of the parasite's subtilisin-like protease 2 (SUB2) (21, 28). Both AMA1 and PTRAMP are transmembrane proteins that are externalized from apical organelles onto the merozoite surface prior to invasion, whereas MSP1 is a resident MSP that is attached by a glycosylphosphatidylinositol (GPI) anchor (17, 26). It is thought that SUB2 cleavage of these proteins is essential for erythrocyte invasion to occur.
In addition to merozoites, there are two other stages of the Plasmodium life cycle that invade host tissue: sporozoites and ookinetes. In all cases these cells must recognize and attach to host receptors as part of this invasion process, which involves the parasite surface and contents of secretory organelles of the invasive zoite. While proteins on the surface of the parasite and some components of the apical organelles are stage specific, some of the secreted proteins as well as components of the motor complex are shared across the stages. We were interested in identifying novel proteins involved in the process of erythrocyte invasion using a bioinformatics approach to define candidate proteins that may be components of the basic invasion machinery that is conserved between merozoites and sporozoites (3), using proteome data for both merozoites and sporozoites (14). Using this approach, we have identified a GPI-anchored micronemal antigen (GAMA), encoded by the gene PF08_0008, which is proteolytically processed during transfer to the micronemes, externalized to allow binding to erythrocytes, and then shed from the surface of merozoites by the action of a protease other than SUB2.
MATERIALS AND METHODS
Identification of GAMA, encoded by PF08_0008, and bioinformatic analyses.
Data from the proteomic study by Florens et al. (14), which identified 2,415 proteins from four developmental stages of P. falciparum, were used as a starting point to identify previously uncharacterized P. falciparum proteins. A set of criteria was developed in order to identify proteins from this data set that may play a role in host cell invasion, based on four features: (i) a predicted signal peptide, (ii) a predicted single transmembrane domain, (iii) “hypothetical protein” status, and (iv) detection in both merozoite and sporozoite fractions. Application of these criteria to the data set resulted in the identification of the gene PF08_0008, which was studied further. Based on the studies described in this paper, this gene codes for a protein we have designated GAMA.
Information on PF08_0008 was gathered from the Plasmodium genome resource, PlasmoDB (http://plasmodb.org), including data from a number of published transcriptome studies (8, 34, 36). Searches for similar protein sequences were carried out using the basic local alignment search tool (BLAST) (2), available at the National Center for BioInformatics (http://ncbi.nlm.nih.gov/BLAST/), with default search parameters. Sequences of relevant hits from other Plasmodium species were downloaded from PlasmoDB and were aligned using the BioEdit sequence alignment software (24). Multiple alignments were carried out using ClustalW. The Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) (46) and ProSite (http://expasy.ch/prosite) (29) searchable databases were used to identify conserved protein domains in GAMA.
Parasite culture and metabolic labeling.
P. falciparum clone 3D7 asexual stages were cultured in vitro as previously described (7) and used to obtain DNA, RNA, and protein. Synchrony of parasite growth was maintained using a combination of sorbitol treatment (32), centrifugation over 70% Percoll (GE Healthcare) (42), and a magnetic activated cell separation depletion column (Miltenyi Biotec) (54). A tightly synchronized culture was sampled at 3-hour intervals throughout the intraerythrocytic cell cycle for RNA preparation as described below. Naturally released merozoites were purified as described previously (5). When merozoites were prepared for use in processing assays, the purification was carried out in the presence of 5 mM EGTA.
Magenetic activated cell separation-purified schizonts were metabolically labeled in methionine- and cysteine-free medium with 100 μCi ml−1 Pro-Mix (35S-labeled methionine and cysteine; GE Healthcare) for 2 h. For metabolic labeling in the presence of brefeldin A (BFA), purified schizonts were first incubated in the presence of 5 μg ml−1 BFA in methanol (0.05% final concentration), or a corresponding amount of methanol alone for control cultures, for 1 h prior to labeling. GPI-anchored proteins were labeled by incubating purified schizonts in glucose-free medium supplemented with 10 mM fructose, 50 μCi ml−1 d-[6-3H(N)]glucosamine hydrochloride (GE Healthcare), and 50 μCi ml−1 d-[2-3H]mannose (GE Healthcare) at 37°C for 2 h (47). All metabolically labeled parasites were washed in phosphate-buffered saline (PBS), harvested, and stored at −70°C until used.
Supernatant was collected from tightly synchronized, purified schizonts that had been returned to culture in the absence of erythrocytes and were closely monitored for rupture and merozoite release. Following removal of cellular material and clarification by sequential centrifugation steps, the supernatant was concentrated 10-fold in a 5-kDa-molecular-mass cutoff centrifugal concentrator (Vivascience) and stored in aliquots at −20°C until used.
Reverse transcription-PCR (RT-PCR) analysis of PF08_0008 expression.
Total RNA was isolated from each of the 3-hour time point samples of P. falciparum parasites using TRIzol (Invitrogen). One microgram of RNA was used to prepare cDNA from each time point, using the reverse transcription system from Promega UK Ltd. Transcriptional analysis was performed using the cDNA time course with oligonucleotide primers F (5′-CGACACCACAAAAAATCAGCA-3′) and R (5′-CCTTTGGTTTTTCTGGATTGAT-3′).
Expression of recombinant GAMA and generation of antisera.
Two fragments of PF08_0008, corresponding to N-terminal domain (NTD) and C-terminal domain (CTD) of the protein, were amplified from genomic DNA with 5′ extensions (underlined) to allow ligation-independent cloning into the pET-30 or pET-32 Xa/LIC expression vector (Merck). Primers pairs NTDf (5′-GGTATTGAGGGTCGCTGTGATATACAAAAAATAGCAG-3′) and NTDr (5′-AGAGGAGAGTTAGAGCCTTGTTCTTCATCTTCCTCAAT-3′) and CTDf (5′-GGTATTGAGGGTCGCGCAAAAGCTTATTGTAAGAA-3′) and CTDr (5′-AGAGGAGAGTTAGAGCCGCCTTTGCATTTGGTCCTT-3′) were used to generate the recombinant N- and C-terminal fragments of GAMA1, respectively.
Recombinant proteins were expressed in Escherichia coli BL21(DE3) cells following induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Harvested cell pellets were solubilized in denaturing lysis buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8 M urea, pH 8.0) and the supernatant applied to a preequilibrated Ni-nitrilotriacetic acid agarose (Qiagen) column. Washing buffer (50 mM NaH2PO4, 300 mM NaCl) supplemented with 10 mM imidazole was then applied to the column. Two further washes were carried out using washing buffer supplemented with 20 mM and 50 mM imidazole. Bound protein was eluted with washing buffer containing 250 mM imidazole. Eluted fractions were extensively dialyzed against PBS using SnakeSkin dialysis tubing (Pierce). Mouse antiserum was generated as described elsewhere (41), and rabbit antiserum was purchased from Harlan Sera Lab. All animal experimentation was carried out under a license issued by the United Kingdom Home Office under the Animals (Scientific Procedures) Act, 1986.
Immunoprecipitation of metabolically labeled parasites.
Pellets of labeled schizont material were lysed in NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 5 mM EGTA, 150 mM NaCl) supplemented with 1× complete protease inhibitors (Roche) on ice for 1 h. The soluble fraction was isolated and preabsorbed with protein G-Sepharose 4 Fastflow (GE Healthcare) for 1 h at 4°C and then incubated overnight with antiserum at 4°C.
Antibody-protein complexes were immunoprecipitated with protein G-Sepharose 4 Fastflow for 1 h at 4°C. Beads were washed extensively (first with 1% NP-40, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 500 mM NaCl, and 1 mg ml−1 bovine serum albumin [BSA] and then with 1% NP-40, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA). Samples were ultimately resuspended in reducing sample buffer, heated to 95°C for 5 minutes, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on either 10% slab gels or precast NuPAGE polyacrylamide gels (Invitrogen). Gels for autoradiography were fixed for 20 min, shaken for 20 min in Amplify (GE Healthcare), dried at 70°C for 2 hours, and exposed to BioMax MR film at −70°C.
Western blot analysis of parasite proteins.
For the analysis of total schizont material, purified parasites were lysed directly in 2× reducing or nonreducing SDS-PAGE sample buffer. For the extraction of membrane-associated proteins, purified schizonts were suspended in high-pH carbonate buffer (0.1 M Na2CO3, pH 11.0), passed through a fine-gauge needle, and centrifuged at 135,240 × g for 30 min at 4°C. Supernatants were retained, and pellets were reextracted with carbonate buffer. Carbonate-soluble and -insoluble material in a similar volume was resuspended in 2× reducing or nonreducing sample buffer. All protein extracts were heated at 95°C for 5 min prior to SDS-PAGE analysis on precast NuPAGE polyacrylamide gels.
Separated proteins were blotted onto Protran nitrocellulose membranes (Schleicher & Schuell). Blots were blocked with 10% (wt/vol) milk powder in washing solution (0.1% [vol/vol] Tween 20 in PBS) and then probed with primary antiserum diluted appropriately in 5% (wt/vol) Marvel in washing solution. Bound antibody was detected by incubation with an appropriate horseradish peroxidase-conjugated secondary antibody, followed by visualization with enhanced chemiluminescence Western blotting detection reagents (GE Healthcare). Blots were also probed with anti-PTRAMP antiserum (21) and anti-MSP3 antiserum (a kind gift from Andrew Pearce, Walter and Eliza Hall Institute) for control purposes.
Immunofluorescence analysis of protein location.
Smears of purified schizont, merozoite, or ring stage parasites were fixed with 4% (vol/vol) formaldehyde in PBS for 30 min and permeabilized in 0.1% (vol/vol) Triton X-100 in PBS for 10 min. Slides were blocked overnight at 4°C in 3% (wt/vol) BSA in PBS and then probed for 60 min at 37°C with primary antibodies diluted appropriately in 1% (wt/vol) BSA and 0.05% (vol/vol) Tween 20 in PBS. Secondary antibodies were used to probe the slides for 30 min. In the case of dual-labeling experiments, slides were additionally probed with monoclonal antibody (MAb) 61.3 (anti-RhopH2) (35), polyclonal mouse anti-AMA1 antibodies (10), or MAb 1E1 (anti-MSP119) (9). Slides were immersed in 0.5 μg ml−1 DAPI (4′,6′-diamidino-2-phenylindole) in PBS to stain nuclei. Slides were mounted in Citifluor (Citifluor, Canterbury, United Kingdom), sealed, and visualized under oil immersion using an Axioplan 2 imaging system (Zeiss, Oberkochen, Germany) and a Plan-Apochromat 100×/1.4 oil immersion objective. Images were captured with Axiovision 3.1 software and prepared for publication with Adobe PhotoShop.
Merozoite protein processing assay.
Processing assays to detect the activity of SUB2 were carried out as described previously (5, 21, 28). Briefly, merozoites prepared in the presence of 5 mM EGTA were washed, resuspended in processing buffer (50 mM Tris-HCl [pH 7.6], 5 mM CaCl2) and divided into equal aliquots on ice. Aliquots were supplemented with processing buffer or EGTA and incubated at 37°C for 2 h. Merozoites were pelleted, and shed proteins were detected in merozoite supernatants by Western blotting using mouse anti-CTD antiserum or rabbit anti-PTRAMP antiserum (21).
Erythrocyte binding assay.
Aliquots (200 μl) of culture supernatant were incubated in the presence or absence of 120 μl washed O+ erythrocytes at 37°C for 1 hour with shaking. Following centrifugation, supernatants were removed to fresh erythrocytes, and the incubation was repeated. Samples were subjected to successive centrifugation steps in order to remove all cellular material, and supernatants were analyzed by Western blotting under reducing conditions. Following extensive washing in PBS, bound proteins were eluted from cellular material by the addition of 0.5 M NaCl in PBS (pH 7.4) and were analyzed by Western blotting.
RESULTS
Identification of PF08_0008, coding for GAMA.
We were interested in identifying previously uncharacterized (“hypothetical”) proteins likely to play a role in the invasion of host cells, and in particular new elements of the basic machinery of invasion that is believed to be conserved between merozoites and sporozoites (3). Proteins identified in a proteomic study of merozoites and sporozoites (14) were filtered according to a set of criteria designed to identify proteins being targeted to either secretory organelles or the parasite surface, as illustrated in Fig. S1A in the supplemental material. Of 2,415 proteins in the data set, 92 were predicted to contain a signal peptide and a single transmembrane sequence (including AMA1 and EBA140, which are known to be important in erythrocyte invasion). Of these, 40 proteins were assigned as “hypothetical,” and only two of this set were detected in both merozoite and sporozoites proteomes, encoded by the genes PF08_0008 and PF14_0016, respectively. Sequence information for GAMA and PF14_0016 retrieved from PlasmoDB (http://www.plasmoDB.org) revealed that PF14_0016 codes for a member of the early-transcribed membrane protein family (51), which are parasitophorous vacuole membrane proteins believed to be important postinvasion (52), whereas PF08_0008 codes for a previously uncharacterized protein. Although this approach may miss other candidate proteins, nevertheless the focus of this study therefore became the protein encoded by PF08_0008.
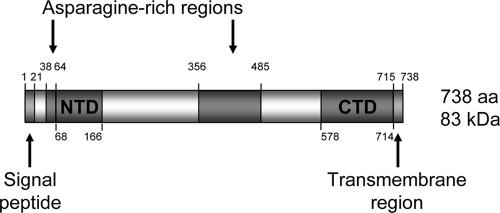
PF08_0008 encodes a 738-amino-acid protein (predicted molecular mass of ∼83 kDa) (Fig. 1) with orthologues identified only in other Plasmodium species, suggesting that it may perform a genus-specific function. No evolutionarily conserved protein domains were identified using the SMART and ProSite databases, other than the previously identified signal peptide (residues 1 to 21) and transmembrane domain (residues 715 to 737). Full-length orthologous sequences are present in the P. vivax (PV088910), P. knowlesi (PKH_050210), and P. yoelii (PY07130) databases (http://www.PlasmoDB.org), Alignment and comparison of these sequences (see Fig. S1B in the supplemental material) revealed two asparagine-rich regions encompassing amino acid residues 38 to 64 and 356 to 485 that are specific to P. falciparum. The second of these is a degenerate repeat based on eight copies of the sequence NNNNNN(Q/N)QV. A second insertion is located elsewhere in the P. vivax sequence, which is also a degenerate repeat based on 21 copies of (A/L)ANAN. Outside of these inserts the sequences are highly conserved, with approximately 40% identity; for example, all eight cysteine residues of the GAMA sequence (disregarding one in the predicted signal peptide) are positionally conserved. Partial sequences are also present in the P. berghei (PB000129.01.0) and P. chabaudi (PC000051.03.0) databases.
FIG. 1.
Schematic of the primary structure of GAMA. Indicated are the predicted signal peptide (residues 1 to 21) and C-terminal transmembrane domain (residues 715 to 738), the two asparagine-rich regions (residues 38 to 64 and 356 to 485), and the two regions at the N- and C-terminal ends of the protein (NTD, residues 68 to 166; CTD, residues 578 to 714) which were expressed in recombinant form and used to raise specific antisera. The protein consists of 738 amino acids (aa) with a calculated molecular mass of ∼83 kDa.
Expression of PF08_0008.
Published transcriptional analyses of PF08_0008 reveal that this gene is expressed in the later stages of the asexual blood cycle, with maximal expression at 40 to 48 h postinvasion (8, 34, 36). PF08_0008 is also transcribed in sporozoites (34), a result which is consistent with the identification of PF08_0008-derived peptides in sporozoite extracts (14). More recently the gene and corresponding protein have also been detected in the P. berghei ookinete transcriptome and proteome (23, 31, 57).
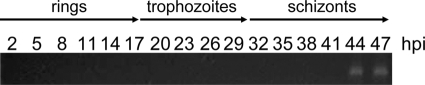
The expression profile of PF08_0008 in asexual blood stages was examined at 3-hour intervals using highly synchronized parasite populations, RT-PCR, and gene-specific primers. (Fig. 2). Expression was detected at only the last two time points, 44 and 47 h postinvasion, confirming that the gene is expressed only at the last stage of the blood cycle just before merozoite release.
FIG. 2.
Transcriptional analysis of PF08_0008 expression by RT-PCR using gene-specific primers. The PF08_0008 transcript was detected in the late schizont stages, between 44 and 47 h postinvasion (hpi). No PCR products were obtained using samples prepared for each time point in the absence of reverse transcriptase (not shown).
GAMA is GPI anchored.
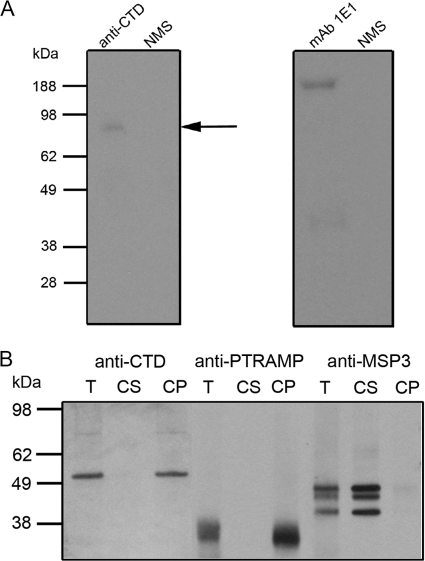
The transmembrane domain predicted by transmembrane prediction using hidden Markov models (30) is located at the extreme C terminus of the protein, encompassing residues 715 to 737, which are predicted to constitute a GPI addition site. Using an algorithm trained on P. falciparum GPI-modified proteins, GAMA is predicted to have a GPI anchor (19), but this modification has not been established experimentally. We therefore raised antibodies to a recombinant protein denoted CTD and comprising the C-terminal sequence (residues 578 to 714) (Fig. 1) and used these to immunoprecipitate GAMA from parasites that had been metabolically labeled with the radioactive GPI precursors [3H]mannose and [3H]glucosamine. Figure 3A shows that full-length, labeled GAMA was immunoprecipitated from these parasites, consistent with it having a GPI anchor. As a control, radiolabeled MSP1, a known GPI-anchored protein (17), could also be immunoprecipitated from the same extract with a specific antibody.
FIG. 3.
Membrane anchorage of GAMA. (A) To provide evidence of a GPI anchor, antibodies were used to immunoprecipitate full-length GAMA (∼83 kDa, arrow) from [3H]glucosamine- and mannose-labeled parasites. As a positive control, the MAb 1E1 was used to immunoprecipitate full-length MSP1 (∼200 kDa) from labeled parasites. The proteins immunoprecipitated with these two antibodies were fractionated on different gels, and the film was subjected to different exposure times. No proteins were immunoprecipitated with normal mouse serum (NMS). (B) To examine whether or not GAMA is membrane bound, total schizont material (T) was treated with a high-pH carbonate buffer to yield carbonate-soluble (CS) and -insoluble (CP) fractions, which were then resolved by SDS-PAGE and analyzed by Western blotting. Anti-CTD antiserum detected two species, both of which were found in the carbonate-insoluble fraction, confirming the membrane association. The larger species is presumably full-length GAMA, and the 49-kDa polypeptide (p49) likely represents the product of a proteolytic cleavage event. Blots were also probed with an antibody raised against the type I transmembrane protein PTRAMP and the soluble MSP3 in order to verify the outcome of the extraction procedure.
GAMA is membrane bound and proteolytically processed.
Antibodies raised to CTD, the C-terminal region of GAMA, detected the full-length protein and a smaller 49-kDa polypeptide (here denoted p49) in Western blot analyses of schizont material, suggesting that the protein is proteolytically cleaved. We therefore produced antibodies to a recombinant protein denoted NTD and comprising the N-terminal sequence (residues 68 to 166) (Fig. 1) and used the antibodies to both the N- and C-terminal regions to investigate the form of the protein in parasite extracts.
In the first set of experiments we wanted to examine whether or not the protein behaved as a membrane protein, consistent with the presence of a GPI anchor. Parasite lysates were prepared in high-pH carbonate buffer in which membrane proteins are expected to be insoluble and other proteins, including peripheral membrane proteins, are expected to be soluble. Both the ∼80-kDa and 49-kDa species were detected with anti-CTD antibodies in carbonate-insoluble schizont material (Fig. 3B), indicating a membrane association. This location is consistent with the presence of a GPI anchor and suggests that p49 is a membrane-bound C-terminal fragment of GAMA that results from a proteolytic cleavage event, occurring prior to schizont rupture. As controls, we examined in parallel the behavior of two proteins, pTRAMP and MSP3, which are a membrane protein and a peripheral membrane protein, respectively. pTRAMP was carbonate insoluble, and MSP3 was carbonate soluble (Fig. 3B).
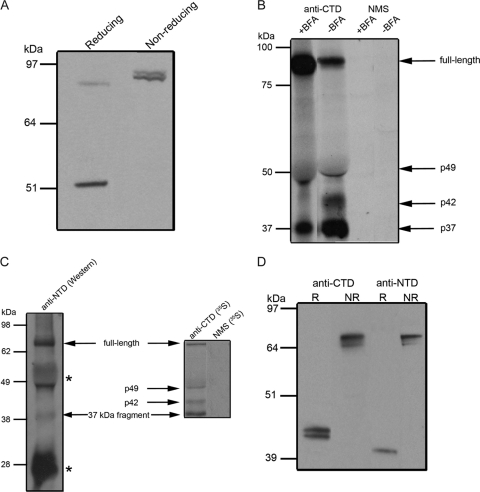
When schizont material was analyzed by Western blotting under nonreducing conditions using the anti-CTD antiserum, the p49 fragment was not detected; instead the antibodies reacted with a protein species similar in mobility to that expected for full-length GAMA (Fig. 4A). This result suggests that p49 is associated by way of a disulfide bond with another polypeptide(s). Since the combined molecular mass of this complex is approximately that of full-length GAMA, it is possible that p49 remains associated with the N-terminal product of the initial processing event.
FIG. 4.
Processing of GAMA. (A) Total schizont material was examined by Western blotting under reducing and nonreducing conditions using the anti-CTD antiserum to provide evidence of a disulfide-linked complex. Under reducing conditions, both full-length GAMA and p49 were evident in the sample, whereas under nonreducing conditions, p49 was absent. An additional band was observed under nonreducing conditions, which effectively comigrated with full-length GAMA and is consistent with p49 remaining associated with the N-terminal product of the initial processing event, by way of a disulfide linkage. (B) To determine the timing of the primary processing event, proteins were immunoprecipitated from BFA-treated (+BFA) or methanol-treated (−BFA) schizont material using anti-CTD antiserum and mouse preimmune serum (NMS). The level of GAMA processing was reduced in BFA-treated parasites, so that full-length GAMA was more abundant in the treated samples than in untreated samples. Although it was difficult to determine any difference in the level of p49 between the samples due to the interference of the comigrating immunoglobulin G heavy chain, the amounts of p42 and p37 fragments were considerably reduced in the BFA-treated samples. (C) A combination of immunoprecipitation and Western blotting was used to identify components of the complex. Proteins immunoprecipitated with anti-CTD antiserum from 35S-labeled NP-40-lysed schizonts were separated by SDS-PAGE, transferred to nitrocellulose, and probed with anti-NTD antiserum on a Western blot (left panel). In the right panel is shown an autoradiograph of 35S-labeled proteins immunoprecipitated from the same preparation of schizonts using either anti-CTD antiserum or mouse preimmune serum. Anti-NTD antiserum reacted in Western blotting with both the full-length GAMA and the 37-kDa polypeptide that were immunoprecipitated by the anti-CTD antiserum (both polypeptides are indicated by arrows and labeled), suggesting that the 37-kDa polypeptide is indeed the N-terminal product of GAMA processing. The labeled secondary antibody also reacted with the heavy and light chains of the immunoglobulin G present in the sample (marked with asterisks). The anti-NTD antiserum did not react with the 42-kDa polypeptide immunoprecipitated by the anti-CTD antiserum, and presumably not with the 49-kDa polypeptide, although the latter was hard to confirm due to reactivity of the secondary antibody with immunoglobulin G heavy chain. (D) The protein is shed from the parasite by proteolysis. Culture supernatant was analyzed by Western blotting under reducing (R) and nonreducing (NR) conditions with the anti-CTD and -NTD antisera. A 42-kDa doublet (p42) was detected by the anti-CTD antiserum, and a 37-kDa polypeptide (p37) was detected by the anti-NTD antiserum. The identical mobilities of the shed fragments recognized by the two different antisera under nonreducing conditions suggest that they are together in a complex.
To examine the proposed processing in more detail, we used BFA, a fungal metabolite that is known to inhibit the transport of proteins from the endoplasmic reticulum (ER) to the Golgi apparatus in P. falciparum (13), to investigate the timing of GAMA processing. Parasites were metabolically radiolabeled in the presence of BFA or a solvent control, and then immunoprecipitations from lysates were carried out using the anti-CTD antiserum. In BFA-treated parasites there was an accumulation of full-length GAMA and a marked reduction in the abundance of the processed forms relative to the control parasites, in which processing appears to have occurred (Fig. 4B). This result indicates that processing of full-length GAMA occurs following transport from the ER to the Golgi apparatus.
Further immunoprecipitation experiments were carried out on extracts of biosynthetically 35S-labeled mature schizonts (42 h postinvasion). The anti-CTD antiserum immunoprecipitated four separate protein species running at sizes of approximately 80 kDa, 49 kDa, 42 kDa, and 37 kDa (Fig. 4C). The largest species presumably correspond to full-length GAMA and the initial processing product p49. The identities of the 42- and 37-kDa species were investigated by separating the proteins immunoprecipitated with the anti-CTD antiserum by SDS-PAGE, transferring them to nitrocellulose, and probing them with the anti-NTD antiserum in a Western blot analysis. The antibodies in this serum reacted with the full-length GAMA as well as a 37-kDa polypeptide, corresponding to the N-terminal region of the protein, designated p37. The NTD-specific antibodies did not react with the 42-kDa species, and this probably represents a truncated form of p49 that is produced late in the cycle (see next section).
GAMA is shed into culture supernatant as a complex of two polypeptides.
At merozoite egress and erythrocyte invasion, some merozoite proteins such as AMA1, MSP1, and EBA175 are shed into the supernatants of parasite cultures, for example, by two distinct mechanisms involving the action of the subtilisin SUB2 or the rhomboid protease ROM4 (reviewed in reference 39). Therefore, schizonts were allowed to rupture, and soluble proteins in the culture supernatant were analyzed by Western blotting with the anti-CTD and anti-NTD antisera following electrophoresis under reducing and nonreducing conditions (Fig. 4D). Both antisera detected what appeared to be a band close to the size of the full-length protein in nonreduced samples and fragments of the protein in reduced samples. A 42-kDa doublet (p42) was detected by the anti-CTD antiserum and a ∼37-kDa polypeptide (presumably p37) was detected by the anti-NTD antiserum. The identical mobilities of the shed fragments under nonreducing conditions detected by the two different sera suggests that they are in a complex. The p42 doublet could arise through the cleavage of p49 at alternative positions close to its membrane anchor to produce a soluble fragment.
Since GAMA may have a GPI anchor, it cannot be the substrate for a rhomboid protease but it could be cleaved by SUB2, which cleaves both integral membrane and GPI-link proteins close to the membrane (28). The activity of SUB2 can be inhibited by chelating agents such as EDTA, because it is dependent on calcium, and by its prodomain, which binds tightly to the active enzyme and blocks its activity. Neither EDTA nor the SUB2 prodomain could inhibit the release of GAMA in a merozoite processing assay (21) (data not shown), suggesting that another, as-yet-uncharacterized protease is involved.
Subcellular location of GAMA: apical end of merozoites in mature schizonts and redistribution on free merozoites and ring stage parasites.
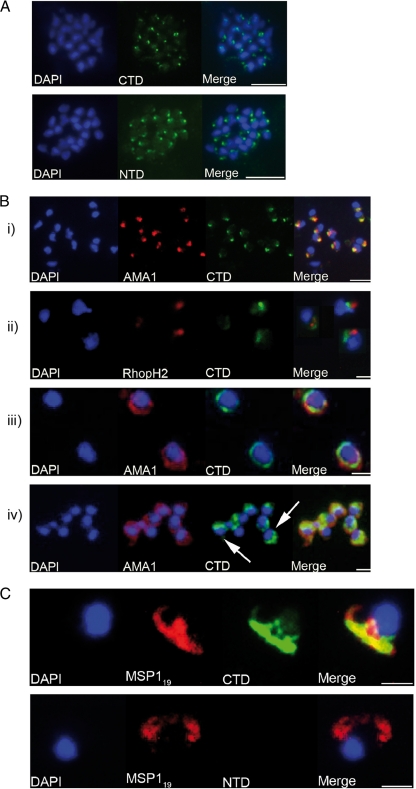
When characterizing a novel P. falciparum protein such as that encoded by PF08_0008, it is useful to determine the subcellular location of the protein in parasites, as this can give clues to its functional role. When formaldehyde-fixed smears of mature schizonts were stained with anti-CTD and anti-NTD antisera, both sets of antibody resulted in a strong punctuate patterning (Fig. 5A). The fluorescence was detected close to a single point away from the nucleus, a pattern that is consistent with an apical organelle-resident protein. When parasites were dual labeled with antibodies specific for the micronemal protein AMA1, some degree of colocalization was evident, suggesting that GAMA may be resident in the micronemes (Fig. 5B, panel i). In contrast, no colocalization was observed when parasites were dual labeled with the MAb 61.3, which recognizes the rhoptry protein RhopH2 (Fig. 5B, panel ii), or an antibody specific for a rhoptry neck protein (data not shown).
FIG. 5.
Analyses of the distribution of GAMA in asexual blood stage parasites by immunofluorescence. (A) Formaldehyde-fixed P. falciparum 3D7 schizonts were probed with mouse anti-CTD (top panel) or anti-NTD (bottom panel) antiserum and an anti-mouse-fluorescein isothiocyanate conjugate (green). Parasite nuclei were stained with DAPI (blue). Staining with both antisera resulted in a strong punctuate pattern, with a single point of fluorescence located away from the nucleus and consistent with a location in the apical organelles. Scale bars represent 2 μm. No signal was detected when parasites were probed with normal mouse serum (not shown). (B) Formaldehyde-fixed free P. falciparum merozoites were probed with rabbit anti-CTD antiserum and either mouse polyclonal anti-AMA1 antibodies (panels i, iii, and iv) or mouse MAb 61.3 (panel ii). Mouse antibodies were detected with an anti-mouse Alexa Fluor 594 conjugate (red) and rabbit antibodies with an anti-rabbit Alexa Fluor 488 conjugate (green). Parasite nuclei were stained with DAPI (blue). Scale bars are 5 μm in panel i and 2 μm in the other panels. In panels i and ii there is a clear apical localization of AMA1, RhopH2, and GAMA, with AMA1 and GAMA appearing to be colocalized; panel iii shows a circumferential localization of both GAMA and AMA1 on the surface of free merozoites; and panel iv shows a “cap”-like distribution of GAMA on the surface of free merozoites (arrows), in contrast to the circumferential distribution of AMA1. (C) Newly invaded ring stage parasites were probed with the mouse anti-MSP119 MAb 1E1 and either rabbit anti-CTD (top panel) or anti-NTD (bottom panel) antiserum. Mouse antibodies were detected with an anti-mouse Alexa Fluor 594 conjugate (red) and rabbit antibodies with an anti-rabbit Alexa Fluor 488 conjugate (green). Parasite nuclei were stained with DAPI (blue). There was no reactivity exhibited by mouse preimmune serum with ring stage parasites (not shown). Scale bars are 2 μm. There is a clear reactivity of anti-CTD antibodies, but not anti-NTD antibodies, with ring stage parasites. Anti-CTD and anti-MSP119 signals appear to colocalize (top panel), suggesting that the “stub” part of GAMA that is produced upon shedding of the p42-p37 complex is carried into the host cell and remains associated with the ring stage parasite.
Some molecules are known to relocate from the apical organelles of segmenting schizonts to the surface of free merozoites and are subsequently shed at the point of invasion. Examples include AMA1, which is shed by SUB2 (25), and the adhesin EBA175, shed by ROM4 (40). In contrast, other apically located molecules do not relocate onto the surface of merozoites (for example, RhopH2). In order to investigate the fate of GAMA following schizont rupture, naturally released merozoites were also examined by indirect immunofluorescence. It was apparent that different merozoites displayed different patterns of GAMA distribution: apical (in approximately 55% of parasites), entire periphery (∼10% of parasites) (Fig. 5B, panel iii), or “cap”-like (∼35% of parasites) (Fig. 5B, panel iv), suggesting that, like some other molecules involved in invasion, GAMA is released onto the parasite surface. Whether or not there is a dynamic relationship between these forms is unclear.
As is the case with MSP1 and AMA1 (6, 27), shedding of GAMA from the merozoite could generate a residual “stub” of protein, which remains anchored to the merozoite and is carried into the next erythrocyte upon invasion. To investigate this possibility, newly invaded (≤4 h postinvasion) ring stage parasites were examined by immunofluorescence with the anti-GAMA antisera (Fig. 5C). The anti-CTD antibodies stained these parasites in a pattern indistinguishable from that observed with the anti-MSP119 MAb 1E1, but no signal was produced by the anti-NTD antiserum. The recombinant protein used to generate the anti-CTD antibodies included the region of GAMA immediately upstream of the GPI anchor addition sequence, and therefore the antibodies were likely to detect any “stub” generated during shedding, explaining the reactivity with ring stage parasites. In contrast, the anti-NTD antiserum would not be expected to react, since all of GAMA except the “stub” is released into the culture supernatant. As yet there is no additional experimental evidence for the existence of this “stub”.
GAMA binds specifically to the surface of erythrocytes.
Relatively few P. falciparum proteins have been shown to have a well-defined erythrocyte binding capability. MSP1 is a putative ligand for the erythrocyte protein band 3 (20); EBA175 is known to bind glycophorin A on the erythrocyte surface; EBA140 binds glycophorin C (37); and EBA181, PfRH1, and PfRH4 bind undefined receptors (16, 18, 44). The presence of GAMA in culture supernatant, as the result of a specific shedding, hints at a potential role for this protein as an adhesin, so this possibility was investigated further. Samples of culture supernatant were analyzed by Western blotting before and after incubation in the presence or absence of washed erythrocytes (Fig. 6A). GAMA was shown to be depleted from culture supernatants incubated in the presence of erythrocytes. As controls EBA175 and MSP3 were also examined, and whereas EBA175 was also depleted, MSP3 was not. To confirm that the depletion of GAMA was due to a specific interaction between this protein and erythrocytes, the protein could be specifically eluted with 0.5 M NaCl from red blood cells incubated with culture supernatant after extensive washing and detection by Western blotting (Fig. 6B).
FIG. 6.
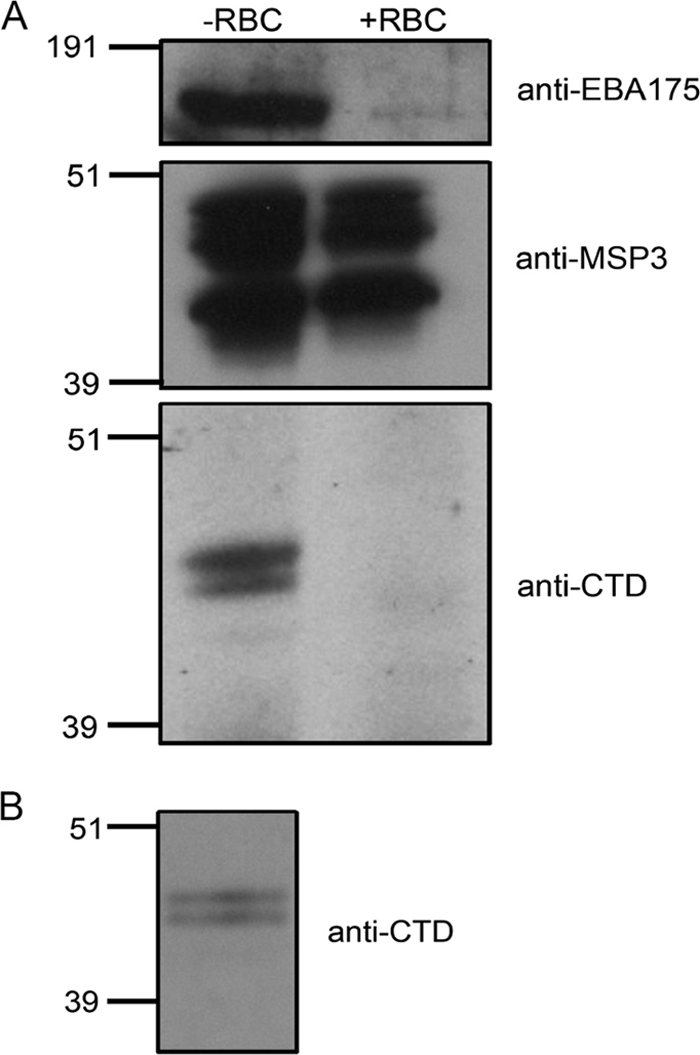
GAMA binds to erythrocytes. (A) Samples of culture supernatant that were incubated in the absence (−RBC) or presence (+RBC) of erythrocytes were analyzed by Western blotting with antibodies specific to EBA175 (top panel), MSP3 (middle panel), and GAMA (bottom panel). The defined erythrocyte adhesin EBA175 is depleted from culture supernatant upon the addition of erythrocytes, as is GAMA. MSP3 is not depleted. (B) Proteins bound to the erythrocyte surface were eluted with 0.5 M NaCl and analyzed by Western blotting with anti-GAMA antibodies. The binding and elution of GAMA indicate a reversible interaction with erythrocytes.
DISCUSSION
We have identified and characterized a novel Plasmodium falciparum protein. We were particularly interested in identifying a protein that may be involved in the basic machinery of host cell invasion, which is conserved between the invasive stages: sporozoites, merozoites, and ookinetes (3). Our starting point was malaria parasite genome, transcriptome, and proteome data, and initially we applied a set of specifically designed criteria to the data set from the proteome study performed by Florens et al. (14). We selected the protein encoded by the PF08_0008 gene for further investigation. We have named this protein GAMA based on its putative characteristics as defined in this study.
The bioinformatic analysis suggested that the protein is expressed in both merozoites and sporozoites, and more recently the corresponding mRNA and protein (encoded by gene PB000129.01.0) have been detected in the P. berghei ookinete transcriptome (57) and proteome (23). The protein is present in the ookinete microneme proteome (31) and the gene identified as coding for putative secreted ookinete protein (PSOP) 9 (12). Interestingly, in the latter study the gene was disrupted, resulting in a striking reduction in infectivity of the mosquito gut and oocyst formation without a reduction in the number of ookinetes formed (12). Despite the fact that the protein does not appear to be essential for P. berghei asexual blood stage development, it is clear from our studies and the earlier studies that the gene and protein are expressed in asexual blood stages. We have confirmed the expression of the gene by carrying out RT-PCR analysis at 3-h intervals on a highly synchronized parasite population. Expression of the gene was detected only at the last two time points, indicating that it is expressed just before merozoite maturation and egress.
The gene is present in all Plasmodium species examined but not in other Apicomplexan parasites. Sequence comparison suggest that the protein is highly conserved across the species, although species-specific insertions (particularly the asparagine-rich region at residues 356 to 485, which is found only in the P. falciparum structure) have been identified.
A hydrophobic sequence at the C terminus of the protein is a potential GPI addition site. Although the prediction is not strong, GAMA is predicted to have a GPI by using an algorithm trained on malarial proteins (19). We demonstrated that the protein could be biosynthetically labeled with [H3]glucosamine and mannose, which is consistent with the presence of a GPI anchor.
Like a number of merozoite proteins such as AMA1, MSP1, MSP7, and apical sushi protein, GAMA is synthesized as a precursor and then subjected to proteolytic cleavage. The first cleavage or primary processing of GAMA occurs after synthesis and is sensitive to the addition of BFA, suggesting that this cleavage occurs post-ER either in the Golgi apparatus, during transport to the micronemes of the parasite or once it has reached this subcellular location at some point prior to schizont rupture. The exact location of the processing site has not been identified, although based on the sizes of the two fragments (N-terminal 37 kDa and C-terminal 49 kDa), it is likely that the site is N terminal to the second asparagine-rich region. The cleavage results in two fragments that are still covalently linked by at least one disulfide bond between them, as evidenced by the mobility of the protein on SDS-PAGE in the presence or absence of a reducing agent. There are seven cysteines and one cysteine in the N- and C-terminal regions of the protein, respectively, but their involvement in cystine formation has not been investigated. The significance of this primary processing and the identity of the parasite protease responsible remain unclear, although it could be hypothesized that such a cleavage could serve to “activate” GAMA by revealing a functional domain. Following merozoite invasion of the red blood cell, GAMA is detected the supernatant, in which the C-terminal fragment is slightly smaller than the form in the merozoite. This suggests that the protein is released by proteolytic cleavage, in a way similar to that for AMA1 and MSP1 from the merozoite surface. However, our results suggest that the enzyme involved is not SUB2, the enzyme that cleaves AMA1 and MSP1, because the shedding is not inhibited by EGTA, which chelates the calcium essential for SUB2 activity, and more specifically the shedding is not inhibited by the prodomain of SUB2, which is a very potent inhibitor of SUB2 activity. The location of the cleavage site for this second enzyme is unknown, but as in the case of MSP1, another GPI-anchored protein that is cleaved at the time of invasion (6), there is a residual “stub” that is detected by the antibodies to the C-terminal region of GAMA and retained and carried into the ring stage parasite. A model for the proteolytic processing of GAMA1 consistent with the results presented here is shown in Fig. 7.
FIG. 7.
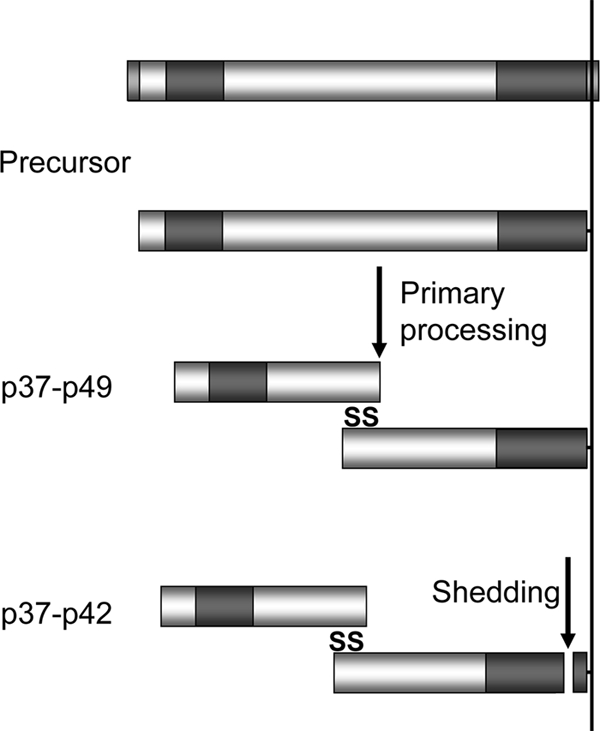
Schematic representation of the processing and shedding of GAMA. Primary processing of full-length protein gives rise to the p49 and p37 fragments. These fragments are held together in a disulfide-linked complex and are released into culture supernatant following alternative cleavage events near the membrane anchor, leaving behind a residual “stub.” The regions of GAMA expressed in a recombinant form in order to raise antisera are indicated in darker shading.
From our immunofluorescence analyses, it appears likely that GAMA is a micronemal protein. The fluorescence signal with antibodies to both the N- and C-terminal regions of the protein colocalizes with the signal from antibodies to AMA1 and EBA175 (data not shown) but not with antibodies to rhoptry proteins. Confirmation of this location requires immunoelectron microscopy studies, but so far these have been unsuccessful. It is interesting that GAMA appears to be located within the micronemes of developing merozoites in schizonts prior to egress, and the vast majority of free merozoites analyzed with anti-GAMA antibodies also displayed a punctuate or “cap”-like apical staining pattern, with few displaying any circumferential distribution. This is consistent with a model in which GAMA is released upon formation of the tight junction with the erythrocyte, rather than before initial attachment, and different from the behavior of AMA1. It has been demonstrated for P. knowlesi that AMA1 is essential for the parasite to reorientate, possibly via a concentration gradient of adhesive interactions, and that it is likely to be distributed on the surface of the merozoite before the formation of the tight junction (38).
GAMA is unusual in that most P. falciparum GPI-anchored proteins are surface antigens, as opposed to apical organellar molecules. Exceptions to this include rhoptry-associated membrane antigen (55), Pf34 (43), and apical sushi protein (19, 41). Interestingly, the GPI-anchored proteins Pf38 (45) and MSP10 (4) have been suggested to have both surface and apical distributions. GAMA appears to be a member of a small subset of GPI-anchored proteins that are initially present in the apical organelles rather than on the merozoite surface.
Our results indicate that GAMA is an erythrocyte binding protein, suggesting that this protein could function as an adhesin. However, although the subcellular location is consistent with this role, the membrane anchorage of GAMA would make it an atypical adhesin. All known adhesive microneme-resident proteins, such as members of the Duffy binding-like and the reticulocyte binding-like erythrocyte binding proteins, are type I integral membrane proteins with a functional cytoplasmic domain. These proteins and others such as the TRAP family (3) may link directly or indirectly to the actin-myosin motor via their cytoplasmic domain. In the case of GAMA, any link to the motor or other functional proteins within the parasite would necessarily be indirect via associations occurring in the protein ectodomain. No recognizable protein motifs were identified in the primary sequence of GAMA, which raises the question as to which regions of the protein are responsible for the demonstrated binding activity. It is possible that the N-terminal region of the protein, which contains seven out of eight of the conserved cysteine residues, is likely to be more structured and hence contain a functional fold. The recombinant proteins made to date do not have erythrocyte binding activity, and it may be important to express these proteins in a system to optimize their conformation before repeating such experiments. Antibodies raised to these recombinant proteins also did not inhibit invasion (data not shown); perhaps this is also because the recombinant protein is not fully correctly folded or because the protein is not accessible or directly involved in invasion. Since the protein is also expressed at sporozoite and ookinete stages, it will be of interest to determine whether or not the protein also binds specifically to host cell surface structures at these stages of the life cycle.
Although the subcellular location, expression profile, and demonstrated erythrocyte binding ability of GAMA are characteristic of known P. falciparum invasion ligands, these results do not preclude the possibility of this protein functioning in another capacity. One alternative hypothesis is that GAMA could function at the moving junction where it serves to exclude a particular host cell molecule from the developing parasitophorous vacuole membrane through its binding activity. The P. falciparum moving junction is believed to consist minimally of AMA1 and the rhoptry neck proteins RON2 and RON4 (1). Although there is no evidence from immunoprecipitation experiments to suggest that GAMA exists in such a complex, it cannot be ruled out at this point. Moreover, GAMA appears to be poorly abundant within parasites, and any protein-protein interactions at the moving junction are likely to be transient and hence difficult to detect. Further binding experiments employing the use of enzyme-treated erythrocytes could serve to shed light on the nature of the GAMA binding partner(s).
GAMA was initially identified as a protein for study partly because it was detected in both merozoite and sporozoite stages in the proteomic study published in 2002 (14). It will be interesting to confirm the proteomic data for GAMA and determine if, like AMA1 (48), it is expressed, processed, and shed in sporozoites as well as merozoites. In summary, we have identified a GPI-anchored microneme protein that is conserved throughout plasmodia. GAMA is proteolytically processed within schizonts and ultimately shed from the surface of merozoites. Initial experiments also implicate GAMA in an erythrocyte binding role, although the erythrocyte binding domain remains to be defined and characterized.
Supplementary Material
Acknowledgments
We thank Mike Blackman for the gift of PfSUB2 prodomain and Andrew Pearce for antibodies to MSP3.
This work was funded in part by the United Kingdom Medical Research Council, The Wellcome Trust Malaria Functional Genomics Initiative (reference 066742), the European Union (through the BioMalPar Network of Excellence), and a grant from the U.S. National Institutes of Health (HL078826). L.H. was in receipt of a United Kingdom Medical Research Council Ph.D. Studentship.
Footnotes
Published ahead of print on 9 October 2009.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alexander, D. L., S. Arastu-Kapur, J. F. Dubremetz, and J. C. Boothroyd. 2006. Plasmodium falciparum AMA1 binds a rhoptry neck protein homologous to TgRON4, a component of the moving junction in Toxoplasma gondii. Eukaryot. Cell 5:1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baum, J., D. Richard, J. Healer, M. Rug, Z. Krnajski, T. W. Gilberger, J. L. Green, A. A. Holder, and A. F. Cowman. 2006. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J. Biol. Chem. 281:5197-5208. [DOI] [PubMed] [Google Scholar]
- 4.Black, C. G., L. Wang, T. Wu, and R. L. Coppel. 2003. Apical location of a novel EGF-like domain-containing protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 127:59-68. [DOI] [PubMed] [Google Scholar]
- 5.Blackman, M. J. 1994. Purification of Plasmodium falciparum merozoites for analysis of the processing of merozoite surface protein-1. Methods Cell Biol. 45:213-220. [DOI] [PubMed] [Google Scholar]
- 6.Blackman, M. J., E. D. Dennis, E. M. Hirst, C. H. Kocken, T. J. Scott-Finnigan, and A. W. Thomas. 1996. Plasmodium knowlesi: secondary processing of the malaria merozoite surface protein-1. Exp. Parasitol. 83:229-239. [DOI] [PubMed] [Google Scholar]
- 7.Blackman, M. J., and A. A. Holder. 1992. Secondary processing of the Plasmodium falciparum merozoite surface protein-1 (MSP1) by a calcium-dependent membrane-bound serine protease: shedding of MSP133 as a noncovalently associated complex with other fragments of the MSP1. Mol. Biochem. Parasitol. 50:307-315. [DOI] [PubMed] [Google Scholar]
- 8.Bozdech, Z., M. Llinas, B. L. Pulliam, E. D. Wong, J. Zhu, and J. L. DeRisi. 2003. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burghaus, P. A., and A. A. Holder. 1994. Expression of the 19-kilodalton carboxy-terminal fragment of the Plasmodium falciparum merozoite surface protein-1 in Escherichia coli as a correctly folded protein. Mol. Biochem. Parasitol. 64:165-169. (Erratum, 67:343, 1994.) [DOI] [PubMed] [Google Scholar]
- 10.Collins, C. R., C. Withers-Martinez, G. A. Bentley, A. H. Batchelor, A. W. Thomas, and M. J. Blackman. 2007. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J. Biol. Chem. 282:7431-7441. [DOI] [PubMed] [Google Scholar]
- 11.Duraisingh, M. T., T. Triglia, S. A. Ralph, J. C. Rayner, J. W. Barnwell, G. I. McFadden, and A. F. Cowman. 2003. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 22:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ecker, A., E. S. Bushell, R. Tewari, and R. E. Sinden. 2008. Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol. Microbiol. 70:209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmendorf, H. G., and K. Haldar. 1993. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. EMBO J. 12:4763-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florens, L., M. P. Washburn, J. D. Raine, R. M. Anthony, M. Grainger, J. D. Haynes, J. K. Moch, N. Muster, J. B. Sacci, D. L. Tabb, A. A. Witney, D. Wolters, Y. Wu, M. J. Gardner, A. A. Holder, R. E. Sinden, J. R. Yates, and D. J. Carucci. 2002. A proteomic view of the Plasmodium falciparum life cycle. Nature 419:520-526. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaur, D., S. Singh, S. Singh, L. Jiang, A. Diouf, and L. H. Miller. 2007. Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion. Proc. Natl. Acad. Sci. USA 104:17789-17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerold, P., L. Schofield, M. J. Blackman, A. A. Holder, and R. T. Schwarz. 1996. Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface proteins-1 and -2 of Plasmodium falciparum. Mol. Biochem. Parasitol. 75:131-143. [DOI] [PubMed] [Google Scholar]
- 18.Gilberger, T. W., J. K. Thompson, T. Triglia, R. T. Good, M. T. Duraisingh, and A. F. Cowman. 2003. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J. Biol. Chem. 278:14480-14486. [DOI] [PubMed] [Google Scholar]
- 19.Gilson, P. R., T. Nebl, D. Vukcevic, R. L. Moritz, T. Sargeant, T. P. Speed, L. Schofield, and B. S. Crabb. 2006. Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol. Cell Proteomics 5:1286-1299. [DOI] [PubMed] [Google Scholar]
- 20.Goel, V. K., X. Li, H. Chen, S. C. Liu, A. H. Chishti, and S. S. Oh. 2003. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc. Natl. Acad. Sci. USA 100:5164-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green, J. L., L. Hinds, M. Grainger, E. Knuepfer, and A. A. Holder. 2006. Plasmodium thrombospondin related apical merozoite protein (PTRAMP) is shed from the surface of merozoites by PfSUB2 upon invasion of erythrocytes. Mol. Biochem. Parasitol. 150:114-117. [DOI] [PubMed] [Google Scholar]
- 22.Guerra, C. A., R. W. Snow, and S. I. Hay. 2006. Mapping the global extent of malaria in 2005. Trends Parasitol. 22:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, N., M. Karras, J. D. Raine, J. M. Carlton, T. W. Kooij, M. Berriman, L. Florens, C. S. Janssen, A. Pain, G. K. Christophides, K. James, K. Rutherford, B. Harris, D. Harris, C. Churcher, M. A. Quail, D. Ormond, J. Doggett, H. E. Trueman, J. Mendoza, S. L. Bidwell, M. A. Rajandream, D. J. Carucci, J. R. Yates III, F. C. Kafatos, C. J. Janse, B. Barrell, C. M. Turner, A. P. Waters, and R. E. Sinden. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307:82-86. [DOI] [PubMed] [Google Scholar]
- 24.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 25.Harris, P. K., S. Yeoh, A. R. Dluzewski, R. A. O'Donnell, C. Withers-Martinez, F. Hackett, L. H. Bannister, G. H. Mitchell, and M. J. Blackman. 2005. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog. 1:241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holder, A. A. 2009. The carboxy-terminus of merozoite surface protein 1: structure, specific antibodies and immunity to malaria. Parasitology 136:1445-1456. [DOI] [PubMed] [Google Scholar]
- 27.Howell, S. A., F. Hackett, A. M. Jongco, C. Withers-Martinez, K. Kim, V. B. Carruthers, and M. J. Blackman. 2005. Distinct mechanisms govern proteolytic shedding of a key invasion protein in apicomplexan pathogens. Mol. Microbiol. 57:1342-1356. [DOI] [PubMed] [Google Scholar]
- 28.Howell, S. A., I. Well, S. L. Fleck, C. Kettleborough, C. R. Collins, and M. J. Blackman. 2003. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J. Biol. Chem. 278:23890-23898. [DOI] [PubMed] [Google Scholar]
- 29.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. De Castro, P. S. Langendijk-Genevaux, M. Pagni, and C. J. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227-D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 31.Lal, K., J. H. Prieto, E. Bromley, S. J. Sanderson, J. R. Yates III, J. M. Wastling, F. M. Tomley, and R. E. Sinden. 2009. Characterisation of Plasmodium invasive organelles; an ookinete microneme proteome. Proteomics 9:1142-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambros, C., and J. P. Vanderberg. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418-420. [PubMed] [Google Scholar]
- 33.Lasonder, E., Y. Ishihama, J. S. Andersen, A. M. Vermunt, A. Pain, R. W. Sauerwein, W. M. Eling, N. Hall, A. P. Waters, H. G. Stunnenberg, and M. Mann. 2002. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature 419:537-542. [DOI] [PubMed] [Google Scholar]
- 34.Le Roch, K. G., Y. Zhou, P. L. Blair, M. Grainger, J. K. Moch, J. D. Haynes, P. De La Vega, A. A. Holder, S. Batalov, D. J. Carucci, and E. A. Winzeler. 2003. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301:1503-1508. [DOI] [PubMed] [Google Scholar]
- 35.Ling, I. T., O. Kaneko, D. L. Narum, T. Tsuboi, S. Howell, H. M. Taylor, T. J. Scott-Finnigan, M. Torii, and A. A. Holder. 2003. Characterisation of the rhoph2 gene of Plasmodium falciparum and Plasmodium yoelii. Mol. Biochem. Parasitol. 127:47-57. [DOI] [PubMed] [Google Scholar]
- 36.Llinas, M., Z. Bozdech, E. D. Wong, A. T. Adai, and J. L. DeRisi. 2006. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 34:1166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maier, A. G., M. T. Duraisingh, J. C. Reeder, S. S. Patel, J. W. Kazura, P. A. Zimmerman, and A. F. Cowman. 2003. Plasmodium falciparum erythrocyte invasion through glycophorin C and selection for Gerbich negativity in human populations. Nat. Med. 9:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell, G. H., A. W. Thomas, G. Margos, A. R. Dluzewski, and L. H. Bannister. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72:154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donnell, R. A., and M. J. Blackman. 2005. The role of malaria merozoite proteases in red blood cell invasion. Curr. Opin. Microbiol. 8:422-427. [DOI] [PubMed] [Google Scholar]
- 40.O'Donnell, R. A., F. Hackett, S. A. Howell, M. Treeck, N. Struck, Z. Krnajski, C. Withers-Martinez, T. W. Gilberger, and M. J. Blackman. 2006. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J. Cell Biol. 174:1023-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Keeffe, A. H., J. L. Green, M. Grainger, and A. A. Holder. 2005. A novel Sushi domain-containing protein of Plasmodium falciparum. Mol. Biochem. Parasitol. 140:61-68. [DOI] [PubMed] [Google Scholar]
- 42.Pasvol, G., R. J. Wilson, M. E. Smalley, and J. Brown. 1978. Separation of viable schizont-infected red cells of Plasmodium falciparum from human blood. Ann. Trop. Med. Parasitol. 72:87-88. [DOI] [PubMed] [Google Scholar]
- 43.Proellocks, N. I., S. Kovacevic, D. J. Ferguson, L. M. Kats, B. J. Morahan, C. G. Black, K. L. Waller, and R. L. Coppel. 2007. Plasmodium falciparum Pf34, a novel GPI-anchored rhoptry protein found in detergent-resistant microdomains. Int. J. Parasitol. 37:1233-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rayner, J. C., E. Vargas-Serrato, C. S. Huber, M. R. Galinski, and J. W. Barnwell. 2001. A Plasmodium falciparum homologue of Plasmodium vivax reticulocyte binding protein (PvRBP1) defines a trypsin-resistant erythrocyte invasion pathway. J. Exp. Med. 194:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders, P. R., P. R. Gilson, G. T. Cantin, D. C. Greenbaum, T. Nebl, D. J. Carucci, M. J. McConville, L. Schofield, A. N. Hodder, J. R. Yates III, and B. S. Crabb. 2005. Distinct protein classes including novel merozoite surface antigens in Raft-like membranes of Plasmodium falciparum. J. Biol. Chem. 280:40169-40176. [DOI] [PubMed] [Google Scholar]
- 46.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarz, R. T., V. Riveros-Moreno, M. J. Lockyer, S. C. Nicholls, L. S. Davey, Y. Hillman, J. S. Sandhu, R. R. Freeman, and A. A. Holder. 1986. Structural diversity of the major surface antigen of Plasmodium falciparum merozoites. Mol. Cell. Biol. 6:964-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silvie, O., J. F. Franetich, S. Charrin, M. S. Mueller, A. Siau, M. Bodescot, E. Rubinstein, L. Hannoun, Y. Charoenvit, C. H. Kocken, A. W. Thomas, G. J. Van Gemert, R. W. Sauerwein, M. J. Blackman, R. F. Anders, G. Pluschke, and D. Mazier. 2004. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J. Biol. Chem. 279:9490-9496. [DOI] [PubMed] [Google Scholar]
- 49.Sim, B. K., C. E. Chitnis, K. Wasniowska, T. J. Hadley, and L. H. Miller. 1994. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264:1941-1944. [DOI] [PubMed] [Google Scholar]
- 50.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434:214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spielmann, T., D. J. Fergusen, and H. P. Beck. 2003. Etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol. Biol. Cell 14:1529-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spielmann, T., D. L. Gardiner, H. P. Beck, K. R. Trenholme, and D. J. Kemp. 2006. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol. Microbiol. 59:779-794. [DOI] [PubMed] [Google Scholar]
- 53.Stubbs, J., K. M. Simpson, T. Triglia, D. Plouffe, C. J. Tonkin, M. T. Duraisingh, A. G. Maier, E. A. Winzeler, and A. F. Cowman. 2005. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science 309:1384-1387. [DOI] [PubMed] [Google Scholar]
- 54.Taylor, H. M., M. Grainger, and A. A. Holder. 2002. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 70:5779-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topolska, A. E., A. Lidgett, D. Truman, H. Fujioka, and R. L. Coppel. 2004. Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J. Biol. Chem. 279:4648-4656. [DOI] [PubMed] [Google Scholar]
- 56.Triglia, T., M. T. Duraisingh, R. T. Good, and A. F. Cowman. 2005. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol. Microbiol. 55:162-174. [DOI] [PubMed] [Google Scholar]
- 57.Vontas, J., I. Siden-Kiamos, G. Papagiannakis, M. Karras, A. P. Waters, and C. Louis. 2005. Gene expression in Plasmodium berghei ookinetes and early oocysts in a co-culture system with mosquito cells. Mol. Biochem. Parasitol. 139:1-13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.