Abstract
Hypocrea jecorina (anamorph: Trichoderma reesei) can grow on plant arabinans by the aid of secreted arabinan-degrading enzymes. This growth on arabinan and its degradation product l-arabinose requires the operation of the aldose reductase XYL1 and the l-arabinitol dehydrogenase LAD1. Growth on arabinan and l-arabinose is also severely affected in a strain deficient in the general cellulase and hemicellulase regulator XYR1, but this impairment can be overcome by constitutive expression of the xyl1 encoding the aldose reductase. An inspection of the genome of H. jecorina reveals four genes capable of degrading arabinan, i.e., the α-l-arabinofuranosidase encoding genes abf1, abf2, and abf3 and also bxl1, which encodes a β-xylosidase with a separate α-l-arabinofuranosidase domain and activity but no endo-arabinanase. Transcriptional analysis reveals that in the parent strain QM9414 the expression of all of these genes is induced by l-arabinose and to a lesser extent by l-arabinitol and absent on d-glucose. Induction by l-arabinitol, however, is strongly enhanced in a Δlad1 strain lacking l-arabinitol dehydrogenase activity and severely impaired in an aldose reductase (Δxyl1) strain, suggesting a cross talk between l-arabinitol and the aldose reductase XYL1 in an α-l-arabinofuranosidase gene expression. Strains bearing a knockout in the cellulase regulator xyr1 do not show any induction of abf2 and bxl1, and this phenotype cannot be reverted by constitutive expression of xyl1. The loss of function of xyr1 has also a slight effect on the expression of abf1 and abf3. We conclude that the expression of the four α-l-arabinofuranosidases of H. jecorina for growth on arabinan requires an early pathway intermediate (l-arabinitol or l-arabinose), the first enzyme of the pathway XYL1, and in the case of abf2 and bxl1 also the function of the cellulase regulator XYR1.
The recently reinitiated interest in second-generation biofuel production (i.e., from renewable plant material whose use does not compete with use for food and feed production) also led to a renaissance in cellulase and hemicellulase production by the ascomycete Hypocrea jecorina (anamorph Trichoderma reesei), the current best producer of these enzymes (27). The efficient use of these enzymes for plant biomass hydrolysis, however, is still limited by several factors, including incomplete knowledge of regulation of production of these enzymes.
The l-arabinose polymer arabinan is another polysaccharide found in plant cell wall heteropolysaccharides as a side chain of pectin (9), and l-arabinose is particularly abundant in the form of glucuronoarabinoxylans in monocotyledon primary cell walls of Commelinoid flowery plants and as arabinoxylans in cereal grains, where it can mount up to 25 to 30% (4, 11). H. jecorina is also able to degrade arabinan to l-arabinose by an arabinanolytic system. To date, two arabinofuranosidases have been characterized: an α-l-arabinofuranosidase (ABF1) (26) and a β-xylosidase which has a separate α l-arabinofuranosidase domain and activity (23). Two further α-l-arabinofuranosidases (ABF2 and ABF3) have been found during genome sequence analysis (14, 27), and a recent analysis of the secretome of two high-producer strains (22).
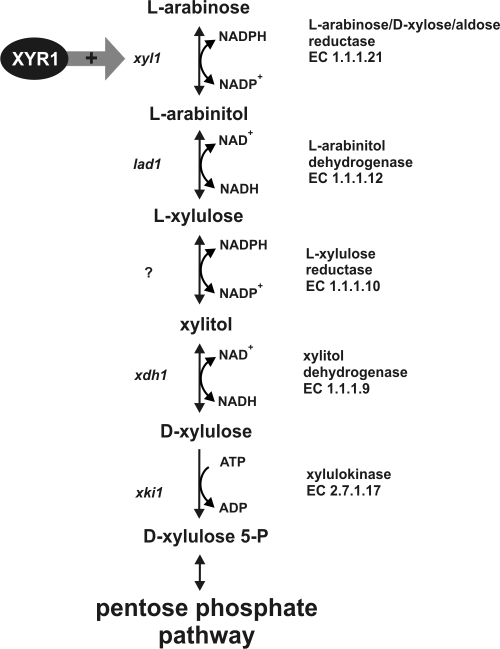
The l-arabinose formed as a consequence of the attack by α-l-arabinofuranosidases is metabolized via the fungal specific l-arabinose catabolic pathway (Fig. 1), which converts l-arabinose to d-xylulose 5-phosphate. The latter is then metabolized via the nonoxidative pentose phosphate pathway. Three genes involved in this pathway have been cloned from H. jecorina: a d-xylose/aldose reductase XYL1, which is not only responsible for d-xylose reduction but also responsible for the reduction of l-arabinose to l-arabinitol (37); an l-arabinitol dehydrogenase LAD1 (30, 33), which converts l-arabinitol to l-xylulose; and a xylitol dehydrogenase XDH1, which oxidizes xylitol to d-xylulose (38). LAD1 can partly replace the function of XDH1 in a Δxdh1 strain (38). A putative l-xylulose reductase has also been cloned (34), but recent investigations showed that its knockout does not affect l-arabinose utilization and that the enzyme is in fact a d-mannitol dehydrogenase (28). The l-xylulose reductase, which is functional in vivo, has not been cloned yet.
FIG. 1.
l-Arabinose catabolic pathway of fungi. The question mark indicates a step for which the respective gene has not been found yet (see, for example, the study by Metz et al. [28]).
Little is thus far known about the regulation of the arabinanase system in H. jecorina. In Aspergillus niger, arabinanolytic genes are specifically and coordinately induced when A. niger is grown on arabinan-containing substrates or the monomeric compounds l-arabinose and l-arabinitol (13, 16, 17, 32, 44, 45). Since the accumulation of intracellular l-arabinitol correlates with higher production of the enzymes involved in arabinan breakdown in A. nidulans, de Vries and Visser (9) hypothesized that l-arabinitol could be the true inducer of this system. de Groot et al. (8) identified two loci (araA and araB) in A. niger, which seem to contain positive-acting genes involved in the expression of arabinanase-encoding genes and the genes encoding the enzymes of the l-arabinose catabolic pathway. However, the genes corresponding to these two loci have not been cloned yet.
The Zn(II)2-Cys6 binuclear cluster protein XYR1/XlnR has been shown to be a transcriptional activator of cellulase and xylanase gene transcription in H. jecorina and different Aspergillus spp. (for a review, see reference 43). However, XlnR has been reported not to regulate arabinan and l-arabinose metabolism in the Aspergilli (8). In analogy, Stricker et al. (42) reported that a xyr1 knock out in H. jecorina did not affect growth of the fungus on l-arabinose and also l-arabinose reductase activities. However, expression of the H. jecorina d-xylose/aldose reductase XYL1 is controlled by XYR1 during growth on d-xylose (41) and a xyl1 knockout results in strongly impaired growth on l-arabinose due to the major involvement of XYL1 in the total l-arabinose reductase activity in this strain (37).
These potentially conflicting data prompted us to perform a more detailed investigation on the possibility of regulation of arabinan and l-arabinose metabolism by XYR1 and the involvement of XYL1 in arabinan and l-arabinose catabolism. We present here clear evidence that XYR1 is essential for the catabolism of arabinan and l-arabinose by directly regulating XYL1 and two α-l-arabinofuranosidases (ABF2 and BXL1), and by indirectly regulating also ABF1 and ABF3 via XYL1. In addition, we show that l-arabinose/l-arabinitol and XYL1 are also essential for induction to occur.
MATERIALS AND METHODS
Strains and culture conditions.
The fungal strains used in the present study were all derived from H. jecorina (T. reesei) QM9414, a moderate cellulase producing mutant strains, and are listed in Table 1. They were maintained on malt extract agar and supplemented with uridine (10 mM) when necessary. Strains were grown in 1-liter flasks on a rotary shaker (250 rpm) at 28°C in 250 ml of the medium described by Mandels and Andreotti (25) with the respective carbon source at a final concentration of 1% (wt/vol). Sugar beet arabinan (Ara:Gal:Rha:GalUA [88:3:2:7]) was purchased from Megazyme (Wicklow, Ireland).
TABLE 1.
Strains used in this study
Escherichia coli strain JM109 (Promega, Madison, WI) was used for plasmid propagation.
Strain constructions.
Construction of strains constitutively expressing the d-xylose reductase gene xyl1 in a xyr1-negative background was performed as follows: 740 bp of the promoter region the H. jecorina tef1 (encoding translation elongation factor 1) were amplified by PCR using the oligonucleotides tef1Xhofw and tef1ClaSalrv. The XhoI/SalI-restricted tef1 fragment was then cloned in the corresponding sites of pLH1hph (20), which contains the E. coli hygromycin B phosphotransferase expression cassette as fungal selection marker. A 1,590-bp xyl1 PCR fragment including the coding and terminator region was amplified with the oligonucleotides xyl1Clafw and xyl1HindIII and inserted downstream of the tef1 promoter region by ClaI/HindIII, resulting in Ptef1-xyl1. Transformation of the Δxyr1 strain with Ptef1-xyl1 was done as described previously (18) by using hygromycin B (50 μg/ml) as a selection agent. The strains were purified twice for mitotic stability, and integration of the Ptef1-xyl1 expression cassette was verified by PCR analysis.
Nucleic acid isolation and hybridization.
Fungal mycelia were harvested by filtration, washed with sterile tap water, shock frozen in liquid nitrogen, and ground to a fine powder. DNA and RNA extraction were done as described previously (39). Standard methods (36) were used for DNA electrophoresis, blotting, and hybridization of RNA. Probes were amplified by PCR (Table 2) and labeled with [α-32P]dCTP by random priming.
TABLE 2.
Oligonucleotides used in this study
| Gene | Primer | Sequence (5′-3′)a | Size (bp) |
|---|---|---|---|
| bxl1 | bXYLfw | GACACTTGCCACGCTCACAC | |
| bXYLrv | CGAAGGTGAAGACGGGAATC | 1,922 | |
| abf1 | aBF1rv | AGCTTCCCTGGCCGTTAATG | |
| ABF1fw | AGGCTCTCTCGTTGCTGCTG | 1,310 | |
| abf2 | aBF2fw | TGCCGTTGTGCTGAGCTTTG | |
| aBF2rv | TGGTCGATAGGGCAAGAGGC | 925 | |
| abf3 | aBF3fw | GTGACATCTACGCATCTGG | |
| aBF3rv | GTTGAAGTGACGCCAGTAG | 1,346 | |
| 18S rRNA | 18sRF | GGTGGAGTGATTTGTCTG | |
| 18sRF | CTTACTAGGGATTCCTCG | 300 | |
| tef1 | Tef1Xhofw | GCCTCGAGGGACAGAATGTAC | |
| Tef1ClaSalrv | TAGTCGACATCGATGACGGTTTGTGTGATGTAGCGTG | 740 | |
| xyl1 | xyl1Clafw | TAATCGATGGCGTCTCCCACGCTCAAG | |
| xyl1HindIII | TGAAGCTTGCAGACAGATTGACCGCACAAC | 1,540 |
Restriction sites are underlined.
Determination of fungal growth.
To determine hyphal growth on agar plates, plates were inoculated with a small piece of agar in the center of an 11-cm plate, and the increase in colony diameter was measured daily twice. To measure growth in submerged cultures, the increase in dry biomass was recorded. To this end, mycelia were harvested after appropriate times, washed extensively with distilled water, and dried to constant weight in an oven at 80°C. The data shown are the average of the three separate biological experiments, which deviated by not more than ±15%.
High-pressure liquid chromatography analysis of intracellular l-arabinose.
The high-pressure liquid chromatography analysis was performed essentially as described previously (28), but with 70% 5 mM sulfuric acid and 30% acetonitrile at 65°C in the eluent to improve the separation of the l-arabinose and l-arabinitol. For estimation of the intracellular concentration, an intracellular volume of 2.4 ml/g of dry biomass was used (41).
Phylogenetic analysis.
DNA and protein sequences were visually aligned by using Genedoc 2.6 (29). Phylogenetic trees were constructed by the neighbor-joining method (35), using the computer program MEGA, version 4.0 (24). Unalignable N- and C-terminal regions in the amino acid sequences were omitted from the analyses, and gaps and missing data were pairwise deleted.
RESULTS
Growth of H. jecorina on arabinan and l-arabinose is dependent on the function of the d-xylose reductase XYL1 and the l-arabinitol dehydrogenase LAD1.
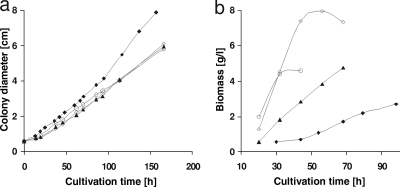
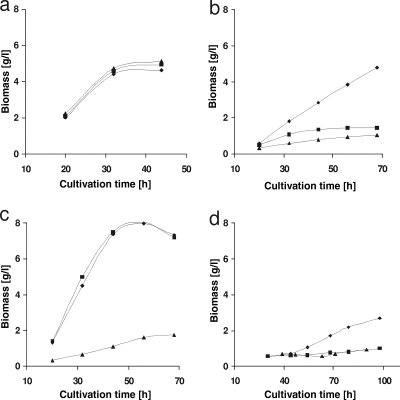
H. jecorina is able to grow on sugar beet arabinan or its major constituent l-arabinose, both on plates and in submerged cultures (Fig. 2). In order to assess that this growth is in fact due to catabolism of the l-arabinose content of arabinan and to assess the involvement of the thus-far-characterized enzymes of the l-arabinose catabolic pathway (see Fig. 1), we compared growth on arabinan with that on l-arabinose in strains deleted in the first two steps of the metabolic pathway: an XYL1-deficient strain (Δxyl1), which is strongly impaired in its ability to convert l-arabinose to l-arabinitol (37), and a strain deficient in the l-arabinitol dehydrogenase LAD1 (Δlad1), which is impaired in its ability to convert l-arabinitol to l-xylulose (30). Growth in submerged cultures of these strains on d-glucose was indistinguishable from that of the parent strain (Fig. 3a). Growth on l-arabinose, however, was different: growth of the Δxyl1 and the Δlad1 strains was strongly reduced (Fig. 3b). Biomass formation in submerged cultures on l-arabinitol was only affected in the Δlad1 strain and occurred normally in the Δxyl1 strain (Fig. 3c). The Δlad1 and Δxyl1 strains were almost unable to grow on arabinan at all (Fig. 3d). These findings are consistent with the assumption that growth on sugar beet arabinan is mainly due to the catabolism of l-arabinose via l-arabinitol and l-xylulose and involves XYL1 and LAD1.
FIG. 2.
Radial growth of H. jecorina on various carbon sources on plates (a) and biomass production in submerged cultures (b). Carbon sources are indicated as follows: d-glucose (○), l-arabinose (▴), l-arabinitol (⋄), and arabinan (⧫), all at 1% (wt/vol).
FIG. 3.
Growth of H. jecorina QM9414 and strains deleted in different steps of the l-arabinose catabolic pathway on d-glucose (a), l-arabinose (b), l-arabinitol (c), and arabinan (d). Strains are indicated as follows: H. jecorina QM9414 (⧫), Δxyl1 (▪), and Δlad1 (▴).
Growth of H. jecorina on arabinan and l-arabinose but not l-arabinitol is dependent on the function of the cellulase regulator XYR1.
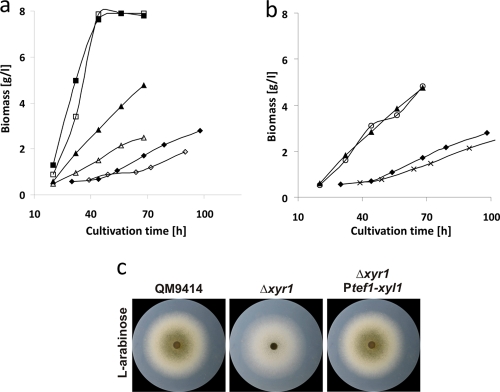
Deletion of xyr1 encoding the Zn(II)2Cys6 regulator XYR1 has recently been shown to strongly affect growth on d-xylose but not on l-arabinose (42). In our experiment, however, the Δxyr1 strain showed strongly reduced growth on l-arabinose and arabinan in submerged cultures, whereas growth of this Δxyr1 strain on l-arabinitol was similar to that of the parent strain QM9414 (Fig. 4a). Cell extracts of the l-arabinose grown Δxyr1 strain showed that l-arabinose reductase activity was reduced to ca. 10% in this strain compared to QM9414. This is consistent with our previous results that XYL1 is responsible for the major l-arabinose reductase activity during growth on l-arabinose and our growth data (presented above) that XYL1 is necessary for rapid growth on l-arabinose. In conclusion, these data suggest that xyr1 regulates the XYL1 step but not later steps in l-arabinose catabolism.
FIG. 4.
Effect of the loss of XYR1 on growth of H. jecorina on l-arabinose and l-arabinitol. (a) Growth of H. jecorina QM9414 (solid symbols) and Δxyr1 (open symbols) on l-arabinose (triangles), l-arabinitol (squares), and arabinan (diamonds) as the carbon source. (b) Growth of H. jecorina QM9414 (▴, ⧫) and Ptef1:xyl1/Δxyr1 (○, ×) strains on l-arabinose and arabinan, respectively. (c) Growth of H. jecorina QM9414, Δxyr1, and Ptef1:xyl1/Δxyr1 strains on plates containing l-arabinose as the sole carbon source.
In order to support our assumption that the growth on l-arabinose is due to a direct regulation of the aldose reductase XYL1 by XYR1, we constructed strains carrying a constitutively expressed xyl1 gene in an xyr1-negative background (see Materials and Methods). The tef1 (translation elongation factor alpha encoding gene) promoter was used for this purpose, because it allows a high level of expression on a wide variety of carbon sources, including l-arabinose and l-arabinitol (unpublished data). The data, shown in Fig. 4b, indicate that wild-type growth was recovered in these strains, indicating that the effect of XYR1 on l-arabinose catabolism is via the regulation of XYL1. In contrast, during growth on arabinan, the Δxyr1 strain with the constitutively expressed xyl1 gene was not able to reach the growth of the parent strain, which points to a regulation of the arabinanolytic system by XYR1.
α-l-Arabinofuranosidase gene expression in H. jecorina.
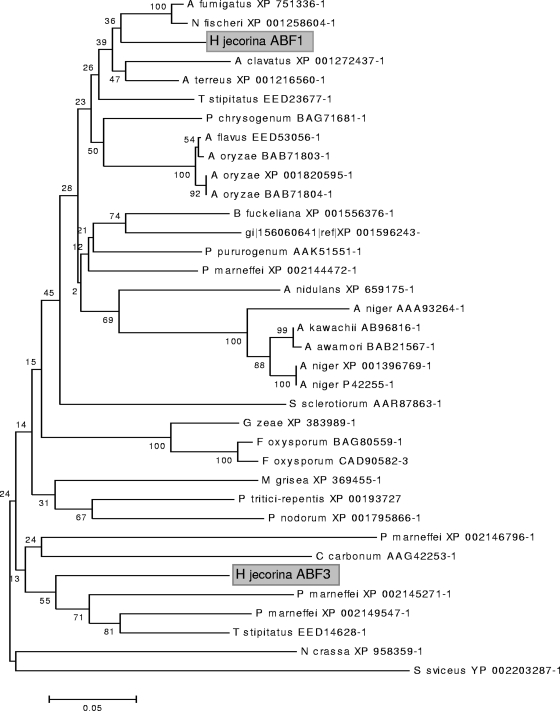
The genome of H. jecorina contains four genes that encode enzymes with α-l-arabinofuranosidase activity, i.e., abf1, abf2, and abf3 and the β-xylosidase-encoding gene bxl1, which has a separate α-l-arabinofuranosidase domain (23, 27). Interestingly, we were not able to detect any orthologue of an endo-arabinanase gene as described for Aspergillus niger (12) in the H. jecorina QM6a genome (http://genome.jgi-psf.org/Trire2/Trire2.home.html). ABF1 and ABF3 are members of glycosyl hydrolase family 54 and contain a carbohydrate-binding module of family 42. They seem to be the result of a gene duplication event which took place early in the evolution of the Hypocreaceae, but the ABF3 orthologues have subsequently been lost from other genera, such as Gibberella/Fusarium (Fig. 5).
FIG. 5.
Phylogenetic analysis of H. jecorina ABF1 and ABF3. The other sequences used were identified as those showing highest similarity in BLAST. Neighbor joining was used, and gaps were not included. Numbers over the branches represent bootstrap coefficients from 1,000 replicas.
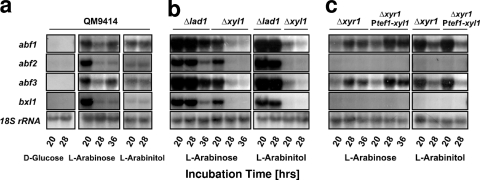
Transcription of all four genes occurred during growth on l-arabinose and l-arabinitol and was absent during growth on glucose (Fig. 6a). This is consistent with the findings that we could not detect any α-l-arabinofuranosidase activity in the culture filtrate or associated with cell walls during growth on glucose (unpublished data). Maximal abundance of transcripts was similar on l-arabinose and l-arabinitol, although initial transcript levels were higher on l-arabinose.
FIG. 6.
α-l-Arabinofuranosidase gene transcription in the parental strain QM9414 (a), strains deleted in the first two steps of the l-arabinose pathway (Δxyl1 and Δlad1) (6), and strains deleted in the regulator XYR1 (Δxyr1) or expressing xyl1 constitutively in an XYR1-negative background (Δxyr1 and Ptef1-xyl1) (c). A total of 20 μg of total RNA was loaded per lane, and α-32P-radiolabeled PCR fragments of the different genes were used as probes.
Metabolic modulation of α-l-arabinofuranosidase gene expression in H. jecorina.
In order to learn whether changes in the metabolism of l-arabinose would influence the transcript levels of abf1, abf2, abf3, and bxl1, we compared their formation in the Δxyl1 and the Δlad1 strains. In the Δxyl1 strain, the expression of abf2 and bxl1 in the presence of l-arabinitol was completely abolished, whereas a very weak expression of abf1 and abf3 still occurred. Transcript formation in the presence of l-arabinose displayed essentially the same picture, with the exception that the transcripts were still present at an earlier time point (20 h) (Fig. 6b).
In the Δlad1 strain, in contrast, all four transcripts were significantly more abundant than in the parent strain QM9414 and were highest when induced by l-arabinitol (Fig. 6b).
Regulation of α-l-arabinofuranosidase gene expression by XYR1.
To test whether the xylanase and cellulase regulator XYR1 would be involved in the regulation of α-l-arabinofuranosidase gene expression in H. jecorina, transcript levels during growth on l-arabinose and l-arabinitol were also investigated in a Δxyr1 and the Δxyr1 Ptef1:xyl1 strains (to rule out indirect effects via XYL1). As shown in Fig. 6c, induction of abf2 and bxl1 by l-arabinose was completely impaired in the Δxyr1 strain, whereas expression of abf1 and abf3 took place, although initial transcript levels were reduced during growth on l-arabinose. When l-arabinitol was used as inducing carbon source, abf2 and bxl1 were again not expressed in the Δxyr1 background, whereas abf1 and abf3 were expressed stronger in the initial phase of growth on l-arabinitol.
In order to test which of these effects is direct (i.e., not caused by the regulation of XYL1), we repeated these experiments with the Δxyr1 Ptef1:xyl1 strain. The expression of abf2 and bxl1 was again completely impaired on both l-arabinose and l-arabinitol, indicating that these effect is due to a loss of XYR1 and can be explained by a direct action of XYR1 on the regulation of both genes. In contrast, the expression of abf1 and abf3 in the Δxyr1 Ptef1:xyl1 strain was higher compared to the Δxyr1 strain on l-arabinose but not affected during growth on l-arabinitol, thus demonstrating that the expression of XYL1 had a positive effect on induction of these two genes.
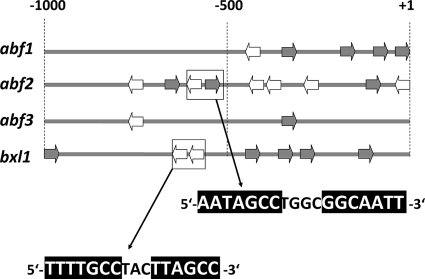
The fact that two of the genes (abf2 and bxl1) were apparently directly regulated by XYR1 prompted us to examine whether we could also detect the respective XYR1 binding sites [5′-GGC(A/T)(3-4)-3′] (15, 40) in the 5′ upstream regions of these two, but not the other two (abf1 and abf3) genes (Fig. 7). abf2 and bxl1 differed in two respects. First, the two exhibited a higher number of consensus binding motifs in the first 1,000 bp upstream of the start ATG (9 and 7 for abf2 and bxl1 versus 5 for abf1 and 2 for abf3, respectively). Second, however, only abf2 and bxl1 contained the consensus binding sites as a divergent (abf2) and tandem (bxl1) repeat, separated by 3 and 4 nucleotides.
FIG. 7.
Occurrence of the XYR1-binding consensus GGC(T/A)4 (15) in the 5′ upstream region of abf1, abf2, abf3, and bxl1. The presence of the consensus is indicated by an arrow (not drawn to scale). Gray arrows indicate the presence of the motif on the sense strand, and open arrows indicate the motif on the antisense strand. The sequences of the two connected motifs are shown below.
DISCUSSION
As a saprobe specialized to feed on decaying plant material, H. jecorina must contain genes whose products enable a synergistic and efficient usage of the available polymers, i.e., cellulose and hemicelluloses. To degrade arabinan, a component belonging to the latter, H. jecorina has three α-l-arabinofuranosidases (ABF1, ABF2, and ABF3) and a single β-xylosidase (BXL1) with a separate α-l-arabinofuranosidase domain. The lack of endo-arabinanase-encoding genes indicates that, in contrast to other fungi such as Aspergillus spp. (9), H. jecorina is likely specialized on the degradation of the single and short l-arabinose side chains present in different hemicelluloses but seems to have problems with longer side chains as present in pectins. The presence of cellulose-binding domains in ABF1 and ABF3 suggests a probable link between cellulose and arabino-oligosaccharide degradation: cellulose microfibrils are known to be tethered through noncovalent interactions with matrix polysaccharides that are involved in the primary cell wall assembly (21). This binding is brought about by the arabinan, xylan, and galactan oligosaccharide side chains of the pectins and, particularly, the xyloglucan contains terminal l-arabinose residues (47). The fact that H. jecorina can only slowly grow on arabinan polymers (e.g., slower than on cellulose or xylan), as shown here, is consistent with this view and illustrates that an exclusive exo-attack is poorly efficient in the degradation of a polymer. It is possible that hemicelluloses with terminal l-arabinose residues are the preferred carbon sources for H. jecorina in its natural habitat.
Although the inducible formation of ABF1, ABF2, and BXL1 by sophorose and lactose has already been reported earlier (14), ABF3 has only recently received attention (22). Interestingly, ABF3 appears to be a relict of an early clade in the evolution of the GH54 α-l-arabinofuranosidases, only present also in Penicillium marneffei and Talaromyces stipitatus. However, the phylogenetic origin of ABF3 is not in accordance with the species phylogeny. This may be due to either a history of horizontal gene transfer or operation of a birth and death mechanism. At this stage, no clear answer can be offered. We note, however, that among the sordariomycetes H. jecorina is the only fungus that contains two genes of the GH54 family.
The l-arabinose, resulting from the operation of ABF1-3 and BXL1, is taken up into the hyphae and metabolized via the reductive pentose catabolic pathway (Fig. 1). In A. niger, de Groot et al. (7) reported on a separate l-arabinose reductase with little activity on d-xylose. Similarly, Stricker et al. (42) provided evidence for the presence of a specific l-arabinose reductase in H. jecorina, albeit using activity assays in cell extracts only. We have provided here clear evidence that the XYL1 aldose reductase, which is primarily responsible for catalyzing the first reductive reaction on d-xylose, l-arabinose, and d-galactose catabolism (37), is also responsible for arabinan degradation. Thus, in contrast to the aspergilli, H. jecorina contains a single enzyme to catalyze the first step in the degradation of different hemicellulose sugars. This appears to be beneficial for the performance of H. jecorina in its habitat, where l-arabinose is not available in isolation but always accompanied by d-xylose.
No α-l-arabinofuranosidase activity was detected during growth on d-glucose (unpublished data), and the respective transcripts of abf1, abf2, abf3, and bxl1 were also absent. We have not investigated whether this is a consequence of carbon catabolite repression or lack of the inducer (or both), but we note that all genes were transcribed during growth on l-arabinose and (less) on l-arabinitol. However, in a Δlad1 strain, which lacks the l-arabinitol dehydrogenase needed for the conversion of l-arabinitol to l-xylulose, the formation by l-arabinitol and l-arabinose was greatly increased. This is consistent with the hypothesis proposed for A. niger (9, 44) that l-arabinitol may be the true intracellular inducer. We note, however, that a possible action of l-arabinose cannot be rejected from our data, which is supported by our findings that l-arabinose can be formed by the reverse reaction of XYL1 during growth on l-arabinitol. Under these conditions, the parent strain QM9414 accumulated an intracellular concentration of 0.18 to 0.4 mM l-arabinose, and this value was decreased to 0.19 to 0.20 mM in the Δxyl1 and elevated to 0.8 to 0.95 mM in the Δlad1 strain (data not shown). Interestingly, the transcription during growth on l-arabinitol (or l-arabinose) was almost completely abolished in the absence of the aldose reductase XYL1 after the initial phase of growth. Since xyl1 is dispensable for growth on l-arabinitol, this effect must therefore be a regulatory one. De Groot et al. (8) reported that the induction of arabinan-degrading enzymes by l-arabinitol in A. niger depends on the function of the products of the araA and araB genes, which positively regulate their expression and also that of the genes encoding the l-arabinose catabolic pathway. Unfortunately, the respective genes have not yet been identified. Compared to the findings of the present study, we cannot rule out that one of them (or both) would indeed encode an l-arabinose reductase, since strongly decreased l-arabinose reductase activities were found in the araA and araB mutants (8). Alternatively, if these genes indeed encode a positive regulator of the expression of an l-arabinose reductase gene, this would result in a deficiency of l-arabinose reductase activity and, when applying our findings of a cross talk by l-arabinitol/l-arabinose and XYL1 to A. niger, this would also, albeit indirectly, block α-l-arabinofuranosidase formation. Although the mechanism of the cross talk of XYL1 and the inducer in H. jecorina still needs to be investigated, the function of metabolic enzymes as transcriptional (co)regulators would not be without precedent (3, 5, 19), and regulatory roles of aldose reductases have been described in mammalian cells (1, 31), albeit without explaining the underlying mechanism. To the best of our knowledge, no similar finding has been made in a filamentous fungus, thus rendering H. jecorina and l-arabinitol regulation an attractive subject for further research in this field.
The Zn(II)2Cys6-type transcription factor XYR1 has previously been shown to be centrally involved in the expression of the cellulolytic and hemicellulolytic enzymes of H. jecorina (42). Although XYR1 is an orthologue of A. niger XlnR, the main transcriptional activator of cellobiohydrolase- and xylanase-encoding genes (16, 46), the molecular mechanisms of transcriptional activation in H. jecorina are different and generally more differentiated (see, for example, reference 43). In A. niger, growth on arabinan and l-arabinose are not regulated by XlnR (9). Growth of H. jecorina on l-arabinose has recently been reported to be unaffected in a Δxyr1 strain of H. jecorina (2). In contrast, we show here that biomass accumulation of H. jecorina on arabinan and l-arabinose is clearly affected in the Δxyr1 strain, although the radial growth is similar. However, this appeared to be due to the positive regulation of the aldose reductase by XYR1 (37, 42) because a strain in which the formation of XYL1 was constitutive and uncoupled from xyr1 regulation no longer showed this reduction on growth. The reason why this effect escaped the detection by Stricker et al. (42) is probably due to the use of agar plates for quantification of growth, which can lead to erroneous interpretations due to overlooking the different density of the mycelial mat formed on the plates (cf. Fig. 2a and 4c).
Expression of two of the α-l-arabinofuranosidase genes (abf1 and abf3) was only slightly affected in a Δxyr1 strain. This indicates that the triggering of their expression by l-arabinitol/l-arabinose and XYL1 requires another transcriptional regulator. In contrast, however, abf2 and bxl1 clearly were under the control of XYR1, and this control even overruled the positive action of l-arabinitol/l-arabinose and XYL1, thus showing that the two require xyr1 for expression. The identification of regulation of these two genes by XYR1 is also consistent with the detection of a higher number of copies of the XYR1-binding consensus GGC(T/A)4 (15, 40) in their promoters than in abf1 and abf3. In addition, only abf2 and bxl1 contained this motif in the form of a divergent and tandem repeat, respectively. Furukawa et al. (15) concluded that both single and double motifs are functional in H. jecorina. Our findings of the regulation by the general cellulase and hemicellulase regulator XYR1 are also supported by the data of Foreman et al. (14), who found elevated transcripts for bxl1 and abf2 (although the latter only very weakly) on sophorose and lactose in H. jecorina QM6a. Thus, in contrast to aspergilli, which have developed more differentiated mechanisms to adapt their regulation of polysaccharide hydrolases to the presence of available substrates (9), H. jecorina also induces at least part of the arabinan-degrading enzymes simultaneously with the other enzymes required for cellulose and hemicellulose degradation. The simplest model which can be designed would therefore postulate a complex of l-arabinitol or l-arabinose, XYL1, and XYR1 to function in the regulation of abf2 and bxl1 gene expression. Further, our data are consistent with the speculation that, for abf1 and abf3, the part played by XYR1 is overtaken by a different transcriptional factor that is as yet unknown.
Acknowledgments
This study was supported by grants from the Austrian Science Foundation (P19690) to C.P.K. and (P19421) to B.S.
We are grateful to R. L. Mach for providing the Δxyr1 strain for these studies.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Alexiou, P., K. Pegklidou, M. Chatzopoulou, I. Nicolaou, and V. J. Demopoulos. 2009. Aldose reductase enzyme and its implication to major health problems of the 21(st) century. Curr. Med. Chem. 16:734-752. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., B. Roger, R. E. Kingston, D. D. Moore, J. G. Seidman, J. Smith, and K. Struhl. 2006. Current protocols in molecular biology. Greene Publishing Associates/Wiley Interscience, New York, NY.
- 3.Bhardwaj, A., and M. F. Wilkinson. 2005. A metabolic enzyme doing double duty as a transcription factor. Bioessays 27:467-471. [DOI] [PubMed] [Google Scholar]
- 4.Carpita, N. C. 1996. Structure and biogenesis of the cell walls of grasses. Annu. Rev. Physiol. Plant Mol. Biol. 47:445-476. [DOI] [PubMed] [Google Scholar]
- 5.Cho, Y. H., S. D. Yoo, and J. Sheen, J. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127:579-589. [DOI] [PubMed] [Google Scholar]
- 6.Reference deleted.
- 7.de Groot, M. J., W. Prathumpai, J. Visser, and G. J. Ruijter. 2005. Metabolic control analysis of Aspergillus niger l-arabinose catabolism. Biotechnol. Prog. 21:1610-1616. [DOI] [PubMed] [Google Scholar]
- 8.de Groot, M. J., P. J. van de Vondervoort, R. P. de Vries, P. A. van Kuyk, G. J. Ruijter, and J. Visser. 2003. Isolation and characterization of two specific regulatory Aspergillus niger mutants shows antagonistic regulation of arabinan and xylan metabolism. Microbiology 149:1183-1191. [DOI] [PubMed] [Google Scholar]
- 9.de Vries, R. P., and J. Visser. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 64:497-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Ebringerová, A., and T. Heinze. 2000. Xylan and xylan derivatives: biopolymers with valuable properties. I. Naturally occurring xylans structures, isolation procedures and properties. Macromol. Rapid Commun. 21:542-556. [Google Scholar]
- 12.Flipphi, M. J., H. Panneman, P. van der Veen, J. Visser, and L. H. de Graaff. 1993. Molecular cloning, expression and structure of the endo-1,5-α-l-arabinanase gene of Aspergillus niger. Appl. Microbiol. Biotechnol. 40:318-326. [DOI] [PubMed] [Google Scholar]
- 13.Flipphi, M. J. A., J. Visser, P. van der Veen, and L. H. Graaff. 1994. Arabinanase gene expression in Aspergillus niger: indications for coordinated gene expression. Microbiology 140:2673-2682. [DOI] [PubMed] [Google Scholar]
- 14.Foreman, P. K., D. Brown, L. Dankmeyer, R. Dean, S. Diener, N. S. Dunn-Coleman, F. Goedegebuur, T. D. Houfek, G. J. England, A. S. Kelley, H. J. Meerman, T. Mitchell, C. Mitchinson, H. A. Olivares, P. J. Teunissen, J. Yao, and M. Ward. 2003. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 278:31988-31997. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa, T., Y. Shida, N. Kitagami, K. Mori, M. Kato, T. Kobayashi, H. Okada, W. Ogasawara, and Y. Morikawa. 2009. Identification of specific binding sites for XYR1, a transcriptional activator of cellulolytic and xylanolytic genes in Trichoderma reesei. Fungal Genet. Biol. 46:564-574. [DOI] [PubMed] [Google Scholar]
- 16.Gielkens, M. M. C., L. Gonzalez-Candelas, P. Sanchez-Torres, P. J. I. van de Vondervoort, L. H. de Graaff, J. Visser, and D. Ramon. 1999. The abfB gene encoding the major l-arabinofuranosidase of Aspergillus nidulans: nucleotide sequence, regulation, and construction of a disrupted strain. Microbiology 145:735-741. [DOI] [PubMed] [Google Scholar]
- 17.Gielkens, M. M. C., J. Visser, and de L. H. Graaff. 1997. Arabinoxylan degradation by fungi: characterisation of the arabinoxylan arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr. Genet. 31:22-29. [DOI] [PubMed] [Google Scholar]
- 18.Gruber, F., J. Visser, C. P. Kubicek, and L. De Graaff. 1990. Cloning of the Trichoderma reesei pyrG gene and its use as a homologous marker for a high-frequency transformation system. Curr. Genet. 18:451-456. [DOI] [PubMed] [Google Scholar]
- 19.Hall, D. A., H. Zhu, X. Zhu, T. Royce, M. Gerstein, and M. Snyder. 2004. Regulation of gene expression by a metabolic enzyme. Science 306:482-484. [DOI] [PubMed] [Google Scholar]
- 20.Hartl, L., C. P. Kubicek, and B. Seiboth. 2007. Induction of the Leloir pathway in the fungus Hypocrea jecorina involves no transcriptional inducer function of the galactokinase. J. Biol. Chem. 282:18654-18659. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi, T., M. P. F. Marsden, and D. P. Delmar. 1987. Pea xyloglucan and cellulose. V. Xyloglucan-cellulose interactions in vitro and in vivo. Plant Physiol. 83:384-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herpoël-Gimbert, I., A. Margeot, A. Dolla, G. Jan, D. Mollé, S. Lignon, H. Mathis, J. C. Sigoillot, F. Monot, and M. Asther. 2008. Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels 23:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrmann, M. C., M. Vrsanska, M. Jurickova, J. Hirsch, P. Biely, and C. P. Kubicek. 1997. The β-d-xylosidase of Trichoderma reesei is a multifunctional β-d-xylan-xylohydrolase. Biochem. J. 321:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 25.Mandels, M. M., and R. E. Andreotti. 1978. The cellulose to cellulase fermentation. Proc. Biochem. 13:6-13. [Google Scholar]
- 26.Margolles-Clark, E., M. Tenkanen, T. Nakari-Setälä, and M. Penttilä. 1996. Cloning of genes encoding α-l-arabinofuranosidase and β-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 10:3840-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, D., R. M. Berka, B. Henrissat, M. Saloheimo, M. Arvas, S. E. Baker, J. Chapman, O. Chertkov, P. M. Coutinho, D. Cullen, E. G. Danchin, I. V. Grigoriev, P. Harris, M. Jackson, C. P. Kubicek, C. S. Han, I. Ho, L. F. Larrondo, A. L. de Leon, J. K. Magnuson, S. Merino, M. Misra, B. Nelson, N. Putnam, B. Robbertse, A. A. Salamov, M. Schmoll, A Terry, N. Thayer, A. Westerholm-Parvinen, C. L. Schoch, J. Yao, R. Barabote, M. A. Nelson, C. Detter, D. Bruce, C. R. Kuske, G. Xie, P. Richardson, D. S. Rokhsar, S. M. Lucas, E. M. Rubin, N. Dunn-Coleman, M. Ward, and T. S. Brettin. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:553-560. [DOI] [PubMed] [Google Scholar]
- 28.Metz, B., R. P. de Vries, S. Polak, V. Seidl, and B. Seiboth. 2009. The Hypocrea jecorina (syn. Trichoderma reesei) lxr1 gene encodes a d-mannitol dehydrogenase and is not involved in l-arabinose catabolism. FEBS Lett. 583:1309-1313. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas, K. B., and H. B. Nicholas, Jr. 1997. Genedoc: a tool for editing and annotating multiple sequence alignments. NSRBC, Pittsburgh, PA. http://www.psc.edu/biomed/genedoc.
- 30.Pail, M., T. Peterbauer, B. Seiboth, C. Hametner, I. Druzhinina, and C. P. Kubicek. 2004. The metabolic role and evolution of l-arabinitol 4-dehydrogenase of Hypocrea jecorina. Eur. J. Biochem. 271:1864-1872. [DOI] [PubMed] [Google Scholar]
- 31.Ramana, K. V., A. Bhatnagar, and S. K. Srivastava. 2004. Inhibition of aldose reductase attenuates TNF-α-induced expression of adhesion molecules in endothelial cells. FASEB J. 18:1209-1218. [DOI] [PubMed] [Google Scholar]
- 32.Ramon, D., P. van der Veen, and J. Visser. 1993. Arabinan degrading enzymes from Aspergillus nidulans: induction and purification. FEMS Microbiol. Lett. 113:15-22. [DOI] [PubMed] [Google Scholar]
- 33.Richard, P., J. Londesborough, M. Putkonen, N. Kalkkinen, and M. Penttilä. 2001. Cloning and expression of a fungal l-arabinitol 4-dehydrogenase gene. J. Biol. Chem. 276:40631-40637. [DOI] [PubMed] [Google Scholar]
- 34.Richard, P., M. Putkonen, R. Väänänen, J. Londesborough, and M. Penttilä. 2002. The missing link in the fungal l-arabinose catabolic pathway, identification of the l-xylulose reductase gene. Biochemistry 41:6432-6437. [DOI] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 37.Seiboth, B., C. Gamauf, M. Pail, L. Hartl, and C. P. Kubicek. 2007. The major d-xylose reductase of Hypocrea jecorina is necessary for efficient pentose and lactose catabolism and for cellulase induction by lactose. Mol. Microbiol. 66:890-900. [DOI] [PubMed] [Google Scholar]
- 38.Seiboth, B., L. Hartl, M. Pail, and C. P. Kubicek. 2003. d-Xylose metabolism in Hypocrea jecorina: loss of the xylitol dehydrogenase step can be partially compensated for by lad1-encoded l-arabinitol-4-dehydrogenase. Eukaryot. Cell 2:867-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seiboth, B., L. Hartl, M. Pail, E. Fekete, L. Karaffa, and C. P. Kubicek. 2004. The galactokinase of Hypocrea jecorina is essential for cellulase induction by lactose but dispensable for growth on d-galactose. Mol. Microbiol. 51:1015-1025. [DOI] [PubMed] [Google Scholar]
- 40.Shida, Y., T. Furukawa, W. Ogasawara, M. Kato, T. Kobayashi, H. Okada, and Y. Morikawa. 2008. Functional analysis of the egl3 upstream region in filamentous fungus Trichoderma reesei. Appl. Microbiol. Biotechnol. 78:515-524. [DOI] [PubMed] [Google Scholar]
- 41.Slayman, C. W., and E. L. Tatum. 1964. Potassium transport in Neurospora. I. Intracellular sodium and potassium concentrations, and cation requirements for growth. Biophys. Acta 88:578-592. [PubMed] [Google Scholar]
- 42.Stricker, A. R., K. Grosstessner-Hain, E. Würleitner, and R. L. Mach. 2006. Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot. Cell 5:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stricker, A. R., R. L. Mach, and L. H. de Graaff. 2008. Regulation of transcription of cellulases- and hemicellulases-encoding genes in Aspergillus niger and Hypocrea jecorina (Trichoderma reesei). Appl. Microbiol. Biotechnol. 78:211-220. [DOI] [PubMed] [Google Scholar]
- 44.van der Veen, P., M. J. Flipphi, A. G. Voragen, and J. Visser. 1991. Induction, purification and characterisation of arabinanases produced by Aspergillus niger. Arch. Microbiol. 157:23-28. [DOI] [PubMed] [Google Scholar]
- 45.van der Veen, P., M. J. Flipphi, A. G. Voragen, and J. Visser. 1993. Induction of extracellular arabinanases on monomeric substrates in Aspergillus niger. Arch. Microbiol. 159:66-71. [DOI] [PubMed] [Google Scholar]
- 46.van Peij, N. N. M. E., J. Visser, and L. H. de Graaff. 1998. Isolation and analysis of xlnR, encoding a transcriptional activator coordinating xylanolytic expression in Aspergillus niger. Mol. Microbiol. 27:131-142. [DOI] [PubMed] [Google Scholar]
- 47.Zykwinska, A., M. C. Ralet, C. Garnier, and J. F. Thibault. 2005. Evidence for in vitro binding of pectic side chains to cellulose. Plant Physiol. 139:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]