Abstract
We wanted to examine the cellular locations of four Neurospora crassa proteins that transport calcium. However, the structure and distribution of organelles in live hyphae of N. crassa have not been comprehensively described. Therefore, we made recombinant genes that generate translational fusions of putative organellar marker proteins with green or red fluorescent protein. We observed putative endoplasmic reticulum proteins, encoded by grp-78 and dpm, in the nuclear envelope and associated membranes. Proteins of the vacuolar membrane, encoded by vam-3 and vma-1, were in an interconnected network of small tubules and vesicles near the hyphal tip, while in more distal regions they were in large and small spherical vacuoles. Mitochondria, visualized with tagged ARG-4, were abundant in all regions of the hyphae. Similarly, we tagged the four N. crassa proteins that transport calcium with green or red fluorescent protein to examine their cellular locations. NCA-1 protein, a homolog of the SERCA-type Ca2+-ATPase of animal cells, colocalized with the endoplasmic reticulum markers. The NCA-2 and NCA-3 proteins are homologs of Ca2+-ATPases in the vacuolar membrane in yeast or in the plasma membrane in animal cells. They colocalized with markers in the vacuolar membrane, and they also occurred in the plasma membrane in regions of the hyphae more than 1 mm from the tip. The cax gene encodes a Ca2+/H+ exchange protein found in vacuoles. As expected, the CAX protein localized to the vacuolar compartment. We observed, approximately 50 to 100 μm from the tip, a few spherical organelles that had high amounts of tagged CAX protein and tagged subunits of the vacuolar ATPase (VMA-1 and VMA-5). We suggest that this organelle, not described previously in N. crassa, may have a role in sequestering calcium.
All cells maintain intracellular concentrations of calcium at precise levels, typically about 0.1 μM in the cytosol. Calcium is often present at high levels in the environment, significantly above the level that is tolerated within the cell. Nevertheless, high concentrations are maintained in some organelles because calcium has an essential role in signaling physiological processes (3, 7, 29). In root hairs, pollen tubes, and the hyphae of filamentous fungi calcium has been postulated to have a central role in directing the growth at the tips of these cells (30, 32, 34, 38, 41, 49). Investigators have reported that in filamentous fungi the concentration of calcium is highest at the hyphal tip (56, 59). Disruption of the calcium gradient by ionophores inhibits growth (52). Mutations in some genes that affect hyphal morphology, e.g., frost and spray, can be suppressed by raising the concentration of calcium in the medium (4, 16). However, the growth of wild-type strains is not significantly affected by the external concentration of calcium, which suggests that cytosolic calcium is controlled by regulating calcium uptake and release from organelles (36, 55, 59).
The proteins that transport calcium into organelles have been studied extensively in Saccharomyces cerevisiae. In this organism, more than 90% of the intracellular calcium is in the vacuole (19, 22), transported there by a protein that facilitates Ca2+/H+ exchange,Vcx1p, and by a calcium-pumping ATPase, Pmc1p (13, 14, 47). Another calcium-pumping ATPase, Pmr1p, can transport calcium or manganese into the Golgi bodies (1, 51, 57). S. cerevisiae has not been reported to have a calcium-pumping ATPase in the plasma membrane or a SERCA-type ATPase in the endoplasmic reticulum (ER). Pmr1p may have a dual function in Golgi body- and ER-associated processes (20).
In plant and animal cells, three types of calcium-pumping ATPases have been described (3, 7, 9, 58). The PMCA type (most closely related to the Pmc1p ATPase of S. cerevisiae) primarily pumps calcium across the plasma membrane, removing excess calcium from the cytosol. The SERCA type, named by its location in the smooth ER, has a major role in transporting calcium in muscle cells but is also present in the ER in many types of cells. S. cerevisiae has no homolog to the SERCA ATPase. The SPCA type (secretory pathway Ca2+-ATPases) is found in the Golgi bodies and is homologous to the Pmr1p ATPase of S. cerevisiae (3, 42). The mitochondria also contain a significant share of intracellular calcium. No calcium-pumping ATPase has been identified in this organelle, and transport has been hypothesized to occur through a channel protein, driven by the same electrochemical gradient that drives the synthesis of ATP (18, 35). In addition, calcium is sequestered in small vesicles and in lysosomelike compartments, presumably transported by Ca2+/H+ exchange proteins (29).
The availability of the complete genomes for N. crassa and other filamentous fungi has allowed us to assess the number and types of calcium transport proteins in these organisms (62). Focusing on N. crassa, we found that all three types of calcium-pumping ATPases are present. These genes had been identified earlier in a PCR-based search for P-type ATPases (2). The N. crassa gene nca-1 encodes a SERCA type ATPase. The nca-2 and nca-3 genes are closely related to each other and appear to encode PMCA type ATPases. The pmr gene is a SPCA type. We also identified the cax gene as a homolog of VCX1, the gene encoding the Ca2+/H+ exchanger that plays a key role in vacuolar transport in S. cerevisiae.
Our long-term goal is to use these five genes (nca-1, nca-2, nca-3, pmr, and cax) to find out where calcium is localized in cells and how it gets there. We first wanted to determine the intracellular location of each transporter, using proteins tagged with green and red fluorescent proteins (GFP and RFP, respectively). For N. crassa and most other filamentous fungi a comprehensive analysis of the structure and distribution of organelles in living cells is lacking. GFP and fluorescent dyes have been used successfully to examine nuclei and mitochondria (24, 26). Several reports have shown that “the vacuole” is far more dynamic and complex than the textbook presentation of a large spherical organelle (10, 24, 31, 33, 54). Our understanding of the structure and abundance of the ER and the Golgi body is limited.
In this report, we have fused GFP and RFP to proteins predicted to be localized to nuclei, mitochondria, the ER, the Golgi body, and the vacuole. Similarly, we have made GFP- and RFP-tagged forms of each of the five calcium transport proteins described above. We have examined the abundance and structures of the organelles and have observed (as have others) that these change with distance from the hyphal tip. We have tried to determine whether each of the calcium transport proteins is associated with a unique organelle or with the plasma membrane. In studies parallel to those reported here, we are measuring the amount of calcium in cell organelles and characterizing the phenotypes of strains in which the calcium transport genes have been deleted.
MATERIALS AND METHODS
Construction of plasmids.
We used plasmid pMF272 to generate proteins with eGFP fused to the C terminus and plasmid pMF334 to generate proteins with tdimer2(12), a variant of dsRED, fused to the N terminus (25-27). To simplify nomenclature, we use “rfp” here when referring to the tdimer2(12) gene (6) or “RFP” when we refer to the protein produced.
Fragments of genomic DNA containing the genes of interest were amplified by PCR from N. crassa strain 74A (FGSC987), using the Pfu Turbo DNA polymerase from Stratagene (now sold by Agilent Technologies). The primers, shown in Table 1, contained restriction endonuclease sites to facilitate cloning into the pMF272 and pMF334 plasmids. Amplified DNA from the PCRs was initially inserted into the vector pJET1.2 (Fermentas Life Sciences), which facilitates cloning of PCR products with blunt ends. The pJET1.2 plasmid containing the insert was digested with the two restriction endonucleases that recognized the sites in the primers. In this way, a DNA fragment of the correct size was obtained with a high likelihood that each end had been cut with the correct enzyme. This fragment was ligated with pMF272 or pMF334 digested with the same pair of restriction endonucleases. All new his-3-targeting plasmids are listed in Table 1. Plasmids are available from the Fungal Genetics Stock Center, Kansas City, MO.
TABLE 1.
Primers and plasmids used in this study
| N. crassa gene | Sequencea |
Host vector | Plasmid | |
|---|---|---|---|---|
| 5′ primer | 3′ primer | |||
| cax | ggtttctagaCCATGGACTCGGAACGCCGC | ccaacccgggGAAAATGGTGGAAAGTTTCTG | pMF272 | pcax-GFP |
| cax | ccttacgcgtATGGACTCGGAACGCCGC | ggttagatctGAAAATGGTGGAAAGTTTCTGAAG | pMF334 | pRFP-cax |
| nca-1 | gcattctagaATATGGAGGCGGCGTTTGCAAAGC | ccaacccgggGAGATCCTTCTTCTTATCTG | pMF272 | pnca-1-GFP |
| nca-1 | ctttacgcgtATGGAGGCGGCGTTTGCAAAG | gtcttctagagGAGATCCTTCTTCTTATCTGAGGACG | pMF334 | pRFP-nca-1 |
| nca-2 | cctatctagaCCATGGACCCCATTGCCAGTC | ccaacccgggGGCCGACTTTTGCCCTGG | pMF272 | pnca-2-GFP |
| nca-2 | ccttacgcgtATGGACCCCATTGCCAGTCAC | ggttagatctGGCCGACTTTTGCCCTGG | pMF334 | pRFP-nca-2 |
| nca-3 | cctctctagaGCTAATGGCGGACCATGAGCCG | cgaacccgggAGACCAACTCCTCCCCAACC | pMF272 | pnca-3-GFP |
| pmr | cctgtctagaCCATCATGGACCGACTCAGC | cataggatcccTACAGCCTGGCTATAGTTAAC | pMF272 | ppmr-GFP |
| pmr | ctttggcgcgcctATGGACCGACTCAGCGCGTG | cccctctagagTACAGCCTGGCTATAGTTAACACC | pMF334 | pRFP-pmr |
| grp-78 | cctttctagaGTAATGGAGCACCGAACGCGAAG | gttttcccgggAAGCTCGTCATGGCCAGCAG | pMF272 | pgrp-GFP |
| grp-78 | ccttacgcgtATGGAGCACCGAACGCGAAG | ggttagatctAAGCTCGTCATGGCCAGCAG | pMF334 | pRFP-grp |
| dpm | cctttctagaCACCATGGCTCCCACCAAG | gttttcccgggCGTGGACCACCAAAGCTGC | pMF272 | pdpm-GFP |
| dpm | ccttacgcgtATGGCTCCCACCAAGACCAC | ggttagatctCGTGGACCACCAAAGCTGC | pMF334 | pRFP-dpm |
| vps-52 | cctttctagaCGCTATGTGGCTTGATCGACGTC | gttttcccgggGATATTCTCTTGTACGCTCGCC | pMF272 | pvps-52-GFP |
| vps-52 | ccttacgcgtATGTGGCTTGATCGACGTC | ggttagatctGATATTCTCTTGTACGCTCGCC | pMF334 | pRFP-vps-52 |
| vam-3 | ccttacgcgtATGTCATTCGATCAGCTCTCC | ggttagatctGCCCAAGAACACCGCCAAC | pMF334 | pRFP-vam-3 |
| vma-1 | cacaggcgcgcctATGGCTCCCGTAAGCCTTG | gttttggccggccCTCATCAATGACAGACGCGAAC | pMF334 | pRFP-vma-1 |
| vma-5 | cacatctagaGACCATGGCACCGACAACTC | ctctcccgggAGGAAATTCAAATTCATAATACAC | pMF272 | pvma-5-GFP |
| arg-4 | ccctacgcgtATGGAGAGCCTCAACGGCTG | cctctctagagCGTGCGGTAGTCTCCATTAATGG | pMF272 | parg-4-GFP |
The region in uppercase letters is the sequence in the N. crassa gene. The region in lowercase letters contains the restriction site used for construction of the plasmid.
Transformation of N. crassa.
The pMF272 and pMF334 vectors contain part of the his-3 gene and are designed to facilitate targeting to the his-3 locus. The N. crassa his-3 A strain was transformed with the plasmids listed in Table 1 by electroporation (25). Typically, for each transformation, eight his+ colonies were transferred to tubes with Vogel's minimal medium. These isolates were examined by laser scanning confocal microscopy (see below). For some transformations, none of the transformed isolates gave a red or green signal that was clearly above the background fluorescence. However, for most strains 50 to 100% gave a strong signal. Because N. crassa conidia used for transformation are typically multinucleate, primary transformants can be heterokaryons. We obtained homokaryons for each strain by isolating microconidia (21). For each of the strains described in this report we obtained two or three independently isolated transformants that were indistinguishable from each other when observed by laser-scanning confocal microscopy. The transformed strains of N. crassa are available from the Fungal Genetics Stock Center.
To verify that the GFP signal came from translational fusions of proteins, we performed Western analysis (data not shown), essentially using the procedure of Garceau et al. (28). Cell extracts were prepared by using phenylmethylsulfonyl fluoride (1 mM) and chymostatin (2 μg/ml) as protease inhibitors. Using an antibody to GFP (Anti-GFP; Roche Applied Science, catalog no. 11 814 460 001), we observed a band of the predicted size for all of the GFP fusion proteins. NCA-2-GFP and NCA-3-GFP also showed some cross-reaction with a band of approximately 27 kDa, the size of GFP itself. As shown in Results, NCA-2-GFP and NCA-3-GFP localize to the vacuole, and the free GFP may arise by proteolysis in that compartment. The only significant anomaly we saw was two bands for ARG-4-GFP, at 77 kDa as predicted and at 50 kDa. Both bands were observed in cell extracts and in purified mitochondria, which indicated that the anomalous 50-kDa band was also targeted to, or generated in, that organelle. We attempted to do Western blots with the RFP fusions using two different polyclonal antibodies to dsRED (dsRED [C-20] and dsRED [L-18]; both from Santa Cruz Biotechnology, Santa Cruz, CA). Neither reacted to the tdimer2(12) protein. dsRED and tdimer2(12) differ in 20 of 238 amino acid residues. We were not able to obtain antibody to tdimer2(12).
Microscopy.
To prepare mycelia for microscopy we inoculated strains on one edge of a 100 mm petri dish containing Vogel's minimal medium with 2% agar and grew them for 16 to 18 h at 30°C. An agar block approximately 1 by 2 cm was cut from the leading edge of the colony and mounted on a glass coverslip as described previously (31). To visualize a GFP-tagged and an RFP-tagged protein in the same hypha, we prepared heterokaryons. Conidia from each strain were suspended in water and counted. Typically, 4 μl or 12 μl of each strain (10,000 conidia per μl) were coinoculated using ratios of 1:1, 1:3, and 3:1. For most samples, the best images were obtained with a 1:1 ratio.
Confocal laser scanning microscopy was performed using a Leica TCS SP5 system with a Leica DM600 inverted microscope. GFP images were obtained by excitation at 488 nm, with emission collected at 500 to 600 nm. RFP images were obtained with excitation at 543 nm and emission at 555 to 700 nm. Lasers were set with a scan speed of 400 Hz, and the camera resolution was 1,024 × 1,024 pixels. The microscope objective was ×63, oil, and images were typically digitally magnified fourfold. To obtain two-color images, the sample was scanned sequentially line by line, and emission was collected at 500 to 535 for GFP and 570 to 640 for RFP. Time-lapse movies for each of the organelles described in the paper can be viewed via the Fungal Genetics Stock Center (http://www.fgsc.net/).
RESULTS
Identification and tagging of calcium transporters and protein markers for organelles.
By analysis of the N. crassa genome, we previously identified genes predicted to encode calcium transport proteins in N. crassa (62). Consistent with an earlier PCR-based study (2), we found homologs of the two Ca2+-ATPases that have been extensively studied in S. cerevisiae, Pmc1p and Pmr1p. In addition, N. crassa and other filamentous fungi have two additional Ca2+-ATPases (Table 2). NCA-1 is most closely related to the SERCA-type ATPase found in the endoplasmic reticula of animal cells. NCA-2 and NCA-3 are closely related to each other and to Pmc1p from S. cerevisiae. A single putative Golgi ATPase, named Pmr1p in yeast and PMR in N. crassa, is found in both kinds of fungi. We also included the cax gene in our study because its homolog, VCX1, has a major role in calcium transport into the vacuole in S. cerevisiae (13, 47).
TABLE 2.
Calcium transport proteins
| N. crassa gene | Locus | Protein | Most similar protein |
|---|---|---|---|
| cax | NCU07075 | Ca2+/H+ transporter | Vcx1p in vacuolar membrane of S. cerevisiae |
| nca-1 | NCU03305 | Ca2+-ATPase | SERCA ATPase in the endoplasmic reticula of animal cells |
| nca-2 | NCU04736 | Ca2+-ATPase | Pmc1p in vacuolar membrane of S. cerevisiae |
| nca-3 | NCU05154 | Ca2+-ATPase | Pmc1p in vacuolar membrane of S. cerevisiae |
| pmr | NCU03292 | Ca2+-ATPase | Pmr1p in Golgi membrane of S. cerevisiae |
To identify organelles in N. crassa, we chose homologs of genes reported to encode organelle-specific proteins in S. cerevisiae (Table 3). grp-78 is the N. crassa homolog of KAR2 from S. cerevisiae (equivalent to bip in animal cells) (43). The gene product facilitates protein folding in the ER. dpm, by analogy to yeast, encodes dolichol-phosphate mannosyltransferase, located in the ER (46). vps-52 encodes a component of the Golgi-associated retrograde protein complex (12). vam-3 encodes a vacuolar-associated SNARE protein (60). Additional markers were two subunits of the vacuolar ATPase, encoded by vma-1 and vma-5 (8). arg-4 encodes acetylornithine-glutamate acetyltransferase, an enzyme of the arginine biosynthetic pathway, localized in the mitochondrion (15).
TABLE 3.
Proteins used as markers for organelles
| N. crassa gene | Locus | Protein | Homolog in S. cerevisiae |
|---|---|---|---|
| grp-78 | NCU03982 | Endoplasmic reticulum-associated HSP | Kar2p |
| dpm | NCU07965 | Endoplasmic reticulum dolichol-phosphate mannosyltransferase | Dpm1p |
| vps-52 | NCU05273 | Golgi body-associated retrograde protein | Vps52p |
| vam-3 | NCU06777 | Vacuole-associated SNARE protein | Vam3p |
| vma-1 | NCU01207 | Subunit A of vacuolar ATPase | Vma1p |
| vma-5 | NCU09897 | Subunit C of vacuolar ATPase | Vma5p |
| arg-4 | NCU10468 | Mitochondrial acetylornithine-glutamate transacetylase | Arg7p |
| H1 | NCU06863 | Histone H1 | Hho1p |
| hpo | NCU04018 | Chromosomal protein HP1 | -a |
See reference 25.
As described in Materials and Methods, we used PCR to isolate each of these genes from the wild-type strain of N. crassa. We constructed recombinant genes to produce proteins with GFP fused to the C terminus or RFP fused to the N terminus. In each case the moderately strong promoter of the N. crassa ccg-1 gene drove expression of the fusion genes. All recombinant genes were targeted to the his-3 locus. To visualize nuclei, we used two markers previously described, histone H1 fused to GFP and the chromosomal heterochromatin protein 1 (HP1) fused to RFP (25-27). These were also targeted to the his-3 locus.
NCA-1, the nuclear envelope, and the ER.
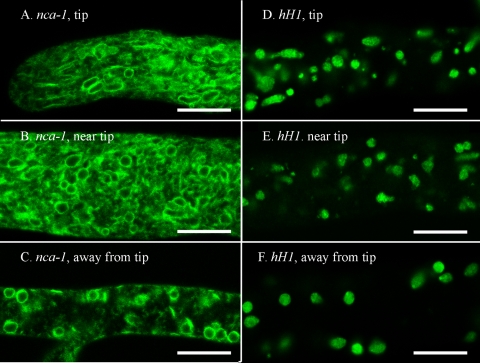
The SERCA-type ATPase encoded by nca-1 was predicted to be in the ER. Using confocal microscopy, we examined a strain of N. crassa transformed with nca-1+-sgfp. We observed the GFP-tagged protein in the nuclear envelope and in other membranes, some of which appeared to be continuous with the nuclear envelope (Fig. 1A, B, and C). For this and all other strains we examined the hyphal tip, the region 100 to 200 μm behind the hyphal tip where the first branch typically appears, and the region 1 to 10 mm farther back, which typically contains large spherical vacuoles. With NCA-1 the GFP-labeled membranes were seen in all regions, from the tip to the older segments. The spherical organelles had the same size and abundance as nuclei (compare panels A to C with panels D to F in Fig. 1). To verify that the GFP was in nuclear envelopes, we coinoculated the NCA-1-GFP strain and the RFP-HP1 strain to obtain a heterokaryon that expressed both green and red proteins (Fig. 2A, B, and C). Merging the two images confirmed the location of NCA-1-GFP in the nuclear envelope. Although we observed fairly uniform diameters in nuclei located in regions of the hypha away from the tip, at the tip itself we saw considerable heterogeneity in the apparent size of nuclei as visualized by tagged histone H1 and NCA-1 (compare panels A and B with panels D and E in Fig. 1).
FIG. 1.
NCA-1-GFP appears in the nuclear envelope and associated membranes. The scale bar in all panels is 10 μm. (A) Tip of a hypha transformed with nca-1+::sgfp. (B) A region approximately 200 μm from the tip in a hypha transformed with nca-1+::sgfp. (C) A region approximately 2 mm from the tip in a hypha transformed with nca-1+::sgfp. (D) Tip of a hypha transformed with hH1+::sgfp. (E) A region approximately 200 μm from the tip in a hypha transformed with hH1+::sgfp. (F) A region approximately 2 mm from the tip in a hypha transformed with hH1+::sgfp.
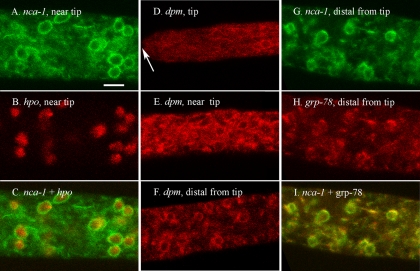
FIG. 2.
NCA-1, DPM, and GRP-78 are located in the nuclear envelope and associated membranes. The scale bar in panel C is 10 μm. All panels are shown at the same magnification. (A to C) A region approximately 200 μm from the tip of a hypha in a heterokaryon formed by coinoculating strains transformed with nca-1+::sgfp and rfp::hpo+. The images in panels A and B were merged in panel C. (D) Tip (marked by an arrow) of a hypha transformed with rfp::dpm+. (E) A region approximately 200 μm from the tip in a hypha transformed with rfp::dpm+. (F) A region approximately 2 mm from the tip of a hypha transformed with rfp::dpm+. (G to I) A region approximately 2 mm from the tip of a hypha in a heterokaryon formed by coinoculating strains transformed with nca-1+::sgfp and rfp::grp-78+. Panels G and H were merged in panel I.
The two presumptive endoplasmic reticulum markers, GRP-78 and DPM, appeared in the same cellular structures as NCA-1 (Fig. 2D to I). The signal from GRP-78 and DPM was not as strong as for NCA-1; however, nuclear envelopes and attached membranes were visible with these constructs (Fig. 2D, E, F, and H). A heterokaryon of NCA-1-GFP plus RFP-GRP-78 showed colocalization of the green and red fluorescent proteins, although RFP-GRP-78 sometimes appeared as a “patch” on one side of the nucleus (Fig. 2G, H, and I). These results suggest that the ER of N. crassa is composed of the nuclear envelope and associated membranes. This observation is consistent with investigations of other organisms, where the nuclear envelope is a specialized region of the ER (23, 44, 48, 61).
The localization of GRP-78 and DPM was the same either with RFP fused to the N terminus (Fig. 2D, E, F, and H) or with GFP fused to the C terminus (DPM-GFP and GRP-78-GFP not shown). Likewise, RFP fused to the N terminus of NCA-1 localized to the same structures as NCA-1-GFP (not shown). With NCA-1-GFP we also compared the results obtained when the host cell for transformation carried the normal wild-type copy of nca-1 or had the endogenous nca-1 gene replaced by the hyg gene (11); NCA-1-GFP appeared in the same intracellular structures, with the same signal intensity (data not shown).
NCA-2, VAM-3, and the vacuolar network.
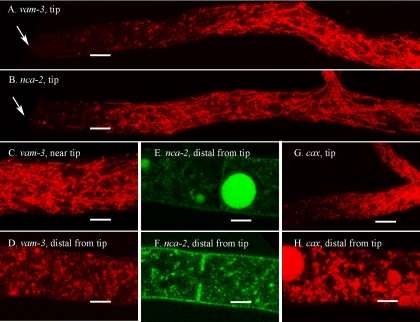
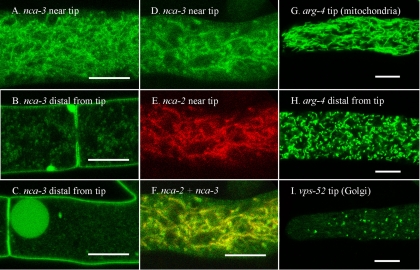
Investigators have previously shown that dyes targeted to the vacuole localize in a tubular network near the tips of hyphae (10, 24, 31, 33). The cellular compartments visualized with RFP-VAM-3 protein, a vacuolar SNARE, looked like components of this vacuolar network. The first 50 to 100 μm at the hyphal tip contained little visible RFP-VAM-3. Next came a short region with a mixture of unorganized vesicles and tubular structures (Fig. 3A and C). The dense tubular network typically started 50 to 100 μm from the tip and continued for 300 to 500 μm in our growth conditions. RFP-NCA-2 (Fig. 3B), NCA-2-GFP (Fig. 3E and F), and RFP-CAX (Fig. 3G and H) localized to the same structures as RFP-VAM-3. In hyphal segments further back from the tip RFP-VAM-3, NCA-2-GFP, and RFP-CAX appeared primarily in small vesicles and also in the large spherical vacuoles that are prominent in distal hyphal segments (Fig. 3D, E, F, and H). In these older segments NCA-2-GFP was also visible in the plasma membrane (Fig. 3E and F, discussed further below).
FIG. 3.
Vacuolar proteins appear in different structures in different regions of the hyphae. The scale bar is 10 μm in all panels. (A) The hyphal tip (marked by an arrow) in a strain transformed with rfp::vam-3+. (B) Hyphal tip (marked by an arrow) in a strain transformed with rfp::nca-2+. (C) A region approximately 200 μm from the hyphal tip in a strain transformed with rfp::vam-3+. (D) A region approximately 2 mm from the hyphal tip in a strain transformed with rfp::vam-3+. (E and F) Regions approximately 2 mm from the hyphal tip in a strain transformed with nca-2+::sgfp. (G) Hyphal tip in a strain transformed with rfp::cax+. (H) A region approximately 2 mm from the hyphal tip in a strain transformed with rfp::cax+.
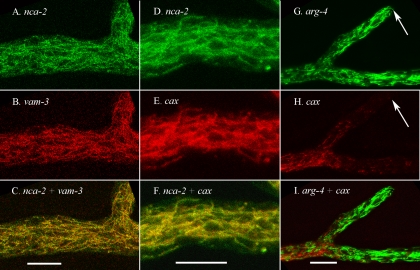
Examination of heterokaryons of NCA-2-GFP plus RFP-VAM-3 or NCA-2-GFP plus RFP-CAX showed that all three of these proteins were in the same vacuolar network. As shown in Fig. 4A to F, the tagged NCA-2 protein colocalized in the same thin tubules and small vesicles as the tagged CAX and VAM-3 proteins.
FIG. 4.
Vacuolar proteins appear in a tubular network near the hyphal tip and are distinct from mitochondria. The scale bar is 10 μm in all panels. (A to C) A region approximately 200 μm from the hyphal tip in a heterokaryon formed by coinoculating strains transformed with nca-2+::sgfp and rfp::vam-3+. Panels A and B were merged in panel C. (D to F) A region approximately 200 μm from the hyphal tip in a heterokaryon formed by coinoculating strains transformed with nca-2+::sgfp and rfp::cax+. Panels D and E were merged in panel F. (G to I) A region approximately 200 μm from the hyphal tip in a heterokaryon formed by coinoculating strains transformed with arg-4+::sgfp and rfp::cax+. Panels G and H were merged in panel I. The hyphal tips are marked with an arrow.
ARG-4 and mitochondria.
Using dyes that localized to the mitochondria other investigators have observed mitochondria in N. crassa as abundant long thin tubules (24). We transformed N. crassa with arg-4+::sgfp and observed tubular mitochondria (Fig. 4G and Fig. 5G), which looked similar to those observed with the dye FM4-64 (24). In hyphal segments 1 to 2 mm from the tip the mitochondria appeared to be shorter but were still abundant (Fig. 5H). The mitochondrial network differed from the vacuolar network by having fewer small bright vesicles associated with the tubular structures. Another obvious difference was the abundance of mitochondria throughout the hyphal tip, where few vacuoles were seen. To look at the vacuolar network and mitochondria in the same hyphae, we examined a heterokaryon expressing both RFP-CAX and ARG-4-GFP proteins (Fig. 4G to I). Although the cytoplasm was densely packed with tubular elements, the vacuolar and mitochondrial proteins clearly localized to different compartments.
FIG. 5.
NCA-3 localizes to the tubular vacuolar network and the plasma membrane. The mitochondrial marker, ARG-4, and the Golgi marker, VPS-52, appear in distinct compartments. The scale bar is 10 μm in all panels. (A) A region approximately 200 μm from the hyphal tip in a strain transformed with nca-3+::sgfp. (B and C) Regions approximately 2 mm from the hyphal tip in a strain transformed with nca-3+::sgfp. (D to F) A region approximately 200 μm from the hyphal tip in a heterokaryon formed by coinoculating strains transformed with nca-3+::sgfp and rfp::nca-2+. (G) Tip of a hypha transformed with arg-4+::sgfp. (H) A region approximately 2 mm from the hyphal tip in a strain transformed with arg-4+::sgfp. (I) Tip of a hypha transformed with vps-52+::sgfp.
NCA-2, NCA-3, and the plasma membrane.
NCA-2 and NCA-3 ATPases share equal sequence similarity to the Pmc1p protein of S. cerevisiae. The PMCA-type ATPases are in the vacuole in S. cerevisiae but in the plasma membrane in most animal cells. In N. crassa these proteins appear to be in both locations (Fig. 3B, E, and F; Fig. 5A, B, and C). The organelles visualized with RFP-NCA-2, NCA-2-GFP, and NCA-3-GFP were indistinguishable from those seen with RFP-VAM-3 and RFP-CAX. There was a dark area immediately behind the hyphal tip, then a region with a network of tubules and small vesicles, and farther back small vesicles and large spherical vacuoles. Unlike VAM-3 and CAX, both NCA-2 and NCA-3 appeared in the plasma membrane. The signal was weak and variable near the tip; however, in hyphal segments that were 200 to 400 μm distal to the tip fluorescent proteins in the plasma membrane became clearly visible. Although the GFP could possibly be in the cell wall, observations of the septum between hyphal compartments (Fig. 5B) showed a dark space in the middle of the septum, indicating that the fusion proteins were in the plasma membrane, not the wall. Tagged NCA-3 gave a particularly strong signal in the plasma membrane in older regions of hyphae (>2 mm from the tip) and, conversely, was weaker than NCA-2-GFP in the tubular network near the tip. None of the other GFP or RFP fusion proteins tested appeared in the plasma membrane. Examination of a heterokaryon of NCA-2-GFP plus RFP-NCA-3 showed that both were in the same intracellular, vacuolar membranes (Fig. 5D, E, and F).
VPS-52 and the Golgi compartment.
In S. cerevisiae the Ca2+-ATPase encoded by the PMR1 gene is localized in the medial Golgi compartment (20). Our preliminary experiments suggest that in N. crassa, as in S. cerevisiae, the cis-, medial, and trans-Golgi bodies may be distinct vesicular structures. This has necessitated the construction and analysis of a number of putative Golgi markers, work that is not presented here. However, to show a more complete set of organelles, we show the localization of VPS-52-GFP (Fig. 5I), which is predicted to be in a late Golgi compartment (12). In N. crassa, VPS-52-GFP does not colocalize with the vacuolar or ER proteins discussed above. PMR-GFP and RFP-PMR appear in vesicles of similar size and abundance to those shown for VPS-52, but PMR and VPS-52 do not colocalize (data not shown).
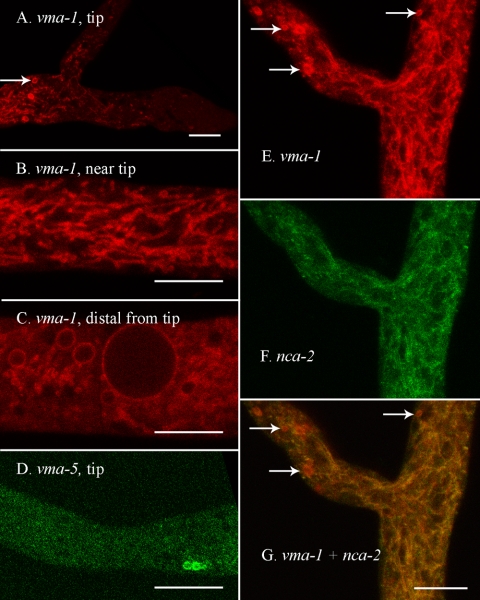
Colocalization of CAX and the vacuolar ATPase.
By transporting protons across the vacuolar membrane the vacuolar ATPase generates an acidic environment within the vacuolar compartment and also generates an electrochemical gradient. The vma-1 gene encodes the ATP-binding “A” subunit of the enzyme. RFP-VMA-1 gave a strong signal in the vacuolar membranes (Fig. 6A, B, C, and E). A heterokaryon composed of RFP-VMA-1 plus NCA-2-GFP showed these two vacuolar markers to colocalize (Fig. 6E, F, and G). In some hyphal compartments with large spherical vacuoles the RFP-VMA-1 signal was predominantly in the vacuolar membrane (Fig. 6C), not in the interior of the vacuole, as observed with other vacuolar proteins (e.g., Fig. 3E and H). The lack of fluorescence in the vacuolar lumen correlated with the degree of cytoplasmic flow in the hyphal segment. In segments with very active flow the interior of the vacuole was not red. For reasons we do not understand, internalization of the vacuolar ATPase may occur at lower rates in hyphae with active cytoplasmic flow.
FIG. 6.
The vacuolar ATPase appears in the vacuolar network, in the membrane of larger spherical vacuoles, and in spherical organelles near the hyphal tip. The scale bar is 10 μm in all panels. (A) Tip of a hypha transformed with rfp::vma-1+. An arrow points to one of several spherical organelles that can be seen near the tip in this strain. (B) A region approximately 200 μm from the hyphal tip in a strain transformed with rfp::vma-1+. (C) A region approximately 2 mm from the hyphal tip in a strain transformed with rfp::vma-1+. (D) The tip of a hypha transformed with vma-5+::sgfp. (E to G) A region approximately 100 μm from the hyphal tip in a heterokaryon formed by coinoculating strains transformed with nca-2+::sgfp and rfp::vma-1+. Panels E and F were merged in panel G. Arrows point to red spherical organelles that can be seen near the tip.
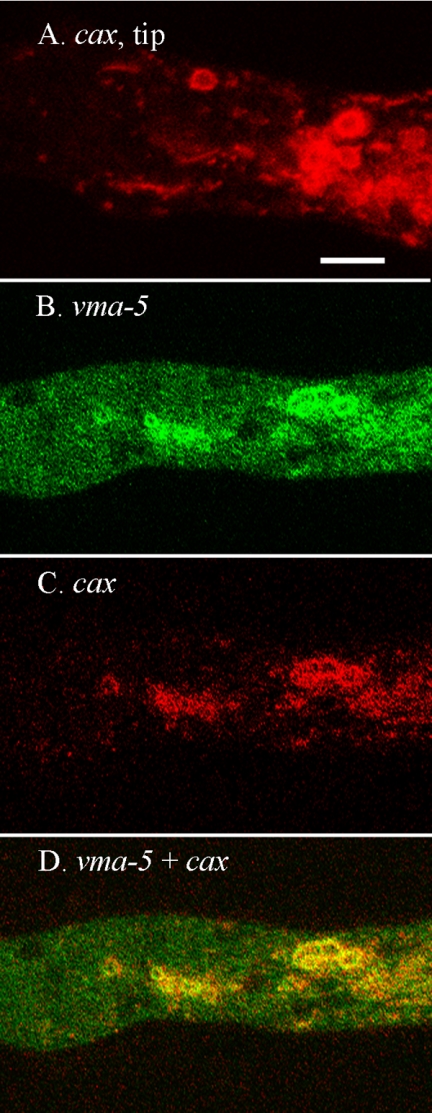
A striking difference between RFP-VMA-1 and the tagged VAM-3 (the vacuolar SNARE), NCA-2, and NCA-3 proteins was its strong appearance in the membrane of unidentified organelles near the hyphal tip (Fig. 6A and E). These organelles, 2 to 5 μm in diameter, were seen only in the region near the tip where the tubular vacuolar network starts to appear. A typical hyphal tip contained 1 to 10 of the novel organelles. We made a GFP fusion with another subunit of the vacuolar ATPase, VMA-5-GFP. Although the signal from this construct was not as strong as for RFP-VMA-1, the unidentified organelle was clearly observable (Fig. 6D). The only other protein that strongly colocalized with the vacuolar ATPase in these intriguing organelles was RFP-CAX (Fig. 7A). A heterokaryon of VMA-5-GFP and RFP-CAX proteins showed that the vacuolar ATPase and the CAX protein were in identical organelles (Fig. 7B, C, and D).
FIG. 7.
Both vacuolar ATPase and CAX proteins localize to spherical organelles near the hyphal tip. The scale bar is 10 μm. All panels are at the same magnification. (A) A region approximately 100 μm from the hyphal tip in a strain transformed with rfp-cax+. (B to D) A region approximately 100 μm from the hyphal tip in a heterokaryon formed by coinoculating strains transformed with vma-5+::sgfp and rfp::cax+.
DISCUSSION
We have found that tagging gene products with green and red fluorescent proteins is an effective and efficient means of examining the structure and abundance of organelles in growing N. crassa hyphae. Using this approach allowed us to determine the organellar location of five calcium transporters.
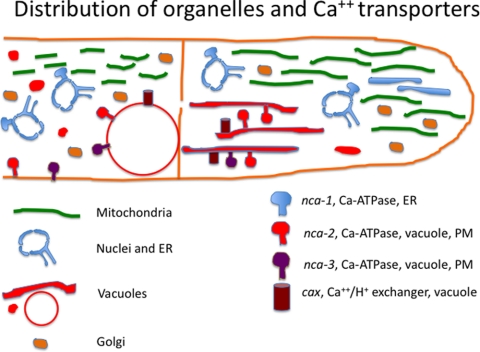
In this report we studied 14 genes: 7 were fused to either GFP or RFP, and 7 were fused to both GFP and RFP. Using both green and red tags, we succeeded in obtaining transformants that expressed fusion proteins with good signals. Nonetheless, some strains had weak GFP signals, which were difficult to distinguish from background green fluorescence. In contrast, virtually no background fluorescence was seen when using this particular RFP, tdimer2(12). Fusion of the fluorescent protein to the N terminus versus the C terminus did not affect the localization of the protein for six of the seven genes we examined. Only for the cax gene, discussed below, did we observe different behavior in GFP-tagged versus RFP-tagged proteins. The organelles in the endomembrane system were dynamic and variable when observed in actively growing hyphae. Our observations are summarized in Fig. 8. We are not aware of previous reports of systematically examining all of these organelles in living filamentous fungi. We limited our observations to young hyphae vigorously growing on the surface of agar medium, comparing the appearance of organelles at the tip, in the region 5 to 200 μm behind the tip and in the region 1 to 5 mm further back. In each of these regions we observed a difference in the structure and abundance of organelles. Some of the structural variability was surprising.
FIG. 8.
Distribution of organelles and calcium transporters in N. crassa hyphae.
As shown in previous examinations of GFP- and RFP-tagged nuclei in N. crassa (25-27, 50), we found abundant nuclei in all regions except for the small zone 20 to 40 μm immediately behind the tip. The structure of nuclei was relatively uniform, with the exception of a surprising heterogeneity of sizes near the hyphal tip. We located proteins predicted to reside in the ER (products of the dpm and grp-78 genes) both in the nuclear envelope and also in other associated, irregular membranes. Thus, our results with N. crassa are consistent with studies done in other fungi showing that the nuclear envelope is a specialized region of the ER (23, 39, 44, 48, 61). The ER was abundant throughout the hyphae from the region immediately behind the tip to the older segments. In older segments of hyphae the fluorescence signals from ER proteins were primarily in the nuclear envelope, with less fluorescence seen in other membranes. Because fungal cells have many nuclei, perhaps the nuclear envelope suffices as the major component of the ER in older hyphal segments.
The product of the nca-1 gene was predicted to be a SERCA-type Ca2+-ATPase, based on DNA sequence similarity and the presence of a canonical ER-retention signal (KKDL) at the C terminus. GFP- and RFP-tagged versions of NCA-1 strongly localized to the nuclear envelope and associated membranes, confirming its ER residence. We speculate that SERCA-type ATPases might play a role in filamentous growth because they are present in the filamentous fungi but not in yeasts.
Visualization of the Golgi compartments in N. crassa has proven to be more complicated than we anticipated. A protein predicted to localize to the Golgi, VPS-52, appeared in small vesicles (0.5 to 2.0 μm) that were distinct from other organelles. In our preliminary analysis we saw the PMR Ca2+-ATPase, which is in the medial Golgi compartment in S. cerevisiae (53), in small vesicles not associated with VPS-52. The data suggest that the Golgi complex in N. crassa may consist of disconnected cis, medial, and trans compartments, as reported for S. cerevisiae (37). We are currently pursuing this hypothesis.
In older textbooks, “the vacuole” was presented as a large spherical organelle, and in N. crassa such vacuoles are often seen in older hyphal segments. However, work pioneered by Anne Ashford and coworkers and since extended by other labs has shown that vacuolar compartments in filamentous fungi can also be in the form of small vesicles or an interconnected network of tubules (10, 24, 31, 33). Using tagged versions of the VAM-3 SNARE protein and the VMA-1 polypeptide of the V-ATPase, we visualized the vacuole in N. crassa. Vacuolar structure varied greatly in different regions of a hypha. Vacuolar markers were not present in the first 20 to 50 μm behind the tip. They appeared as small vesicles and tubules in the zone between about 20 to 50 and 200 μm behind the tip. In the region before the first septum the vacuolar markers were abundant in an interconnected network of thin (<1 μm) tubules, often with small, attached vesicles. These results with N. crassa are generally consistent with observations made using GFP-tagged VAM-3 in Aspergillus oryzae (45, 54). N. crassa grows faster, and the diameter of hyphae can be fivefold larger. The network of vacuolar tubules near the tips is denser near the tip, and fast-growing hyphae have fewer of the large spherical vacuoles. Shoji et al. (54) reported that small punctate structures tagged with GFP-VAM-3 associated with large vacuoles. These authors tentatively identified these as endosomes or prevacuolar compartments. We observed VAM-3 in N. crassa only in compartments that also contained other vacuolar proteins - VMA-1, CAX, NCA-2, or NCA-3.
The tubular network was highly dynamic, moving rapidly and changing shape as the hyphae grew. In older hyphal segments, typically more than 300 μm from the tip, we observed spherical vacuoles of widely ranging sizes, plus numerous small vesicles. This region, of course, represents the majority of the hyphal colony. The dramatic dynamic activity occurs in the growing tip cells.
The tubular network seen with tagged VAM-3 and VMA-1 in some ways resembled the structure of mitochondria, as observed previously with dyes (24). Using ARG-4-GFP we obtained images of mitochondria that agreed well with the images obtained with dyes. Abundant in all regions examined, the mitochondria were seen as thin tubes with a more uniform thickness, lacking the punctate vesicular structures that appear to be part of the vacuolar network. Mitochondrial structure also varied with position in the hypha, the organelles appearing significantly shorter in older hyphal segments.
The Ca2+-H+ transporter encoded by the cax gene is located in the vacuole in S. cerevisiae (13, 47). We have found that, consistent with the results obtained from yeast, vacuoles isolated from cax-deficient strains of N. crassa have less than 10% of the calcium found in vacuoles from the wild-type strain (unpublished results). The CAX protein tagged with RFP localized to all components of the vacuole, both the tubular network and the larger spherical structures. Surprisingly, CAX-GFP appeared in the ER. A likely explanation is that fusion of the GFP protein to the C terminus of CAX interfered with a vacuolar targeting signal, causing the protein to be retained in the ER immediately after synthesis. Retention in the ER has been reported for other modified membrane proteins, e.g., the plasma membrane ATPase, PMA1 (40).
As described earlier, nca-2 and nca-3 encode PMCA-type Ca2+-ATPases. These enzymes are located in the plasma membrane in animal cells but in the vacuolar membrane in S. cerevisiae. Interestingly, in N. crassa these Ca2+-ATPases appeared in both the vacuolar and the plasma membranes. In the region within 200 to 500 μm of the hyphal tip neither NCA2 nor NCA3 was visible in the plasma membrane, but the fluorescent signal was clearly visible in the plasma membrane in older segments of the hyphae. The one difference noted with NCA-2 versus NCA-3 is that tagged NCA-2 was relatively more abundant in the vacuolar compartments, whereas tagged NCA-3 was strongly incorporated into the plasma membrane, especially in regions 1 mm or more away from the tip. These data suggest that in N. crassa NCA-2 and NCA-3 may serve to pump calcium out of the cell, as do the homologs of these enzymes in animal cells.
Although the genes we examined—nca-2, nca-3, cax, vam-3, and vma-1—encode membrane proteins, all of the proteins appeared to be internalized to some extent in the large spherical vacuoles. Because of the small size of the tubular components in the vacuolar network we could not determine whether the tagged proteins were inside the tubules. Many types of proteins can be targeted to the vacuole for degradation, but of the 14 different gene products we examined only those predicted to have their primary function in the vacuole localized to the vacuolar interior. The internalization seen in the larger vacuoles could be due to normal turnover of vacuolar membrane proteins. Only for RFP-VMA-1 did we observe hyphae with the tagged protein predominantly in the membrane of larger vacuoles. This is interesting because VMA-1 encodes the A subunit of the vacuolar ATPase and has no membrane-spanning region, unlike the other proteins we examined. VMA-1, as part of the V1 sector of the enzyme, lies on the cytosolic side of the membrane and can dissociate from the integral membrane sector of the enzyme into the cytosol (5).
From the analysis of these 14 gene products our most surprising finding was an apparently novel organelle. The vacuolar ATPase subunits and CAX allowed visualization of a type of organelle not seen with any of the other tagged marker proteins. RFP-VMA-1, VMA-5-GFP, and RFP-CAX appeared in the membranes of roughly spherical organelles, 2 to 5 μm in diameter. These unidentified structures occurred only in a small region of each hypha, 100 to 200 μm behind the tip. Conceivably, they are compartments of the vacuole specialized for sequestration of calcium. We speculate that they could be related to organelles called acidocalcisomes, described primarily in trypanosomes but also identified in other types of organisms (17). Acidocalcisomes are approximately the same size, have membranes enriched in vacuolar ATPase, and contain high concentrations of calcium bound to polyphosphate.
Acknowledgments
This study was supported by Public Health Service grant GM058903 from the Institute of General Medicine to B.J.B. and American Cancer Society grant RSG-08-030-01-CCG to M.F.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Antebi, A., and G. R. Fink. 1992. The yeast Ca2+-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol. Biol. Cell 3:633-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benito, B., B. Garciadeblas, and A. Rodriguez-Navarro. 2000. Molecular cloning of the calcium and sodium ATPases in Neurospora crassa. Mol. Microbiol. 35:1079-1088. [DOI] [PubMed] [Google Scholar]
- 3.Berridge, M. J., M. D. Bootman, and H. L. Roderick. 2003. Calcium signalling: dynamics, homeostasis and remodeling. Nat. Rev. Mol. Cell Biol. 4:517-529. [DOI] [PubMed] [Google Scholar]
- 4.Bok, J. W., T. Sone, L. B. Silverman-Gavrila, R. R. Lew, F. J. Bowring, D. E. Catcheside, and A. J. Griffiths. 2001. Structure and function analysis of the calcium-related gene spray in Neurospora crassa. Fungal Genet. Biol. 32:145-158. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, B. J., W. J. Dschida, T. Harris, and E. J. Bowman. 1989. The vacuolar ATPase of Neurospora crassa contains an F1-like structure. J. Biol. Chem. 264:15606-15612. [PubMed] [Google Scholar]
- 6.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carafoli, E. 2002. Calcium signaling: a tale for all seasons. Proc. Natl. Acad. Sci. USA 99:1115-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavez, C., E. J. Bowman, J. C. Reidling, K. H. Haw, and B. J. Bowman. 2006. Analysis of strains with mutations in six genes encoding subunits of the V-ATPase: eukaryotes differ in the composition of the V0 sector of the enzyme. J. Biol. Chem. 281:27052-27062. [DOI] [PubMed] [Google Scholar]
- 9.Clapham, D. E. 2007. Calcium signaling. Cell 131:1047-1058. [DOI] [PubMed] [Google Scholar]
- 10.Cole, L., D. A. Orlovich, and A. E. Ashford. 1998. Structure, function, and motility of vacuoles in filamentous fungi. Fungal Genet. Biol. 24:86-100. [DOI] [PubMed] [Google Scholar]
- 11.Colot, H. V., G. Park, G. E. Turner, C. Ringelberg, C. M. Crew, L. Litvinkova, R. L. Weiss, K. A. Borkovich, and J. C. Dunlap. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103:10352-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conibear, E., and T. H. Stevens. 2000. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 11:305-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham, K. W., and G. R. Fink. 1996. Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2226-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham, K. W., and G. R. Fink. 1994. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol. 124:351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cybis, J., and R. H. Davis. 1975. Organization and control in the arginine biosynthetic pathway of Neurospora. J. Bacteriol. 123:196-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dicker, J. W., and G. Turian. 1990. Calcium deficiencies and apical hyperbranching in wild-type and ‘frost’ and ‘spray’ morphological mutants of Neurospora crassa. J. Gen. Microbiol. 136:1413-1420. [Google Scholar]
- 17.Docampo, R., W. de Souza, K. Miranda, P. Rohloff, and S. N. Moreno. 2005. Acidocalcisomes: conserved from bacteria to man. Nat. Rev. Microbiol. 3:251-261. [DOI] [PubMed] [Google Scholar]
- 18.Duchen, M. R., A. Verkhratsky, and S. Muallem. 2008. Mitochondria and calcium in health and disease. Cell Calcium 44:1-5. [DOI] [PubMed] [Google Scholar]
- 19.Dunn, T., K. Gable, and T. Beeler. 1994. Regulation of cellular Ca2+ by yeast vacuoles. J. Biol. Chem. 269:7273-7278. [PubMed] [Google Scholar]
- 20.Durr, G., J. Strayle, R. Plemper, S. Elbs, S. K. Klee, P. Catty, D. H. Wolf, and H. K. Rudolph. 1998. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell 9:1149-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebbole, D., and M. S. Sachs. 1990. A rapid and simple method of isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37:17-18. [Google Scholar]
- 22.Eilam, Y., H. Lavi, and N. Grossowicz. 1985. Cytoplasmic Ca2+ homeostasis maintained by a vacuolar Ca2+ transport system in the yeast Saccharomyces cerevisiae. J. Gen. Microbiol. 131:623-629. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Abalos, J. M., H. Fox, C. Pitt, B. Wells, and J. H. Doonan. 1998. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol. Microbiol. 27:121-130. [DOI] [PubMed] [Google Scholar]
- 24.Fischer-Parton, S., R. M. Parton, P. C. Hickey, J. Dijksterhuis, H. A. Atkinson, and N. D. Read. 2000. Confocal microscopy of FM4-64 as a tool for analyzing endocytosis and vesicle trafficking in living fungal hyphae. J. Microsc. 198:246-259. [DOI] [PubMed] [Google Scholar]
- 25.Freitag, M., P. C. Hickey, T. K. Khlafallah, N. D. Read, and E. U. Selker. 2004. HP1 is essential for DNA methylation in neurospora. Mol. Cell 13:427-434. [DOI] [PubMed] [Google Scholar]
- 26.Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker, and N. D. Read. 2004. GFP as a tool to analyze the organization, dynamics, and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897-910. [DOI] [PubMed] [Google Scholar]
- 27.Freitag, M., and E. U. Selker. 2005. Expression and visualization of red fluorescent protein (RFP) in Neurospora crassa. Fungal Genet. Newsl. 52:14-17. [Google Scholar]
- 28.Garceau, N. Y., Y. Liu, J. J. Loros, and J. C. Dunlap. 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89:469-476. [DOI] [PubMed] [Google Scholar]
- 29.Gerasimenko, O., and A. Tepikin. 2005. How to measure Ca2+ in cellular organelles? Cell Calcium 38:201-211. [DOI] [PubMed] [Google Scholar]
- 30.Harold, F. M. 2002. Force and compliance: rethinking morphogenesis in walled cells. Fungal Genet. Biol. 37:271-282. [DOI] [PubMed] [Google Scholar]
- 31.Hickey, P. C., S. R. Swift, and M. G. Roca. 2004. Live-cell imaging of filamentous fungi using vital fluorescent dyes and confocal microscopy. Methods Microbiol. 34:63-87. [Google Scholar]
- 32.Holdaway-Clarke, T. L., J. A. Feijo, G. R. Hackett, J. G. Kunkel, and P. K. Hepler. 1997. Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9:1999-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde, G. J., D. Davies, L. Cole, and A. E. Ashford. 2002. Regulators of GTP-binding proteins cause morphological changes in the vacuole system of the filamentous fungus, Pisolithus tinctorius. Cell Motil. Cytoskel. 51:133-146. [DOI] [PubMed] [Google Scholar]
- 34.Jackson, S. L., and I. B. Heath. 1993. Roles of calcium ions in hyphal tip growth. Microbiol. Rev. 57:367-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirichok, Y., G. Krapivinsky, and D. E. Clapham. 2004. The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427:360-364. [DOI] [PubMed] [Google Scholar]
- 36.Lew, R. R. 1999. Comparative analysis of Ca2+ and H+ flux magnitude and location along growing hyphae of Saprolegnia ferax and Neurospora crassa. Eur. J. Cell Biol. 78:892-902. [DOI] [PubMed] [Google Scholar]
- 37.Losev, E., C. A. Reinke, J. Jellen, D. E. Strongin, B. J. Bevis, and B. S. Glick. 2006. Golgi maturation visualized in living yeast. Nature 441:1002-1006. [DOI] [PubMed] [Google Scholar]
- 38.Malho, R., and A. J. Trewavas. 1996. Localized apical increases of cytosolic free calcium control pollen tube orientation. Plant Cell 8:1935-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruyama, J., S. Kikuchi, and K. Kitamoto. 2006. Differential distribution of the endoplasmic reticulum network as visualized by the BipA-EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet. Biol. 43:642-654. [DOI] [PubMed] [Google Scholar]
- 40.Mason, A. B., K. E. Allen, and C. W. Slayman. 2006. Effects of C-terminal truncations on trafficking of the yeast plasma membrane H+-ATPase. J. Biol. Chem. 281:23887-23898. [DOI] [PubMed] [Google Scholar]
- 41.McGillviray, A. M., and N. A. R. Gow. 1987. The transhyphal electrical current of Neurospora crassa is carried principally by protons. J. Gen. Microbiol. 133:2875-2881. [Google Scholar]
- 42.Missiaen, L., L. Dode, J. Vanoevelen, L. Raeymaekers, and F. Wuytack. 2007. Calcium in the Golgi apparatus. Cell Calcium 41:405-416. [DOI] [PubMed] [Google Scholar]
- 43.Monnerjahn, C., D. Techel, U. Meyer, and L. Rensing. 2001. The grp78 promoter of Neurospora crassa: constitutive, stress and differentiation-dependent protein-binding patterns. Curr. Genet. 39:319-326. [DOI] [PubMed] [Google Scholar]
- 44.Newport, J. W., and D. J. Forbes. 1987. The nucleus: structure, function, and dynamics. Annu. Rev. Biochem. 56:535-565. [DOI] [PubMed] [Google Scholar]
- 45.Ohneda, M., M. Arioka, H. Nakajima, and K. Kitamoto. 2002. Visualization of vacuoles in Aspergillus oryzae by expression of CPY-EGFP. Fungal Genet. Biol. 37:29-38. [DOI] [PubMed] [Google Scholar]
- 46.Orlean, P., C. Albright, and P. W. Robbins. 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263:17499-17507. [PubMed] [Google Scholar]
- 47.Pozos, T. C., I. Sekler, and M. S. Cyert. 1996. The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol. Cell. Biol. 16:3730-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prinz, W. A., L. Grzyb, M. Veenhuis, J. A. Kahana, P. A. Silver, and T. A. Rapoport. 2000. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prokisch, H., O. Yarden, M. Dieminger, M. Tropschug, and I. B. Barthelmess. 1997. Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol. Gen. Genet. 256:104-114. [DOI] [PubMed] [Google Scholar]
- 50.Ramos-Garcia, S. L., R. W. Roberson, M. Freitag, S. Bartnicki-Garcia, and R. R. Mourino-Perez. 2009. Cytoplasmic bulk flow propels nuclei in mature hyphae of Neurospora crassa. Eukaryot. Cell 8:1880-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudolph, H. K., A. Antebi, G. R. Fink, C. M. Buckley, T. E. Dorman, J. LeVitre, L. S. Davidow, J. I. Mao, and D. T. Moir. 1989. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell 58:133-145. [DOI] [PubMed] [Google Scholar]
- 52.Schmid, J., and F. M. Harold. 1988. Dual roles for calcium ions in apical growth of Neurospora crassa. J. Gen. Microbiol. 134:2623-2631. [DOI] [PubMed] [Google Scholar]
- 53.Schroder, S., F. Schimmoller, B. Singer-Kruger, and H. Riezman. 1995. The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in alpha-COP. J. Cell Biol. 131:895-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2006. Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryot. Cell 5:411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silverman-Gavrila, L., and R. R. Lew. 2000. Calcium and tip growth in Neurospora crassa. Protoplasma 213:203-217. [Google Scholar]
- 56.Silverman-Gavrila, L. B., and R. R. Lew. 2001. Regulation of the tip-high [Ca2+] gradient in growing hyphae of the fungus Neurospora crassa. Eur. J. Cell Biol. 80:379-390. [DOI] [PubMed] [Google Scholar]
- 57.Sorin, A., G. Rosas, and R. Rao. 1997. PMR1, a Ca2+-ATPase in yeast Golgi, has properties distinct from sarco/endoplasmic reticulum and plasma membrane calcium pumps. J. Biol. Chem. 272:9895-9901. [DOI] [PubMed] [Google Scholar]
- 58.Sze, H., F. Liang, I. Hwang, A. C. Curran, and J. F. Harper. 2000. Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:433-462. [DOI] [PubMed] [Google Scholar]
- 59.Torralba, S., and I. B. Heath. 2001. Cytoskeletal and Ca2+ regulation of hyphal tip growth and initiation. Curr. Top. Dev. Biol. 51:135-187. [DOI] [PubMed] [Google Scholar]
- 60.Wada, Y., Y. Ohsumi, and Y. Anraku. 1992. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J. Biol. Chem. 267:18665-18670. [PubMed] [Google Scholar]
- 61.Wedlich-Soldner, R., I. Schulz, A. Straube, and G. Steinberg. 2002. Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 13:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zelter, A., M. Bencina, B. J. Bowman, O. Yarden, and N. D. Read. 2004. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet. Biol. 41:827-841. [DOI] [PubMed] [Google Scholar]