Abstract
Background
Inorganic arsenic is an environmental carcinogen that may act through multiple mechanisms including formation of methylated derivatives in vivo. Sodium arsenite (up to 5.0 μM) renders arsenic methylation–competent TRL1215 rat liver epithelial cells tumorigenic in nude mice at 18 weeks of exposure and arsenic methylation-deficient RWPE-1 human prostate epithelial cells tumorigenic at 30 weeks of exposure. We assessed the role of arsenic biomethylation in oxidative DNA damage (ODD) using a recently developed immuno-spin trapping method.
Methods
Immuno-spin trapping was used to measure ODD after chronic exposure of cultured TRL1215 vs RWPE-1 cells, or of methylation-competent UROtsa/F35 vs methylation-deficient UROtsa human urothelial cells, to sodium arsenite. Secreted matrix metalloproteinase (MMP)-2 and -9 activity, as analyzed by zymography, cellular invasiveness by using a transwell assay, and colony formation by using soft agar assay were compared in cells exposed to arsenite with and without selenite, an arsenic biomethylation inhibitor, to assess the role of ODD in the transition to an in vitro cancer phenotype.
Results
Exposure of methylation-competent TRL1215 cells to up to 1.0 μM sodium arsenite was followed by a substantial increase in ODD at 5–18 weeks (eg, at 16 weeks with 1.0 μM arsenite, 1138% of control, 95% confidence interval [CI] = 797% to 1481%), whereas exposure of methylation-deficient RWPE-1 cells to up to 5.0 μM arsenite did not increase ODD for a 30-week period. Inhibition of arsenic biomethylation with sodium selenite abolished arsenic-induced ODD and invasiveness, colony formation, and MMP-2 and -9 hypersecretion in TRL1215 cells. Arsenic induced ODD in methylation-competent UROtsa/F35 cells (eg, at 16 weeks, with 1.0 μM arsenite 225% of control, 95% CI = 188% to 262%) but not in arsenic methylation-deficient UROtsa cells, and ODD levels corresponded to the levels of increased invasiveness, colony formation, and hypersecretion of active MMP-2 and -9 seen after transformation to an in vitro cancer phenotype.
Conclusion
Arsenic biomethylation appears to be obligatory for arsenic-induced ODD and appears linked in some cells with the accelerated transition to an in vitro cancer phenotype.
CONTEXT AND CAVEATS
Prior knowledge
Metabolic conversion of inorganic arsenic to methylated arsenic compounds was suspected to increase its carcinogenicity, but the mechanism was unclear.
Study design
Two arsenic biomethylation–competent cell lines (TRL1215, UROtsa/F35) and two arsenic biomethylation–deficient cell lines (RWPE-1, UROtsa) were continuously exposed to up to 5.0 μM inorganic sodium arsenite for up to 55 weeks. Oxidative DNA damage was measured by a new immuno-spin trapping procedure, and arsenic-treated cells were subjected to invasion, colony formation, and matrix metalloproteinase secretion assays as in vitro measures of transformation.
Contribution
Exposure of methylation-competent cells, but not methylation-deficient cells, was followed by a sharp rise in oxidative DNA damage. Subsequent to the peak of oxidative DNA damage, methylation-competent cells, more than methylation-deficient cells, acquired the in vitro characteristics of transformed cells, including growth in soft agar, increased invasiveness, and increased secretion of matrix metalloproteinases. This coincided with the time at which the cells became tumorigenic in nude mice.
Implications
Biomethylation of arsenic compounds appears to cause oxidative DNA damage and to increase their carcinogenicity. Alternate mechanisms appear to work less efficiently in methylation-deficient cells.
Limitations
Methylated arsenicals were not directly measured. All work was done in vitro in four cell lines, and in vivo correlates rested on previous experiments. In vivo testing in arsenic methylation-deficient mice has not yet been performed.
From the Editors
Inorganic arsenic is a common contaminant of human drinking water, and chronic arsenicosis affects tens of millions of people worldwide (1,2). Arsenic is clearly carcinogenic in humans and has multiple in vivo targets that include the skin, lung, bladder, prostate, and liver, although the carcinogenic mechanisms remain incompletely defined (1,2). Chronic low-level exposure to arsenic compounds induces in vitro malignant transformation of human and rodent cells derived from similar organs (3–7). Inorganic arsenic also undergoes multistep biomethylation in some, but not in all cells, through specific methyltransferases, notably arsenic–(+3 oxidation state)–methyltransferase (AS3MT), using S-adenosyl-l-methionine as the methyl donor (8–10). Inorganic arsenic was once believed to be detoxified by biomethylation (11,12), but more recent work has convincingly shown that it is not (13–18). In fact, trivalent methylated arsenic compounds, such as methylarsonous acid, are much more reactive and toxic than their unmethylated forms and consequently have been hypothesized to be the ultimate carcinogenic metabolites of inorganic arsenic (15–19). Because of their reactivity, if the methylated forms of arsenic were to be the ultimate carcinogens, methylation of arsenic would need to occur at, or near, the target cells of carcinogenic initiation in vivo.
Arsenic has genotoxic potential and is associated with DNA damage, such as chromosomal aberrations and sister chromatid exchange (13,20–22). Several in vivo and in vitro studies have shown that inorganic arsenic or methylarsonous acid induced the formation of reactive oxygen species and increased 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG) generation in vitro or in vivo (13,22–24). Widely used as a biomarker of oxidative DNA damage (ODD), 8-oxo-dG has been used as an indicator of genotoxicity by many chemicals, including arsenic (22–24). However, it is difficult to reliably measure 8-oxo-dG levels by standard methods, such as mass spectrometry, chromatography, or immunohistochemistry, because artifactual adventitious oxidation generated during DNA extraction and sample preparation results in high background readings (25,26) that often exceed any chemically induced increases in 8-oxo-dG, creating major questions about the relevance of relatively small chemically induced increases. Furthermore, 8-oxo-dG–based methods do not directly measure DNA radicals.
Recently, a new method was developed for detection of oxidative DNA radicals using immuno-spin trapping (IST; 27,28). IST directly traps DNA radicals by formation of 5,5-dimethyl 1-pyrroline N-oxide (DMPO)–DNA radical adducts, which are then converted to stable DNA–nitrone adducts before DNA isolation and immunochemical quantification for a simple, sensitive, and reliable means of ODD measurement. DNA radicals induced by reactive oxygen species are short-lived species, with half-lives in the nanoseconds to milliseconds because of their high reactivity, but when DMPO–DNA radical adducts are formed and converted to DNA–nitrone adducts, the half-lives are extended to months or years (27,28). Thus, the measurement of ODD by the IST method circumvents many of the potential errors of other methods heretofore in common use.
To address recent suspicions that arsenic biomethylation could be important to its mechanism of carcinogenesis, we used the IST method to assess the ability of chronic arsenite exposure to produce ODD in a rat liver epithelial cell line, TRL1215, which methylates arsenic (7), and in a human prostate cell line, RWPE-1, which methylates arsenic only very poorly (3,29). We used levels of chronic arsenite exposure (0.5–5.0 μM) that are known to induce malignant transformation in these cells by the appearance of a cancer phenotype in vitro (as defined by increased invasiveness, anchorage-independent growth in soft agar, and hypersecretion of matrix metalloproteinase [MMP]-2 and -9) and the ability to form tumors in mice (3,7). We also directly tested the hypothesis that arsenic-induced ODD and arsenic-induced acquisition of an in vitro cancer cells phenotype are mediated by arsenic methylation by comparing these features in human urinary bladder epithelial cells (UROtsa) that do not methylate arsenic vs UROtsa-derived cells expressing AS3MT (UROtsa/F35) that do methylate arsenic (30).
Methods
Cell Lines and Culture Conditions
The TRL1215 cell line is a rat liver epithelial cell line and was cultured in William E medium (Gibco/Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (Gibco/Invitrogen), 31 μg/mL penicillin, and 50 μg/mL streptomycin. The RWPE-1 cell line (passage 14) is a human prostate epithelial cell line kindly provided by Dr Mukta M. Webber (Michigan State University) and was cultured in keratinocyte serum-free medium (Gibco/Invitrogen) containing 50 μg/mL bovine pituitary extract (Gibco/Invitrogen) and 5 ng/mL epidermal growth factor (Gibco/Invitrogen) supplemented with 1% antibiotic/antimycotic mixture (100 U/mL penicillin and 100 μg/mL streptomycin). The UROtsa cell line is a human urothelial cell line and the UROtsa/F35 cell line is the UROtsa cell line transduced with the rat As3mt gene (30; both cell lines obtained from laboratory of Dr M. Stýblo); both were cultured in Eagle minimum essential medium (Gibco/Invitrogen) containing 10% fetal bovine serum supplemented with 1% antibiotic/antimycotic mixture. All cells were maintained in a humidified atmosphere of 5% CO2 and 95% air at 37°C. Untreated TRL1215, RWPE-1, and UROtsa cells are normally nontumorigenic (3,7,31). Phenotypically, TRL1215, RWPE-1, UROtsa, and UROtsa/F35 all have typical epithelial cell morphology in culture as previously reported (3,7,31). Genotypically, TRL1215 cells are periodically verified as liver epithelial cells with hepatocyte markers (P450 enzymes, metallothionein), RWPE-1 cells are verified as prostate epithelial cells via prostate-specific antigen, and UROtsa and UROtsa/F35 cells were recently extensively verified via genomics (32).
Chronic Arsenic Exposure and Cancer Phenotype
TRL1215, UROtsa, and UROtsa/F35 cells were continuously exposed to 0.25, 0.5, or 1.0 μM sodium arsenite (Sigma Chemical Co, St Louis, MO), and RWPE-1 cells to 1.0, 2.5, or 5.0 μM sodium arsenite for various times in their respective media in 75-cm2 tissue culture flasks. The medium was changed at half-weekly intervals, and cells were passaged weekly with new cultures seeded at 1 × 106 cells/flask. The concentrations of arsenite used had no effect on cell growth. Acquisition of a transformed phenotype in vitro was defined by increased invasiveness, anchorage-independent growth in soft agar, and hypersecretion of MMP-2 and -9, whereas malignant transformation was defined as all of these changes in addition to the ability to produce malignant tumors upon injection into nude mice.
DNA Isolation and ODD Measurement
ODD was measured in all cell lines by the previously established IST method (26,27), which directly measures the formation of DNA-centered radicals by adduction with DMPO, conversion to stable nitrone adducts, and immunochemical quantification. After growth in medium with or without sodium arsenite (0–5 μM) for up to 55 weeks, cells were incubated with 20 mM DMPO (Alexis Biochemicals, San Diego, CA) for 30 minutes at 37°C, harvested by incubation in trypsin–EDTA, and washed three times with calcium- and magnesium-free phosphate-buffered saline (CMF-PBS). Cells (approximately 1 × 106) were then incubated in 500 μL of digestion buffer (1% sodium dodecyl sulfate, 100 mM NaCl, 25 mM diethylenetriaminepentaacetic acid, and 10 mM Tris–HCl; pH 8.0) containing 25 μL proteinase K (Sigma Chemical Co.; from a 20 mg/mL stock solution of proteinase K in 50 mM Tris–HCl containing 1 mM CaCl2) for 1 hour at 52°C. After the addition of 10 μL RNase A (20 mg/mL), the incubation continued for 1 hour at 37°C. DNA was purified by a three-step procedure that used successive extractions in ultrapure buffer-saturated phenol (Invitrogen), 1 mM diethylenetriaminepentaacetic acid, phenol:chloroform:isoamyl alcohol (25:24:1), and chloroform:isoamyl alcohol (24:1) (27,28). DNA was resuspended in 100 μL TE buffer (10 mM Tris–HCl [pH 8.0], 1 mM EDTA), and its purity and concentration were determined by absorbance at 260 and 280 nm. This method produced DNA preparations with an Abs260/280 ratio between 1.8 and 2.0 (27,28). The purified DNA containing the nitrone adducts was then diluted to 5 μg/mL in CMF-PBS, and 25 μL of DNA solution were mixed with 25 μL of React-bind DNA coating solution (Pierce Chemicals, Rockford, IL) in each well of flat-bottom 96-well microtiter plates (PGC Scientifics, San Diego, CA). The reactions were incubated for 4 hours at 37°C to allow the DNA to bind to the wells, after which wells were washed once with 300 μL washing buffer (CMF-PBS containing 0.05% nonfat dry milk and 0.1% Tween-20). To block nonspecific binding of antisera, 120 μL of blocking solution (CMF-PBS containing 3% nonfat dry milk) was added to each well and incubated for 1.5 hours at 37°C. Each well was then washed once for 5 minutes with 300 μL washing buffer on an orbital shaker at room temperature. Next, 100 μL of rabbit anti-DMPO polyclonal serum (1:10 000 dilution in washing buffer; Cayman Chemical, Ann Arbor, MI) was added to each well and incubated for 1 hour at 37°C. Plates were washed three times with 300 μL washing buffer, and 100 μL of goat anti-rabbit IgGFc conjugated to horseradish peroxidase (1:10 000; Pierce) was added and incubated for 1 hour at 37°C. After washing the plates three times with 300 μL washing buffer, 50 μL of LumiGLO chemiluminescent substrate (Upstate, Temecula, CA) was added to each well and incubated for 30 seconds, and luminescence was read using Xfluor4 Software (Tecan, Männedorf, Switzerland). ODD measurements represent the mean of nine samples from three independent experiments.
Assessment of Acquired Cancer Phenotype
Several common assays were used to assess whether the cells acquired an in vitro cancer phenotype during arsenic exposure and were applied to all cell lines used. All assays represent the mean of nine samples from three independent experiments. The assays included zymographic analysis of secreted MMP-2 and -9 activity after the method of Achanzar et al. (3), in which cells were cultured in basal medium (without serum or supplements) for 48 hours, the conditioned medium was collected, and secreted MMP-2 and -9 activity was measured by a standard zymographic method. Enhanced MMP-2 and -9 activity is common for cancer cells and frequently found in cells transformed by arsenic (3,29,33–36).
Cellular invasiveness was assessed as described previously (37). Culture medium containing 10% fetal bovine serum served as a chemoattractant and was loaded in the lower well of a blind-well chamber (NeuroProbe, Inc, Gaithersburg, MD). A polycarbonate 8.0-μm filter (Fisher Scientific, Hanover Park, IL) coated with Matrigel (BD Bioscience, San Jose, CA) was layered on top of the lower well. Cells (2 × 105 cells/chamber) were suspended in basal medium and added on top of the filter and incubated for 48 hours. Cell invasion was then quantified as the number of cells that penetrated the Matrigel and filter. Cells were fixed and stained (HEMA 3 Manual Staining System; Fisher Scientific), stain was extracted, and absorbance determined at 630 nm and quantified as percent control.
Colony formation in soft agar was performed as previously described method (38) to assess the cells’ capacity for anchorage-independent growth. Cell line–dependent medium (see above) containing 0.5% agar was first added to 35-mm culture plates, and 1.25 × 104 cells were then suspended in medium containing 0.33% agar and layered on top. Cultures were incubated at 37°C in 5% CO2 for 3 weeks, and then, the number of colonies (all sizes) was counted with an automated colony counter.
Real-Time Reverse Transcription–Polymerase Chain Reaction Analysis
Expression levels for glutathione-S-transferase-π (GSTP1; accession number: NM_000852; primers, forward: 5′-AGAGCTGGAGGAGGAGGTG-3′; reverse: 5′-AGGTCTCCGTCCTGG-AACTT-3′), ATP-Binding Cassette C1 (ABCC1; accession number: L05628; primers, forward: 5′-GAGGAGGTGGAGGCTTTGATC-3′; reverse: 5′-AAGTAGGGCCCAAAGGTCTTG-3′), nuclear factor (erythroid-derived 2)-like 2 (NFE2L2; accession number: NM_006164; primers, forward: 5′-AACCAGTGGATCTGCCAACTACTC-3′; reverse: 5′-CTGCGCCAAAAGCTGCAT-3′), and heme oxygenase-1 (HMOX1; accession number: NM_002133; forward: 5′-GCCTGGAAGACACCCTAATGTG-3′; reverse: 5′-GGCCGTGTCAACAAGGATACTT-3′) were quantified by real-time reverse transcription–polymerase chain reaction analysis by the reaction conditions described in Liu et al. (39). The selected genes were first normalized with β-actin levels within the same sample and then expressed as percent control (0 μM arsenite in each case), which was set to 100%. Results are expressed as the mean of nine samples from three independent experiments.
Influence of Selenite on Arsenite-Induced Acquired Cancer Phenotype
TRL1215 cells were continuously exposed to 0.5 μM sodium arsenite in the presence or absence of 1.0 μM sodium selenite (Sigma) for up to 24 weeks in 75-cm2 tissue culture flasks. The medium was changed at half-weekly intervals, and cells were passaged weekly with new cultures seeded at 1 × 106 cells/flask. The levels of arsenite and selenite had no effect on cell growth. ODD, MMP-2 and -9 secretion, cellular invasiveness, and colony formation in these cells were measured approximately biweekly as above. The activity of glutathione peroxidase was measured using a Glutathione Peroxidase Assay Kit (Cayman) in control and selenite-treated cells at 0, 4, 8, 16, and 24 weeks of exposure. Control cells and cells exposed to 1.0 μM selenite for 20 weeks were further treated with 0.0, 1.0, 2.5, and 5.0 mM hydrogen peroxide for 24 hours, and then, ODD in these cells was measured. Results are expressed as the mean of nine samples from three independent experiments.
Statistics
Data are expressed as the mean with 95% confidence intervals (CIs). Statistical significance was determined by Student t test or analysis of variance followed by Dunnett multiple comparison test, as appropriate. In some cases, correlations were calculated by linear regression and further assessed by Pearson correlation. A P value of less than .05 from two-sided tests was considered to be statistically significant in all cases.
Results
ODD and Chronic Arsenite Exposure at Levels Known to Induce Malignant Transformation
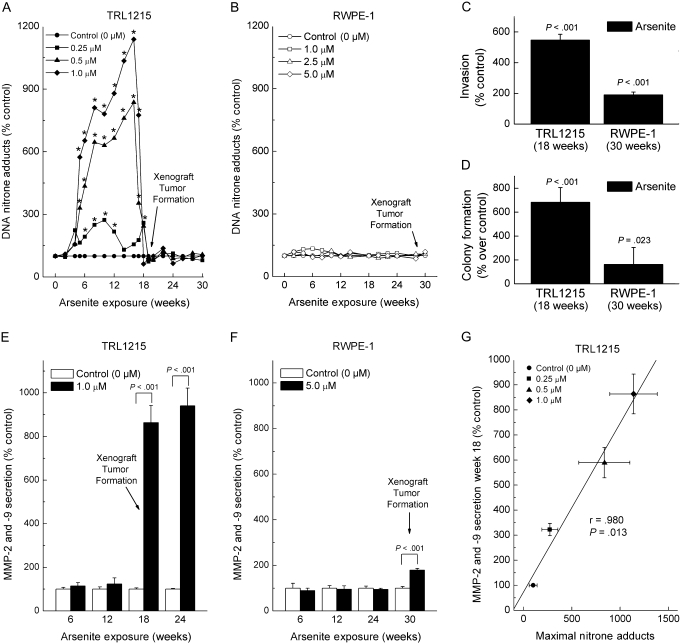
In our initial experiments, arsenic biomethylation–competent TRL1215 rat liver cells and arsenic methylation–deficient RWPE-1 human prostate cells were continuously exposed to sodium arsenite concentrations known to induce malignant transformation as defined by the acquisition of a transformed phenotype in vitro and the ability to produce malignant xenograft tumors in immunodeficient mice (3,7). Cellular ODD was measured during exposure to arsenite at approximately biweekly intervals for up to 30 weeks (Figure 1). Exposure of TRL1215 cells to arsenite induced a remarkable, but delayed, increase in ODD in an arsenite concentration–dependent manner, starting at 5 weeks of exposure (Figure 1, A). For example, after growth in medium with 0.5 μM arsenite for 16 weeks, ODD reached 836% compared with control cells without arsenite (95% CI = 368% to 1304%, P < .01), and after growth in medium with 1.0 μM arsenite for 16 weeks, ODD reached 1138% of control (95% CI = 797% to 1481%, P < .01). The ODD levels then precipitously declined to baseline after about 18–20 weeks of exposure. Previous work showed that malignant transformation of TRL1215 cells, as established by production of malignant xenograft tumors in mice, occurs at 18 weeks of exposure to similar levels of arsenite as used in this study and that tumor formation rate is directly related the arsenic concentration used for in vitro transformation (7). In sharp contrast, after chronic exposure of RWPE-1 cells to arsenite, there was no evidence of ODD (Figure 1, B). For example, for cells grown in medium with 5.0 μM arsenite for 30 weeks, ODD was 119% compared with that in control cells grown without arsenite (95% CI = 103% to 135%, P > .05). RWPE-1 cells undergo malignant transformation with these arsenic levels, as established by production of malignant xenograft tumors, although it requires approximately 30 weeks of continuous arsenic exposure in vitro, they are able to do so after inoculation into mice (3).
Figure 1.
Oxidative DNA damage (ODD) and acquisition of an in vitro transformed phenotype upon chronic sodium arsenite exposure in arsenic methylation–competent TRL1215 cells and arsenic methylation–deficient RWPE-1 cells. A) TRL1215 cells were exposed to various concentrations of sodium arsenite for up to 30 weeks, and ODD was assessed by the formation of DNA–nitrone adducts using the immuno-spin trapping method (27,28). B) RWPE-1 cells were exposed to sodium arsenite for up to 30 weeks, and ODD was assessed as in (A). C) Cellular invasiveness, as an in vitro marker of malignant transformation, was measured in a transwell assay with a Matrigel-coated membrane for TRL1215 cells (0.5 μM) after 18 weeks of arsenite exposure and for RWPE-1 cells (5.0 μM) at 30 weeks of arsenite exposure. D) Anchorage independence, as an in vitro marker of malignant transformation, was measured by colony formation in soft agar for TRL1215 cells (0.5 μM) after 18 weeks of arsenite exposure and for RWPE-1 cells (5.0 μM) after 30 weeks of arsenite exposure. Colonies (all sizes) were scored. E) Secreted matrix metalloproteinase (MMP)-2 and -9 activity during chronic arsenite exposure in TRL1215 cells. Cells were exposed to 1 μM sodium arsenite for 6–24 weeks, and MMP-2 and -9 in conditioned media were assayed by zymography. F) Secreted MMP-2 and -9 activity during chronic arsenite exposure in RWPE-1 cells. Cells were exposed to 5 μM and otherwise treated as in (F). G) Correlation between arsenite-induced, concentration-response ODD (from A) and secreted MMP-2 and -9 activity at 18 weeks at the various concentrations of chronic arsenite exposure. Statistical analysis performed by using linear regression and further assessed by Pearson correlation. Results in all panels (A–G) are expressed as the mean of nine samples from three independent experiments. The 95% confidence intervals have been included in panels (C–G) but have been omitted for clarity in (A) and (B) and can be found in Supplementary Tables 1 and 2, available online. In panels (A), (B), (E), and (F), an arrow marked “Xenograft Tumor Formation” marks the time point at which the specific cells in question acquired the ability to form malignant tumors after inoculation into nude mice in our prior work with these same cells under similar conditions (3,7). All P values are from two-sided tests.
Both the TRL1215 and the RWPE-1 cell lines acquired an in vitro transformed phenotype after arsenite exposure, based on invasiveness, colony formation, and hypersecretion of MMP-2 and -9 (Figure 1, C–F). All three parameters were increased over the course of 18 weeks of arsenite exposure for TRL1215 cells, or over 30 weeks of arsenite exposure for RWPE-1 cells, which were time frames similar to those reported for these cell lines to acquire the ability to form xenograft tumors in mice (3,7). For example, more than 18 weeks of arsenite exposure invasiveness of TRL1215 cells increased to 547% of unexposed control cells (95% CI = 509% to 585%, P < .001), and colony formation increased 719% above that in control cells (95% CI = 595% to 845%, P < .001). MMP-2 and -9 secretion after 18 weeks of arsenite exposure increased 836% above that in control cells (95% CI = 796% to 876%, P < .001). By contrast, RWPE-1 cells exposed to 5 μM arsenite for 30 weeks showed a more modest 191% increase in invasion compared with control cells (95% CI = 172% to 209%, P < .001). Likewise, at 30 weeks of arsenite exposure, RWPE-1 cells showed a comparatively modest increase in colony formation at 260% of control (95% CI = 104% to 415%, P = .023), and an increase of MMP-2 and -9 secretion of 179% of control (95% CI = 172% to 186%, P < .001). MMP-2 and -9 are frequently secreted by cancer cells (33–35), including arsenic-transformed cells (3,29,36). Levels of increased MMP-2 and -9 secretion in TRL1215 cells at the approximate time of malignant transformation (week 18) showed a robust positive correlation (linear regression followed by Pearson correlation, r = .98, P = .013) with the levels of maximal ODD for all of the various concentrations of arsenite used for treatment, suggesting that ODD was positively linked to the acquired cancer phenotype (Figure 1, G). In fact, when MMP-2 and -9 secretion was measured in TRL1215 cells that had acquired a cancer phenotype in vitro after 18 weeks of arsenite exposure in present work, and correlated with either in vitro arsenite concentration-related xenograft tumor formation rate or the rate of metastasis seen previously (7), both showed robust positive correlations (linear regression followed by Pearson correlation, P < .001; Supplementary Figure 1, A and B, available online). From these results, it appeared that arsenic methylation might be obligatory for ODD, and although cells could acquire a cancer phenotype in its absence, the presence of ODD might hasten the process. However, TRL1215 and RWPE-1 cells differ widely in terms of genetic background, which made direct comparisons of time to acquired cancer phenotype in vitro somewhat problematic. Because arsenic methylation can vary widely with target site in vivo, the relative role of ODD formation in vivo will also require direct study.
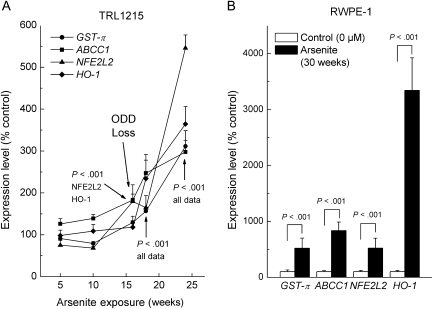
ODD in TRL1215 cells rapidly diminished with time after transformation. We initially suspected that this decline was because of progressive arsenic adaptation. Various genes could participate in this adaptation including those related to oxidative stress, glutathione metabolism, and arsenic transport. Therefore, we looked at expression of GSTP1, ABCC1, NFE2L2, and HMOX1 using real-time reverse transcription–polymerase chain reaction.
ABCC1 encodes for a transport protein that is responsible for arsenic efflux in complex with a glutathione trimer produced by GST-π and is a key to arsenic tolerance (40). In TRL1215 cells, arsenic increased expression of GSTP1 and ABCC1 (Figure 2, A). Glutathione levels were also increased by arsenite (at 24 weeks of arsenite exposure, maximal levels = 264%,of unexposed control cells, 95% CI = 184% to 344%, P < .001).
Figure 2.
Expression of adaptive response genes during chronic arsenite exposure in TRL1215 and RWPE-1 cells. A) Expression of the genes for glutathione-S-transferase-π (GSTP1, here shown as GST-π), the ATP-Binding Cassette Transporter C1 (ABCC1), an antioxidant response transcription factor (NFE2L2), and heme oxygenase-1 (HMOX1, here shown as HO-1) in TRL1215 cells exposed to arsenite. Expression was quantified by reverse transcription–polymerase chain reaction and normalized to β-actin expression levels within each sample. An arrow points to the approximate time of rapid decline in oxidative DNA damage (ODD) (ODD Loss; see Figure 1, A). Arrows with P values indicate statistical significance of specific data at specific time points. B) Adaptive response gene expression in RWPE-1 cells at their time of malignant transformation (30 weeks). Assays were performed as in (A). Results are expressed as the mean of nine samples from three independent experiments with 95% confidence interval. All P values are from two-sided tests.
Arsenic also enhanced the expression of oxidative stress-related genes like NFE2L2, HMOX1 (Figure 2, A), superoxide dismutase 1 (eg, at 18 weeks, 211% of control, 95% CI = 162% to 260%, P < .001), and metallothionein-1 (eg, at 18 weeks, 1727% of control, 95% CI = 1315% to 2138%, P < .001). However, these genes were also induced at 30 weeks of arsenic exposure in RWPE-1 cells (Figure 2, B), which suggests that cell-specific increases in expression as an adaptive phenomenon are unlikely to explain the precipitous loss of ODD in TRL1215 cells.
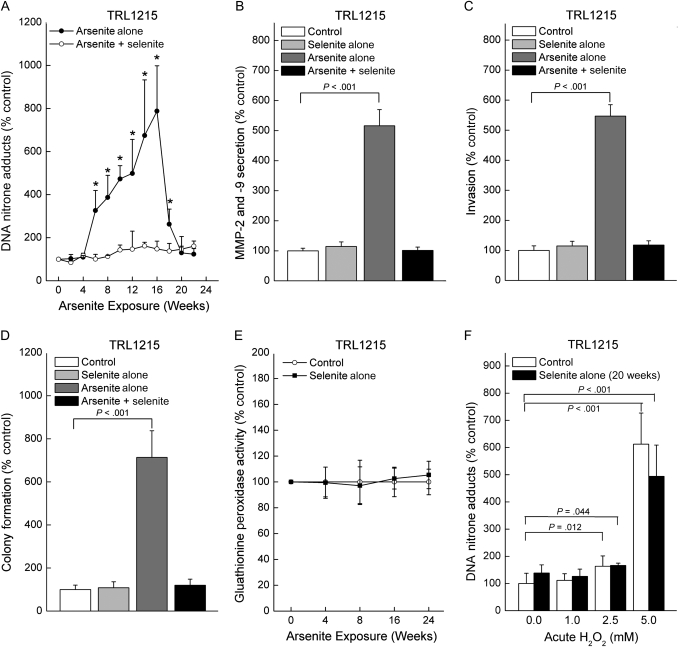
Treatment of cells with sodium selenite very effectively inhibits arsenic methylation (41). Therefore, to further test whether arsenic methylation might be responsible for ODD, TRL1215 cells were exposed to a concentration of sodium arsenite, 0.5 μM, that induces ODD and is known to produce malignant transformation (7), together with or without a nontoxic level of sodium selenite, 1.0 μM, for up to 24 weeks. Co-exposure to selenite abolished arsenite-induced ODD (Figure 3, A). Furthermore, at 18 weeks exposure to arsenite [the approximate time of malignant transformation for TRL1215 cells (7)], arsenite-induced increases in MMP-2 and -9 secretion, invasion, and colony formation were all abolished by selenite (Figure 3, B–D). Selenite could potentially have antioxidant effects, most likely as a component of antioxidant enzymes, the most prominent being glutathione peroxidase; suboptimal selenium nutritional status in humans reduces glutathione peroxidase activity, which can increase cellular reactive oxygen species (42). However, chronic selenite exposure had no effect on glutathione peroxidase activity in TRL1215 cells (Figure 3, E). Furthermore, if selenite treatment were blocking oxidative stress to eliminate ODD, rather than inhibiting arsenic methylation, it would be expected to block ODD from externally applied oxidants. However, when cells chronically treated with selenite were exposed to hydrogen peroxide, they showed the same level of ODD as in control (Figure 3, F), indicating that selenite treatment had no effect on ODD induced by reactive oxygen species or at least from the species generated by hydrogen peroxide. These data support a non-antioxidative mechanism for selenite-mediated inhibition of arsenite-induced ODD. This conclusion is consistent with the scenario that biomethylated arsenicals could be key factors in arsenite-induced ODD, and the ability of selenite to reduce ODD appears to be related to a blockade of acquired cancer phenotype in vitro.
Figure 3.
Influence of exposure to sodium selenite (1.0 μM) on arsenite-induced (0.5 μM) oxidative DNA damage (ODD) and acquired in vitro cancer phenotype in TRL1215 cells. A) ODD in cells exposed to sodium arsenite in the presence or absence of sodium selenite for up to 24 weeks, as assessed by the immuno-spin trapping method. Control cells and those grown in the presence of selenite alone showed no ODD and are omitted for clarity. B) Influence of selenite on secreted matrix metalloproteinase (MMP)-2 and -9 activity induced by chronic arsenite exposure at 18 weeks, the time of malignant transformation as assessed by arsenic-induced xenograft tumor formation in a prior study (7). Assays were performed as in Figure 1, E. C) Influence of selenite on invasiveness induced by 18 weeks of chronic arsenite exposure. Assays were performed as in Figure 1, C. D) Influence of selenite on colony formation induced by 18 weeks of chronic arsenite exposure. Assays were performed as in Figure 1, D. E) Influence of selenite on glutathione peroxidase activity for more than 24 weeks of exposure. F) Influence of 20 weeks of chronic selenite treatment on hydrogen peroxide–induced ODD. Hydrogen peroxide was added to cells at indicated levels for 24 hours before ODD measurement. Untreated cells served as a control. Results in all cases are expressed as the mean with 95% confidence intervals of nine samples from three independent experiments. In panel (A), an asterisk indicates a statistically significant (P < .05) difference from control. All P values are from two-sided tests. P values were calculated by using analysis of variance followed by Dunnett multiple comparison test (B–D) or Student t test (A and F).
Association of Arsenic-Induced ODD With Acquisition of a Cancer Phenotype in UROtsa/F35 Cells but not UROtsa Cells
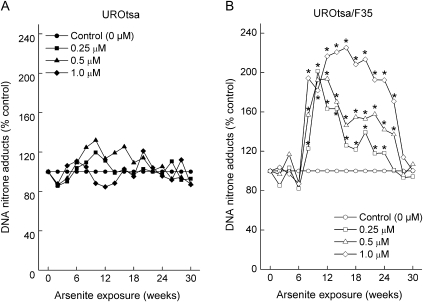
To test the role of arsenic biomethylation in ODD in an isogenic pair of cell lines that differed only in their ability to methylate arsenic, we also did experiments in UROtsa human urothelial cells and in UROtsa/F35 cells, which are stably transduced with As3mt and methylate arsenic (30). Chronic exposure of arsenic methylation–deficient UROtsa human urothelial cells to 0.25–1.0 μM sodium arsenite did not induce ODD (Figure 4, A). However, exposure of UROtsa/F35 cells to the same arsenite concentrations induced a remarkable, but delayed, increase in ODD starting after 6 weeks of exposure that rose to 192% of control (10 weeks, 95% CI = 180% to 205%, P < .01) for cells exposed to 0.5 μM or to 225% of control (16 weeks, 95% CI = 198% to 252%, P < .01) for cells exposed to 1.0 μM (Figure 4, B). ODD, as measured by levels of DNA–nitrone adducts, then precipitously returned to baseline after approximately 28 weeks of arsenic exposure. This pattern of delayed ODD onset and then precipitous loss was remarkably similar to that seen in methylation-competent TRL1215 cells (Figure 1, A).
Figure 4.
Oxidative DNA damage (ODD) induced by chronic low-level sodium arsenite exposure in UROtsa cells and UROtsa/F35 cells. UROtsa cells (A) and UROtsa/F35 cells (B) were exposed to 0.25, 0.5, or 1.0 μM sodium arsenite for up to 30 weeks, and ODD was assessed by the immuno-spin trapping method. Results are expressed as mean of nine samples from three independent experiments (the 95% confidence intervals have been omitted for clarity but can be found in the Supplementary Tables 3 and 4, available online). An asterisk indicates a statistically significant (P < .05) difference from control (Supplementary Tables 3 and 4, available online, for complete data). P values were calculated by analysis of variance followed by Dunnett multiple comparison test.
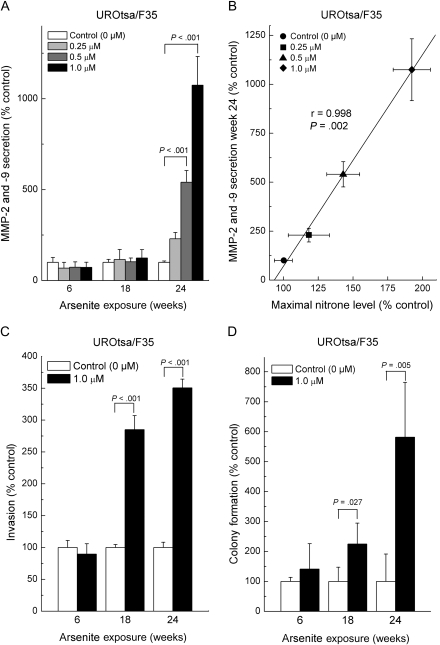
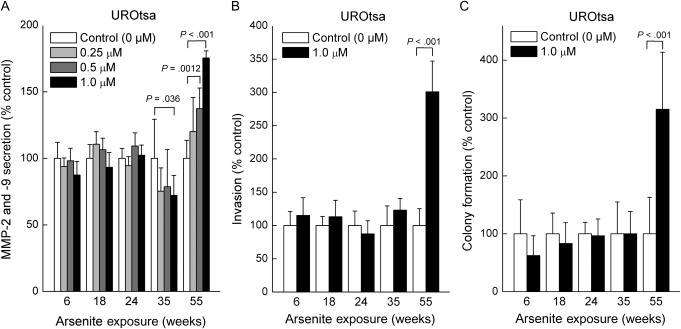
We next tested our hypothesis that arsenite methylation mediated arsenite-induced ODD and accelerated the acquisition of a cancer cell phenotype in vitro by using arsenic biomethylation–deficient UROtsa vs methylation–competent UROtsa/F35 cells. Arsenite exposure in biomethylation-competent UROtsa/F35 cells markedly increased MMP-2 and -9 secretion, with dramatic increases by 24 weeks (510% of control at 0.5 μM arsenite, 95% CI = 323% to 698%, P < .01; 1055% of control at 1.0 μM arsenite, 95% CI = 634% to 1476%, P < .01; Figure 5, A). Enhanced MMP secretion is common in urothelial cancer and in arsenic-transformed cells (3,29,33–36). Indeed, MMP-2 and -9 secretion, a marker of an acquired cancer phenotype in vitro and in vivo (3,29,33–36), was strongly correlated (r = .998, P = .002, linear regression followed by Pearson correlation) with ODD in arsenite-exposed UROtsa/F35 cells (Figure 5, B). Chronic exposure to 1 μM sodium arsenite also increased invasiveness and colony formation in arsenic methylation–competent UROtsa/F35 cells by 18 weeks of exposure, at which time, invasion was 285% of control (95% CI = 263% to 307%, P < .001) and colony formation was 224% of control (95% CI = 95% to 355%, P = .027) (Figure 5, C and D). By contrast, exposure of methylation-deficient UROtsa cells to arsenite only showed increases in MMP-2 and -9 secretion, invasiveness, or colony formation beginning at 55 weeks of arsenite exposure. For example, invasiveness was increased 300%, compared with control, in UROsta/F35 cells exposed to 1.0 μM arsenic for 55 weeks (95% CI = 254% to 346%, P < .001; Figure 6, C). Because UROtsa/F35 cells are isogenic with UROtsa cells except that the former contains the AS3MT transgene, which allows arsenic methylation (30), it seems reasonable to attribute the more rapid acquisition of an in vitro transformed phenotype in arsenite-exposed UROtsa/F35 cells to their ability to methylate arsenic and thereby acquire ODD.
Figure 5.
Evidence for acquired cancer phenotype in vitro and correlation with oxidative DNA damage (ODD) in UROtsa/F35 cells. UROtsa/F35 cells were exposed to sodium arsenite for up to 30 weeks, and secreted matrix metalloproteinase (MMP)-2 and -9 activity by zymography, cellular invasiveness by Boyden chamber assay, and colony formation by agar assay were assessed. Secreted MMP-2 and -9 activity was also correlated with ODD as assessed by the immuno-spin trapping method. A) Secreted MMP-2 and -9 activity during chronic arsenite exposure. B) Correlation between arsenite-induced, concentration-response ODD and secreted MMP-2 and -9 activity (24 weeks). Statistical analysis done by linear regression and further assessed by Pearson correlation. C) Cellular invasiveness. D) Colony formation. Results are expressed as the mean with 95% confidence intervals (nine samples from three independent experiments). All statistical tests were two-sided. P values were calculated by using analysis of variance followed by Dunnett multiple comparison test (A) or Student t test (C and D).
Figure 6.
Evidence for acquired cancer phenotype in vitro in UROtsa cells. UROtsa cells were exposed to sodium arsenite for up to 55 weeks, and secreted matrix metalloproteinase (MMP)-2 and -9 activity by zymography, cellular invasiveness by Boyden Chamber assay, and colony formation by agar assay were assessed. A) MMP-2 and -9 secreted activity during chronic arsenite exposure. B) Cellular invasiveness. C) Colony formation. Results are expressed as the mean with 95% confidence intervals of nine samples from three independent experiments. All P values are from two-sided tests. P values were calculated using analysis of variance followed by Dunnett multiple comparison test (A) or Student t test (B and C).
Discussion
In this study, we used the IST method, which measures DNA radicals directly, to investigate the ability of inorganic arsenite to induce ODD in arsenite methylation–deficient and –competent cells. A major advantage of the IST method is that it converts unstable DNA radicals induced by reactive oxygen species into stable nitrone adducts allowing for direct measurement of DNA radicals and avoiding sample preparation–induced artifactual ODD, making results more reliable than previous methods (27,28). Furthermore, we used biologically relevant levels of arsenite exposure: chronic exposure to inorganic arsenic levels in the 5.0 μM range in drinking water has been associated with oxidative stress in humans (43).
In our initial experiments, we found ODD to occur in TRL1215 cells, which can methylate arsenic, but not in RWPE-1 cells, which cannot, even though both cell lines were exposed to arsenite concentrations and durations sufficient to cause an acquired malignant phenotype in vitro (in the present work) and instill the ability when inoculated to cause formation of malignant tumors in immunodeficient mice (3,7). An inhibitor of arsenic methylation, sodium selenite (40), abolished arsenite-induced ODD and the acquisition of an in vitro cancer phenotype in arsenic methylation–competent TRL1215 cells. These observations were fortified by our observation that arsenic methylation–competent (As3mt-transduced) UROtsa/F35 cells showed much more ODD upon arsenic exposure than its parental UROtsa cell line and acquired a transformed phenotype in vitro in much less time. Hence, it appears that arsenic methylation is obligatory for ODD in some cells and hastens the acquisition of cancer phenotype. However, because cells unable to methylate arsenic can still acquire an arsenic-induced cancer phenotype in vitro over time, it is likely that arsenic is also carcinogenic by mechanisms that neither require biomethylation nor ODD. It is possible that multiple mechanisms may account for carcinogenesis by arsenic even within the same cell line and that the speed with which a cancer phenotype is acquired depends on the number of mechanisms at play.
Chronic exposure to low levels of sodium arsenite caused a remarkable, but delayed, increase in ODD only in arsenic methylation–competent cells. Accumulating evidence indicates that the methylation of arsenic is not a detoxifying event and that arsenic biomethylation may produce highly toxic compounds with genotoxic potential (13–19). Although inorganic arsenic can stimulate reactive oxygen species production, trivalent methylated arsenic compounds appear to stimulate the production of reactive oxygen species more efficiently than inorganic arsenic compounds (13). Indeed, ex vivo evidence indicates that dimethylarsinous acid is an indirect genotoxin that forms hydroxyl radicals through an unknown mechanism (44). In addition to directly increasing reactive oxygen species, arsenic compounds might also induce ODD indirectly by inhibiting important detoxifying enzymes. Generally speaking, trivalent methylated arsenicals are more potent inhibitors of enzymes when compared with inorganic forms, possibly because of higher affinities for critical thiol groups (45). It is important to note that high concentrations of inorganic arsenic that are lethal to the cells can acutely induce ODD in cells without arsenic biomethylation, like RWPE-1 cells (data not shown), but the relevance of finding ODD in dead or dying cells to cancer is undoubtedly limited.
Several striking, mutually supportive similarities were evident in the temporal pattern of ODD generation in the arsenic methylation–competent cells (TRL1215 and UROtsa/F35) in this study. First, both the human and the rat cells showed a delay of several weeks before the onset of ODD [Figures 1, A and 4, B (7)]. Second, both cell lines showed a rapid drop-off in levels of ODD at about the point in time they acquired a cancer phenotype [Figures 1, A and 4, B (7)]. The reasons for both phenomena are unclear. The delay may involve buildup of a key toxic metabolite, such as a methylated arsenic compound. Initially, we suspected that the precipitous loss of ODD might be attributed to adaptation to arsenic; adaptive increases in arsenic transport–related gene expression (eg, GSTP1 and ABCC1) and in glutathione levels are not uncommon, all of which promote arsenic efflux from exposed cells (40,46,47). Chronic arsenic exposure also typically increases the expression of stress-response genes such as NFE2L2, HMOX1, and metallothionein (39,48,49). We had suspected that a differential adaptive response might account for the loss of ODD in TRL1215 cells, but we found these genes to also be overexpressed in RWPE-1 cells (which show no ODD) at about the time of arsenic-induced malignant transformation, so this gene expression pattern is not likely to be an adaptation caused by ODD loss. In any event, by the time of the precipitous drop in ODD, it appears that sufficient damage had already occurred to hasten malignant transformation in methylation-competent cells.
Some limitations of this study have already been discussed. It is unknown why arsenic biomethylation–competent cells show a delayed onset of ODD with arsenic exposure or why there is a precipitous drop near the point of acquired cancer phenotype. Furthermore, the observed ODD, although consistently linked to more rapid acquisition of cancer phenotype, is not linked to precise precipitating events (ie, mutations) that would drive cells more rapidly toward malignancy. This is an unproven assumption. Only four cell lines were used in this work, two of which are isogenic with the exception of the transduction of As3mt. Methylated arsenicals were never actually measured, and it is only assumed that they were differentially produced based on prior work. Cell lines exposed here to arsenite in vitro were not directly used in in vivo xenograft studies but were assumed to be transformed based on in vitro phenotype and timing relative to earlier experiments. In vivo testing in arsenic methylase knockout mice would be critical to confirm our hypothesis. In vitro cell models of cancer may not be strictly concordant in timing, and so on, with much more complicated formation of tumors in vivo.
In summary, chronic exposure to inorganic arsenic consistently caused a delayed increase in ODD, but only in cells able to methylate arsenic, which was then precipitously lost at about the same time that these methylation-competent cells acquired a cancer phenotype in vitro. Arsenite-induced ODD appeared to accelerate acquisition of cancer phenotype in vitro and only required the transduction of a single gene, As3mt. Human data are emerging that clearly show that polymorphisms in AS3MT exist that affect arsenical methylation patterns (50) and thereby may alter susceptibility to carcinogenesis. AS3MT-null humans are not known, but As3mt knockout mice have very recently been introduced (51). Although inorganic arsenicals have not yet been tested for carcinogenic effects in these genetically altered mice, this clearly should be a high priority. AS3MT polymorphism analysis may one day provide a metric of human susceptibility to arsenical carcinogenesis at critical target sites.
Funding
Intramural Research Program of the National Cancer Institute, Center for Cancer Research (C.K., E.J.T., M.P.W.); National Institute of Environmental Health Sciences (D.C.R., R.P.M.).
Supplementary Material
Footnotes
The authors are solely responsible for the design of the study, the collection and analysis of data and the interpretation of the results, the preparation of the manuscript, and the decision to submit the manuscript for publication.
Present address: Free Radical Biology and Aging Research Program, Oklahoma Medical Research Foundation, Oklahoma City, OK (D. C. Ramirez).
We thank Drs J. Liu and L. Keefer for careful review of this material and Mr M. Bell for his help with the graphics.
References
- 1.International Agency for Research on Cancer (IARC) Arsenic in Drinking Water. International Agency for Research on Cancer Monographs on the Evaluation of Carcinogenic Risk to Humans. Vol. 84. Lyon, France: IARC Press; 2004. pp. 269–477. [Google Scholar]
- 2.National Research Council. Arsenic in Drinking Water. Washington, DC: National Academy Press; 1999. [Google Scholar]
- 3.Achanzar WE, Brambila EM, Diwan BA, Webber MM, Waalkes MP. Inorganic arsenite-induced malignant transformation of human prostate epithelial cells. J Natl Cancer Inst. 2002;94(24):1888–1891. doi: 10.1093/jnci/94.24.1888. [DOI] [PubMed] [Google Scholar]
- 4.Sens DA, Park S, Gurel V, Sens MA, Garrett SH, Somji S. Inorganic cadmium- and arsenic-induced malignant transformation of human bladder epithelial cells. Toxicol Sci. 2004;79(1):56–63. doi: 10.1093/toxsci/kfh086. [DOI] [PubMed] [Google Scholar]
- 5.Bredfeldt TG, Jagadish B, Eblin KE, Mash EA, Gandolfi AJ. Monomethylarsonous acid induces transformation of human bladder cells. Toxicol Appl Pharmacol. 2006;216(1):69–79. doi: 10.1016/j.taap.2006.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pi J, He Y, Bortner C, et al. Low level, long-term inorganic arsenite exposure causes generalized resistance to apoptosis in cultured human keratinocytes: potential role in skin co-carcinogenesis. Int J Cancer. 2005;116(1):20–26. doi: 10.1002/ijc.20990. [DOI] [PubMed] [Google Scholar]
- 7.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci U S A. 1997;94(20):10907–10912. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: arsenite-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005;79(4):183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- 9.Thomas DJ, Li J, Waters SB, et al. Arsenic (+3 oxidation state) methyltransferase and the methylation of arsenicals. Exp Biol Med. 2007;232(1):3–13. [PMC free article] [PubMed] [Google Scholar]
- 10.Waters SB, Devesa V, Del Razo LM, Styblo M, Thomas DJ. Endogenous reductants support catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem Res Toxicol. 2004;17(3):404–409. doi: 10.1021/tx0342161. [DOI] [PubMed] [Google Scholar]
- 11.Sakurai T, Kaise T, Matsubara C. Inorganic and methylated arsenic compounds induce cell death in murine macrophages via different mechanisms. Chem Res Toxicol. 1998;11(4):273–283. doi: 10.1021/tx9701384. [DOI] [PubMed] [Google Scholar]
- 12.Sakurai T, Qu W, Sakurai MH, Waalkes MP. A major human arsenic metabolite, dimethylarsinic acid, requires reduced glutathione to induce apoptosis. Chem Res Toxicol. 2002;15(5):629–637. doi: 10.1021/tx0101604. [DOI] [PubMed] [Google Scholar]
- 13.Gomez SE, del Razo LM, Munoz Sanchez JL. Induction of DNA damage by free radicals generated either by organic or inorganic arsenic (AsIII, MMAIII, and DMAIII) in cultures of B and T lymphocytes. Biol Trace Elem Res. 2006;108(1–3):115–126. doi: 10.1385/bter:108:1-3:115. [DOI] [PubMed] [Google Scholar]
- 14.Kojima C, Qu W, Waalkes MP, Himeno S, Sakurai T. Chronic exposure to methylated arsenicals stimulates arsenic excretion pathways and induces arsenic tolerance in rat liver cells. Toxicol Sci. 2006;91(1):70–81. doi: 10.1093/toxsci/kfj117. [DOI] [PubMed] [Google Scholar]
- 15.Kojima C, Sakurai T, Waalkes MP, Himeno S. Cytolethality of glutathione conjugates with monomethylarsenic or dimethylarsenic compounds. Biol Pharm Bull. 2005;28(10):1827–1832. doi: 10.1248/bpb.28.1827. [DOI] [PubMed] [Google Scholar]
- 16.Mass MJ, Tennant A, Roop BC, et al. Methylated trivalent arsenic species are genotoxic. Chem Res Toxicol. 2001;14(4):355–361. doi: 10.1021/tx000251l. [DOI] [PubMed] [Google Scholar]
- 17.Sakurai T, Kojima C, Kobayashi Y, et al. Toxicity of a trivalent organic arsenic compound, dimethylarsinous glutathione in a rat liver cell line (TRL 1215) Br J Pharmacol. 2006;149(7):888–897. doi: 10.1038/sj.bjp.0706899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Styblo M, del Razo LM, Vega L, et al. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch Toxicol. 2000;74(6):289–299. doi: 10.1007/s002040000134. [DOI] [PubMed] [Google Scholar]
- 19.Petrick JS, Ayala-Fierro F, Cullen WR, Carter DE, Vasken Aposhian H. Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in Chang human hepatocytes. Toxicol Appl Pharmacol. 2000;163(2):203–207. doi: 10.1006/taap.1999.8872. [DOI] [PubMed] [Google Scholar]
- 20.Nakamuro K, Sayato Y. Comparative studies of chromosomal aberration induced by trivalent and pentavalent arsenic. Mutat Res. 1981;88(1):73–80. doi: 10.1016/0165-1218(81)90091-4. [DOI] [PubMed] [Google Scholar]
- 21.Rossman TG. Mechanism of arsenic carcinogenesis: an integrated approach. Mutat Res. 2003;533(1–2):37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Hei TK, Filipic M. Role of oxidative damage in the genotoxicity of arsenic. Free Radic Biol Med. 2004;37(5):574–581. doi: 10.1016/j.freeradbiomed.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Eblin KE, Bowen ME, Cromey DW, et al. Arsenite and monomethylarsonous acid generate oxidative stress response in human bladder cell culture. Toxicol Appl Pharmacol. 2006;217(1):7–14. doi: 10.1016/j.taap.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi H, Aminaka Y, Yoshida K, Sun G, Pi J, Waalkes MP. Evaluation of DNA damage in patients with arsenic poisoning: urinary 8-hydroxydeoxyguanine. Toxicol Appl Pharmacol. 2004;198(3):291–296. doi: 10.1016/j.taap.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Collins AR, Cadet J, Möller L, Poulsen HE, Viña J. Are we sure we know how to measure 8-oxo-7,8-dihydroguanine in DNA from human cells? Arch Biochem Biophys. 2004;423(1):57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 26.Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol. 2004;142(2):231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez DC, Mejiba SE, Mason RP. Immuno-spin trapping of DNA radicals. Nat Methods. 2006;3(2):123–127. doi: 10.1038/nmeth852. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping analyses of DNA radicals. Nat Protoc. 2007;2(3):512–522. doi: 10.1038/nprot.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benbrahim-Tallaa L, Waterland RA, Styblo M, Achanzar WE, Webber MM, Waalkes MP. Molecular events associated with arsenic-induced malignant transformation of human prostatic epithelial cells: aberrant genomic DNA methylation and K-ras oncogene activation. Toxicol Appl Pharmacol. 2005;206(3):288–298. doi: 10.1016/j.taap.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Drobná Z, Waters SB, Devesa V, Harmon AW, Thomas DJ, Stýblo M. Metabolism and toxicity of arsenic in human urothelial cells expressing rat arsenic (+3 oxidation state)-methyltransferase. Toxicol Appl Pharmacol. 2005;207(2):147–159. doi: 10.1016/j.taap.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petzoldt JL, Leigh IM, Duffy PG, Sexton C, Masters JR. Immortalisation of human urothelial cells. Urol Res. 1995;23(6):377–380. doi: 10.1007/BF00698738. [DOI] [PubMed] [Google Scholar]
- 32.Hester SD, Drobna Z, Andrews DMK, et al. Expression of AS3MT alters transcriptional profiles in human urothelial cells exposed to arsenite. Hum Exp Toxicol. 2009;28(1):49–61. doi: 10.1177/0960327109102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durkan GC, Nutt JE, Marsh C, et al. Alteration in urinary matrix metalloproteinase-9 to tissue inhibitor of metalloproteinase-1 ratio predicts recurrence in nonmuscle-invasive bladder cancer. Clin Cancer Res. 2003;9(7):2576–2582. [PubMed] [Google Scholar]
- 34.Papathoma AS, Petraki C, Grigorakis A, et al. Prognostic significance of matrix metalloproteinases 2 and 9 in bladder cancer. Anticancer Res. 2000;20(3B):2009–2013. [PubMed] [Google Scholar]
- 35.Wallard MJ, Pennington CJ, Veerakumarasivam A, et al. Comprehensive profiling and localisation of the matrix metalloproteinases in urothelial carcinoma. Br J Cancer. 2006;94(4):569–577. doi: 10.1038/sj.bjc.6602931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cooper KL, Myers TA, Rosenberg M, Chavez M, Hudson LG. Roles of mitogen activated protein kinases and EGF receptor in arsenite-stimulated matrix metalloproteinase-9 production. Toxicol Appl Pharmacol. 2004;200(3):177–185. doi: 10.1016/j.taap.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 37.Albini A, Benelli R. The chemoinvasion assay: a method to assess tumor and endothelial cell invasion and its modulation. Nat Protoc. 2007;2(3):504–511. doi: 10.1038/nprot.2006.466. [DOI] [PubMed] [Google Scholar]
- 38.Hamburger AW, Salmon SE. Primary bioassay of human tumor stem cells. Science. 1977;197(4302):461–463. doi: 10.1126/science.560061. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Benbrahim-Tallaa L, Qian X, et al. Further studies on aberrant gene expression associated with arsenic-induced malignant transformation in rat liver TRL1215 cells. Toxicol Appl Pharmacol. 2006;216(3):407–415. doi: 10.1016/j.taap.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Leslie EM, Haimeur A, Waalkes MP. Arsenic transport by the human multidrug resistance protein 1 (MRP1/ABCC1). Evidence that a tri-glutathione conjugate is required. J Biol Chem. 2004;279(31):32700–32708. doi: 10.1074/jbc.M404912200. [DOI] [PubMed] [Google Scholar]
- 41.Walton FS, Waters SB, Jolley SL, LeCluyse EL, Thomas DJ, Styblo M. Selenium compounds modulate the activity of recombinant rat AsIII-methyltransferase and the methylation of arsenite by rat and human hepatocytes. Chem Res Toxicol. 2003;16(3):261–265. doi: 10.1021/tx025649r. [DOI] [PubMed] [Google Scholar]
- 42.Drake EN. Cancer chemoprevention: selenium as a prooxidant, not an antioxidant. Med Hypotheses. 2006;67(2):318–322. doi: 10.1016/j.mehy.2006.01.058. [DOI] [PubMed] [Google Scholar]
- 43.Pi J, Yamauchi H, Kumagai Y, et al. Evidence for induction of oxidative stress caused by chronic exposure of Chinese residents to arsenic contained in drinking water. Environ Health Perspect. 2002;110(4):331–336. doi: 10.1289/ehp.02110331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nesnow S, Roop BC, Lambert G, et al. DNA damage induced by methylated trivalent arsenicals is mediated by reactive oxygen species. Chem Res Toxicol. 2002;15(12):1627–1634. doi: 10.1021/tx025598y. [DOI] [PubMed] [Google Scholar]
- 45.Thomas DJ, Styblo M, Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicol Appl Pharmacol. 2001;176(2):127–144. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Chen H, Miller DS, et al. Overexpression of glutathione S-transferase II and multidrug resistance transport proteins is associated with acquired tolerance to inorganic arsenic. Mol Pharmacol. 2001;60(2):302–309. doi: 10.1124/mol.60.2.302. [DOI] [PubMed] [Google Scholar]
- 47.Romach EH, Zhao CQ, del Razo LM, Cebrián ME, Waalkes MP. Studies on the mechanisms of arsenic-induced self tolerance developed in liver epithelial cells through continuous low-level arsenite exposure. Toxicol Sci. 2000;54(2):500–508. doi: 10.1093/toxsci/54.2.500. [DOI] [PubMed] [Google Scholar]
- 48.Pi J, Qu W, Reece JM, Kumagai Y, Waalkes MP. Transcription factor Nrf2 activation by inorganic arsenic in cultured keratinocytes: involvement of hydrogen peroxide. Exp Cell Res. 2003;290(2):234–245. doi: 10.1016/s0014-4827(03)00341-0. [DOI] [PubMed] [Google Scholar]
- 49.Scandalios JG. Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Braz J Med Biol Res. 2005;38(7):995–1014. doi: 10.1590/s0100-879x2005000700003. [DOI] [PubMed] [Google Scholar]
- 50.Schläwicke Engström K, Nermell B, Concha G, Strömberg U, Vahter M, Broberg K. Arsenic metabolism is influenced by polymorphisms in genes involved in one-carbon metabolism and reduction reactions. Mutat Res. 2009;667(1-2):4–14. doi: 10.1016/j.mrfmmm.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 51.Drobna Z, Naranmandura H, Kubachka KM, et al. Disruption of arsenic (+3 oxidation state) methyltransferase gene in the mouse alters the phenotype for methylation of arsenic and affects distribution and rentention of orally administered arsenate. Chem Res Toxicol. 2009;22(10):1713–1720. doi: 10.1021/tx900179r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.